Abstract
Background
Emergence of rmtB-positive Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae (KPC-KP) poses a great threat to antimicrobial treatment options.
Methods
From January 2010 to December 2010, non-duplicate KPC-KP isolates from our hospital were screened for rmtB and multiple other resistance determinants with PCR. Subsequent studies included MIC determination, PFGE, and multilocus sequence typing. Records from patients with KPC-KP isolated were retrospectively reviewed. Comparisons of molecular and clinical characteristics between rmtB-positive and rmtB–negative isolates were systematically performed, as well as the environmental colonization study in ICU wards.
Results
A total of 84 KPC-KP strains were collected, including 48 rmtB-positive KPC-KP (RPKP) and 36 rmtB-negative KPC-KP (RNKP) isolates. All KPC-KP isolates were multidrug resistant, with colistin and tigecycline being the most active agents. Compared with RNKP, RPKP displayed a much severer resistance phenotype. Susceptibility rates for amikacin (0% for RPKP versus 88.9% for RNKP, p < 0.01), fosfomycin (8.5% for RPKP versus 88.9% for RNKP, p < 0.01), and minocycline (6.7% for RPKP versus 52.8% for RNKP, p < 0.01), were all significantly lower in RPKP strains. Isolates belonging to PFGE pulsetype A and sequence type 11 were predominant in both groups, including 39 (81.3%) RPKP and 22 (61.1%) RNKP isolates. Nevertheless, RNKP showed more complex genetic backgrounds compared with RPKP. Diverse clinical characteristics were found in both cohorts, however, no significant differences were observed between RPKP and RNKP patients.
Conclusions
RPKP strains have spread widely and gradually replaced RNKP in our hospital. They seemed to show much severer resistance phenotypes compared with RNKP and had a bigger dissemination potential. Prudent use of available active agents combined with good control practices is therefore mandatory.
Similar content being viewed by others
Background
During the past 10 years, Chinese clinicians have witnessed a dramatic increase in the rates of carbapenem resistance among clinical isolates of Klebsiella pneumoniae. According to Mohnarin and CHINET, two leading antimicrobial resistance surveillance networks in China that cover all regions except Tibet, carbapenem resistance rates among K. pneumoniae escalated from 0.7% in 2000 to 2.9% in 2009 [1]. Moreover, in a university hospital in Shanghai, the incidence of carbapenem-resistant K. pneumoniae in 2009 reached 12.9% [2]. In China, carbapenem resistance in K. pneumoniae has mainly resulted from the rapid dissemination of Klebsiella pneumoniae carbapenemase (KPC) [2–4], and ST11 was demonstrated to be the dominant clone of KPC-producing K. pneumoniae (KPC-KP) [4].
KPC-KP isolates were usually multidrug resistant, but might remain susceptible to one or more aminoglycoside agents as well as to colistin and/or tigecycline [4–6]. Nevertheless, 16S rRNA methylases (ArmA and RmtB) were reported in a few KPC-producing Enterobacteriaceae recently, conferring high level resistance to almost all clinically important aminoglycosides [7–9]. Co-production of 16S rRNA methylases in KPC-producing pathogens and their potential spread could leave few choices for antimicrobial treatment.
Since its first isolation in 2007 [10], KPC-KP has been increasingly emerging at our hospital. Most of these isolates were still susceptible to one or more aminoglycoside antibiotics until 2009; nevertheless, high-level aminoglycoside resistance has emerged and gradually become prevalent in the last two years (data not shown). In May 2010, an Enterobacter amnigenus as well as a K. pneumoniae isolate, both co-producing bla KPC and rmtB on a single plasmid, were simultaneously identified from a patient in our neurosurgery ward. And a small outbreak of KPC-RmtB-producing K. pneumoniae was revealed in the neurosurgery department [7]. This promoted us to perform a hospital-wide screening of rmtB gene in KPC-KP strains. Our study focused on the comparisons of microbiological, molecular and clinical characteristics between rmtB-positive KPC-KP (RPKP) and rmtB-negative KPC-KP (RNKP) isolates.
Methods
Bacterial isolates and patients
Between January 2010 and December 2010, all clinical K. pneumoniae isolates exhibiting non-susceptibility to carbapenems (inhibition diameter <16 mm for at least one of meropenem and imipenem with the help of disc diffusion method) were obtained in the First Affiliated Hospital, School of Medicine, Zhejiang University. This hospital is a 2500-bed tertiary-care academic medical center receiving about 2 560 000 outpatients and 71 000 inpatients annually. Detection of bla KPC and rmtB genes was conducted by PCR and subsequent sequencing based on established methods [4, 11]. A case patient was defined as patient with KPC-KP (including RPKP and RNKP) isolated from clinical specimens during our study period. If more than one isolate were obtained from a patient, only the initial isolate was submitted.
Cohort study
A retrospective observational cohort study was conducted between RPKP and RNKP patients. Detailed clinical information of case patients was extracted from their medical records and follow-up was possible until discharge from our hospital or death. Isolates identified during the first 72 h after admission were characterized as imported, whereas those identified >72 h after admission were determined as nosocomial transmission during the current hospitalization. Infection was defined according to the criteria from Centers for Disease Control and Prevention (CDC)/National Healthcare Safety Network [12]. Patients without infection but with KPC-KP isolated were considered as colonized. The study was approved by the Institutional Review Board of our hospital.
Bacterial identification and susceptibility testing
Species identification was performed using the Vitek 2 system (bioMerieux, France). MICs of various antimicrobials were determined by agar dilution method and results were interpreted according to the criteria recommended by Clinical and Laboratory Standards Institute (CLSI) 2010 [13]. With regard to tigecycline, breakpoint for Enterobacteriaceae based on FDA was used (MIC ≤2 μg/mL as susceptible). And concentration of 4 μg/mL was used as the breakpoint of resistance for colistin [14].
Molecular typing
Pulse field gel electrophoresis (PFGE) was performed using XbaΙ restriction enzyme on all clinical isolates and PFGE profiles were interpreted by the criteria proposed by Tenover [15]. Multilocus sequence typing (MLST) was carried out according to protocols on the MLST website for K. pneumoniae (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html).
PCR testing and DNA sequencing
PCR analysis was conducted to detect a variety of resistance determinants as described previously, including bla OXA, bla TEM, bla SHV, and bla CTX genes [4, 16], six more 16S rRNA methylase-encoding genes (armA, rmtA, rmtC, rmtD, rmtE and npmA) [11], and plasmid-mediated quinolone resistance genes (qnrA, qnrB, qnrS and aac(6’)-Ib-cr) [16]. The PCR products were subsequently sequenced by dideoxynucleotide chain-termination method by ABI 377 (ABI, U.S.A.) and sequences were compared with the nucleotide sequences from GenBank (http://www.ncbi.nlm.nih.gov/blast/).
Environmental colonization study
A point prevalence survey of environmental colonization was conducted in the general ICU wards in July 2010 to probe the potential environmental reservoirs of RPKP. Environmental surfaces including medical equipments in the immediate vicinity of patients, communal areas, and contaminated hands of medical staff on shift, were under screening. Swab samples were obtained by repeatedly rubbing designated sites with premoistened swabs. The swabs were then placed in brain and heart infusion (BHI) containing meropenem (1 μg/mL) and incubated overnight at 37°C. Isolated strains were subjected to further analysis as clinical isolates as described above.
Statistical analysis
Comparative analyses were performed using the χ2 test or the Fischer’s exact test for categoric variables, and Student’s t test for the continuous variables, as appropriate. A 2-tailed P value of <0.05 was considered to indicate statistical significance. Analyses were done with SPSS 17.0 (SPSS Inc, Chicago, USA).
Results
A total of 84 KPC-KP isolates were identified during the study period. Among them, 48 were confirmed to be RPKP, and the rest RNKP. Medical records were available for 35 RPKP patients and 29 RNKP patients. Comparisons of various clinical characteristics between RPKP and RNKP patients are presented in Table 1. Most patients were severely ill and half were from ICU departments. Respiratory tract was the most common site of infection in both cohorts. Although a majority of KPC-KP isolates were acquired at our hospital, 7 RNKP (24.1%) and 4 RPKP isolates (11.4%) were imported. Another notably feature was that bed transfers were rather common among these patients (2.54 times for RPKP versus 2.52 times for RNKP), even after isolation of KPC-KP (1.57 times for RPKP versus 1.66 times for RNKP). Crude mortality was 22.9% among RPKP patients and 27.6% among RNKP patients, and attributable mortality rates were 19.4% and 17.9%, respectively.
The epidemic curve revealed three phases (Figure 1): period 1 (January to April), during which sporadic cases of KPC-KP were identified and RPKP initiated to emerge; period 2 (May to July), during which a hospital-wide outbreak of KPC-KP was observed, with dramatic increase in both RNKP and RPKP isolates, and RPKP became predominant gradually; period 3 (August to December), during which high-level epidemic of KPC-KP persisted, and RPKP overtook RNKP to become the dominant KPC-KP.
KPC-KP isolates were multidrug resistant (Table 2). They only showed sufficient susceptibility to colistin and tigecycline (susceptibility rate: 98.8% for colistin and 96.3% for tigecycline). However, RPKP displayed much severer multidrug resistance phenotypes compared with RNKP. With regard to aminoglycoside antibiotics, all RPKP undoubtedly showed high-level resistance, whereas a majority of RNKP were still susceptible to at least one of them (88.9% for amikacin; 72.2% for netilmicin; 47.2% for gentamicin). In addition, RPKP isolates demonstrated significantly lower susceptibility rates to fosfomycin (8.5% for RPKP versus 88.9% for RNKP, P < 0.01) and minocycline (6.7% for RPKP versus 52.8% for RNKP, P < 0.01).
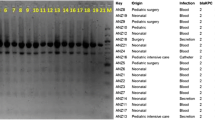
Molecular typing results and resistance determinant profiles are shown in Table 3. PFGE analysis revealed 10 different pulsetypes (PTs), designated as A-J (Figure 2). The most widely disseminated PT was A, consisting of 63 clinical isolates (75%). By MLST, 10 distinct sequence types (STs) were identified, including 3 novel STs (ST689, ST690, and ST691). ST11 was the predominant ST, with 68 isolates (80.9%). Most of MLST data were consistent with results generated by PFGE. ST11-PTA was the dominant molecular type in both RPKP and RNKP. However, a greater diversity of STs and more complicated relatedness between the results of PFGE and MLST were observed in RNKP, indicating more diverse genetic backgrounds in RNKP.
Apart from bla KPC-2 in all isolates and rmtB in RPKP, bla CTX-M-14, bla SHV-12 and bla TEM-1 were revealed in most isolates. In addition, four RNKP belonging to ST412 or ST692 also encoded bla OXA-1–type and bla OXA-10–type β-lactamases and aac(6’)-Ib-cr, and a single ST689 RNKP encoded bla OXA-1. Only one RPKP and three RNKP strains carried qnrS. No other 16S rRNA methylase-encoding genes were identified.
In total, 210 environmental samples were obtained from the general ICU department. Among the 210 samples, only one was RPKP positive, which was identified from a folder of patient chart from an empty bed with a RPKP patient discharged. The environmental RPKP was assigned to ST11-PTA clone, and carried bla CTX-M-14, bla SHV-12 and bla TEM-1 in addition to bla KPC and rmtB genes. It also showed similar antibiogram as its clinical counterparts (data not shown).
Discussion
Nowadays, KPC-KP isolates have become major hospital pathogens, and their worldwide spread makes them a great threat to currently available antibiotic-based treatments. Tigecycline, colistin, and aminoglycosides if active, are agents recommended for severe infections caused by carbapenemase-producing Enterobacteriaceae[17]. However, despite of good in vitro activity of colistin and tigecycline against most of these strains, clinical data on their in vivo efficacy are quite limited [17]. Emergence of tigecycline resistance and colistin resistance during therapy with these agents is another issue to be considered [18–20]. A colistin-resistant RNKP isolate was also identified from a patient in our study, even without history of colistin therapy. Fortunately, a high proportion of KPC-KP isolates still showed in vitro susceptibility to aminoglycosides [6, 21, 22], including most of recently emerging colistin-resistant or tigecycline-resistant KPC-KP strains [18–20]. The colistin-resistant isolate found in our study was susceptible to aminoglycosides as well. In addition, a more recent study on KPC-KP bacteriuria demonstrated that treatments including aminoglycosides achieved a significantly higher rate of microbiologic clearance than did treatments with polymyxin B or tigecycline [23]. These findings together indicated that aminoglycosides may serve as effective antimicrobials in treating KPC-KP infections. Emergence of aminoglycoside-resistance in KPC-KP isolates therefore threatens the viability of this therapeutic option and further limits choices of available active agents.
In the present study, we documented the emergence and rapid spread of rmtB-positive KPC-KP (RPKP) in a Chinese university hospital. Due to the presence of rmtB gene, RPKP isolates showed high level resistance to almost all clinical available aminoglycosides. In addition, RPKP were usually resistant to many more clinically useful antimicrobials compared with RNKP, including minocycline and fosfomycin, which could serve as salvage remedies alone or in combination for infections due to carbapenem-resistant K. pneumoniae[21, 24]. The fact that RPKP only exhibited sufficient susceptibility to colistin and tigecycline has led to the dead-end of antimicrobial therapy in our hospital, since both colistin and tigecycline were unavailable in China during the study period.
Our results are concordant with the observation that ST11 was the dominant clone of KPC-KP in Mainland China [4]. ST11 is a single-locus variant of the international hyper-epidemic lineage ST258, and has also been identified in Singapore and more recently in Taiwan [25, 26]. It is increasingly recognized that ST258 is the predominant clone of KPC-KP across the whole world and has caused outbreaks in many countries [5, 6, 22]. Furthermore, ST258 has extended to the recently reported colistin-resistant KPC-KP [18, 27]. In the present study, ST11 KPC-KP also seemed to show dissemination advantage over other clones and be good colonizers to capture and accumulate resistance determinants. Of particular concern is that increasing prevalence of RPKP coincides with decreasing prevalence of RNKP in our hospital, which might suggest a bigger dissemination advantage of ST11 RPKP over ST11 RNKP, even in the absence of selective pressure. Another particular concern is the fact that 3 RPKP isolates (all belonged to ST11-PTA) were imported from other local hospitals, suggesting the presence of inter-hospital dissemination of this extremely multi-drug resistant pathogen. This indicates a dangerous possibility that ST11 RPKP clone might spread widely outside our hospital in the future.
The fact that most RPKP isolates belonged to ST11-PTA and were acquired in our hospital indicates the possibility that monoclonal cross transmission is the main mode of spreading. However, the failure to find a common environmental reservoir indicated patient-to-patient transmission as the main mechanism of RPKP spread in our study. Frequent bed transfers of case patients, particularly after isolation of RPKP, combined with lack of adequate preventive measures might have facilitated this process. It is also worth mentioning that the only one environmental RPKP isolate was identified from a decontaminated empty bed, which obliged us to enhance the effectiveness of decontamination procedures in our hospital.
Infections with pathogens resistant to more antibiotics have been reported to be associated with poorer functional status and worse outcomes [28, 29]. However, in our study, despite of diverse clinical characteristics, no significant differences were observed between the two cohorts. The similar clinical characteristics between RPKP and RNKP patients were likely multifactorial. Firstly, patients in both cohorts were at poor functional status, with severe underlying diseases and highly vulnerable to the colonization or infection of multidrug resistant pathogens. Secondly, RPKP and RNKP were both extremely drug-resistant pathogens. They showed sufficient susceptibility only to colistin and tigecycline. However, neither of them was readily available in China during our study period. Finally, although most RNKP (88.9%) were susceptible to at least one aminoglycoside in vitro, treatment containing aminoglycoside agents was very limited in our hospital, due to limited data on their in-vivo efficacy on KPC-KP infections and their nephrotoxicity [30]. Only three patients with RNKP were treated with antimicrobial regimens containing aminoglycosides in our study, but all of them had good clinical outcomes (data not shown).
Conclusions
In summary, we documented the emergence and wide dissemination of rmtB-positive KPC-KP in a Chinese university hospital. These isolates showed much severer resistance phenotypes compared with their rmtB-negative counterparts and seemed to disseminate more easily, representing a major threat for hospitalized patients. Moreover, patients suffered from RPKP infections were usually severely ill and ended up with rather high attributable mortality. These results, together with the fact that discovery of new antibiotics is reduced in recent years, suggest that prudent and conservative use of available active agents combined with good control practices are mandatory.
Abbreviations
- KPC:
-
Klebsiella pneumoniae carbapenemase
- KPC-KP:
-
Klebsiella pneumoniae carbapenemase-producing K. pneumoniae
- RPKP:
-
rmtB-positive Klebsiella pneumoniae carbapenemase-producing K. pneumoniae
- RNKP:
-
rmtB-negative Klebsiella pneumoniae carbapenemase-producing K. pneumoniae
- CLSI:
-
Clinical and Laboratory Standards Institute
- PFGE:
-
Pulse field gel electrophoresis
- MLST:
-
Multilocus sequence typing
- BHI:
-
Brain and heart infusion
- PTs:
-
Pulsetypes
- STs:
-
Sequence types
- MIC:
-
Minimal inhibition concentration
- PCR:
-
Polymerase chain reaction
- ICU:
-
Intensive care unit.
References
Xiao YH, Giske CG, Wei ZQ, Shen P, Heddini A, Li LJ: Epidemiology and characteristics of antimicrobial resistance in China. Drug Resist Updat. 2011, 14 (4–5): 236-250.
Chen S, Hu F, Xu X, Liu Y, Wu W, Zhu D, Wang H: High prevalence of KPC-2-type carbapenemase coupled with CTX-M-type extended-spectrum beta-lactamases in carbapenem-resistant Klebsiella pneumoniae in a teaching hospital in China. Antimicrob Agents Chemother. 2011, 55 (5): 2493-2494. 10.1128/AAC.00047-11.
Cai JC, Zhou HW, Zhang R, Chen GX: Emergence of Serratia marcescens, Klebsiella pneumoniae, and Escherichia coli Isolates possessing the plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC-2 in intensive care units of a Chinese hospital. Antimicrob Agents Chemother. 2008, 52 (6): 2014-2018. 10.1128/AAC.01539-07.
Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y: ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011, 66 (2): 307-312. 10.1093/jac/dkq431.
Andrade LN, Curiao T, Ferreira JC, Longo JM, Climaco EC, Martinez R, Bellissimo-Rodrigues F, Basile-Filho A, Evaristo MA, Del Peloso PF, et al: Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother. 2011, 55 (7): 3579-3583. 10.1128/AAC.01783-10.
Giakkoupi P, Papagiannitsis CC, Miriagou V, Pappa O, Polemis M, Tryfinopoulou K, Tzouvelekis LS, Vatopoulos AC: An update of the evolving epidemic of blaKPC-2-carrying Klebsiella pneumoniae in Greece (2009–10). J Antimicrob Chemother. 2011, 66 (7): 1510-1513. 10.1093/jac/dkr166.
Sheng JF, Li JJ, Tu S, Sheng ZK, Bi S, Zhu MH, Shen XM, Li LJ: blaKPC and rmtB on a single plasmid in Enterobacter amnigenus and Klebsiella pneumoniae isolates from the same patient. Eur J Clin Microbiol Infect Dis. 2012, 31 (7): 1585-1591. 10.1007/s10096-011-1481-x.
Wu Q, Zhang Y, Han L, Sun J, Ni Y: Plasmid-mediated 16S rRNA methylases in aminoglycoside-resistant Enterobacteriaceae isolates in Shanghai, China. Antimicrob Agents Chemother. 2009, 53 (1): 271-272. 10.1128/AAC.00748-08.
Zacharczuk K, Piekarska K, Szych J, Zawidzka E, Sulikowska A, Wardak S, Jagielski M, Gierczynski R: Emergence of Klebsiella pneumoniae coproducing KPC-2 and 16S rRNA methylase ArmA in Poland. Antimicrob Agents Chemother. 2011, 55 (1): 443-446. 10.1128/AAC.00386-10.
Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, Li LJ: Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2007, 51 (2): 763-765. 10.1128/AAC.01053-06.
Zhou Y, Yu H, Guo Q, Xu X, Ye X, Wu S, Guo Y, Wang M: Distribution of 16S rRNA methylases among different species of Gram-negative bacilli with high-level resistance to aminoglycosides. Eur J Clin Microbiol Infect Dis. 2010, 29 (11): 1349-1353. 10.1007/s10096-010-1004-1.
Horan TC, Andrus M, Dudeck MA: CDC/NHSN surveillance definition of health care-associated infection and criteria for specific types of infections in the acute care setting. Am J Infect Control. 2008, 36 (5): 309-332. 10.1016/j.ajic.2008.03.002.
Clinical and Loboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing: 20th informational supplement, CLSI document M100-S20. 2010, Wayne: CLSI
Gales AC, Reis AO, Jones RN: Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J Clin Microbiol. 2001, 39 (1): 183-190. 10.1128/JCM.39.1.183-190.2001.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B: Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995, 33 (9): 2233-2239.
Luo Y, Yang J, Zhang Y, Ye L, Wang L, Guo L: Prevalence of beta-lactamases and 16S rRNA methylase genes amongst clinical Klebsiella pneumoniae isolates carrying plasmid-mediated quinolone resistance determinants. Int J Antimicrob Agents. 2011, 37 (4): 352-355. 10.1016/j.ijantimicag.2010.12.018.
Peleg AY, Hooper DC: Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010, 362 (19): 1804-1813. 10.1056/NEJMra0904124.
Bogdanovich T, Adams-Haduch JM, Tian GB, Nguyen MH, Kwak EJ, Muto CA, Doi Y: Colistin-Resistant, Klebsiella pneumoniae Carbapenemase (KPC)-Producing Klebsiella pneumoniae Belonging to the International Epidemic Clone ST258. Clin Infect Dis. 2011, 53 (4): 373-376. 10.1093/cid/cir401.
Daly MW, Riddle DJ, Ledeboer NA, Dunne WM, Ritchie DJ: Tigecycline for treatment of pneumonia and empyema caused by carbapenemase-producing Klebsiella pneumoniae. Pharmacotherapy. 2007, 27 (7): 1052-1057. 10.1592/phco.27.7.1052.
Elemam A, Rahimian J, Mandell W: Infection with panresistant Klebsiella pneumoniae: a report of 2 cases and a brief review of the literature. Clin Infect Dis. 2009, 49 (2): 271-274. 10.1086/600042.
Souli M, Galani I, Antoniadou A, Papadomichelakis E, Poulakou G, Panagea T, Vourli S, Zerva L, Armaganidis A, Kanellakopoulou K, et al: An outbreak of infection due to beta-Lactamase Klebsiella pneumoniae Carbapenemase 2-producing K. pneumoniae in a Greek University Hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis. 2010, 50 (3): 364-373. 10.1086/649865.
Samuelsen O, Naseer U, Tofteland S, Skutlaberg DH, Onken A, Hjetland R, Sundsfjord A, Giske CG: Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J Antimicrob Chemother. 2009, 63 (4): 654-658. 10.1093/jac/dkp018.
Satlin MJ, Kubin CJ, Blumenthal JS, Cohen AB, Furuya EY, Wilson SJ, Jenkins SG, Calfee DP: Comparative effectiveness of aminoglycosides, polymyxin B, and tigecycline for clearance of carbapenem-resistant Klebsiella pneumoniae from urine. Antimicrob Agents Chemother. 2011, 55 (12): 5893-5899. 10.1128/AAC.00387-11.
Michalopoulos A, Virtzili S, Rafailidis P, Chalevelakis G, Damala M, Falagas ME: Intravenous fosfomycin for the treatment of nosocomial infections caused by carbapenem-resistant Klebsiella pneumoniae in critically ill patients: a prospective evaluation. Clin Microbiol Infect. 2010, 16 (2): 184-186. 10.1111/j.1469-0691.2009.02921.x.
Balm MN, Ngan G, Jureen R, Lin RT, Teo J: Molecular characterization of newly emerged blaKPC-2-producing Klebsiella pneumoniae in Singapore. J Clin Microbiol. 2012, 50 (2): 475-476. 10.1128/JCM.05914-11.
Lee CM, Liao CH, Lee WS, Liu YC, Mu JJ, Lee MC, Hsueh PR: Outbreak of Klebsiella pneumoniae Carbapenemase-2- producing K. pneumoniae Sequence Type 11 in Taiwan, 2011. Antimicrob Agents Chemother. 2012, 56 (10): 5016-5022. 10.1128/AAC.00878-12.
Mezzatesta ML, Gona F, Caio C, Petrolito V, Sciortino D, Sciacca A, Santangelo C, Stefani S: Outbreak of KPC-3-producing, and colistin-resistant, Klebsiella pneumoniae infections in two Sicilian hospitals. Clin Microbiol Infect. 2011, 17 (9): 1444-1447.
Zarkotou O, Pournaras S, Voulgari E, Chrysos G, Prekates A, Voutsinas D, Themeli-Digalaki K, Tsakris A: Risk factors and outcomes associated with acquisition of colistin-resistant KPC-producing Klebsiella pneumoniae: a matched case–control study. J Clin Microbiol. 2010, 48 (6): 2271-2274. 10.1128/JCM.02301-09.
Zarkotou O, Pournaras S, Tselioti P, Dragoumanos V, Pitiriga V, Ranellou K, Prekates A, Themeli-Digalaki K, Tsakris A: Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin Microbiol Infect. 2011, 17 (12): 1798-1803. 10.1111/j.1469-0691.2011.03514.x.
Drusano GL, Louie A: Optimization of aminoglycoside therapy. Antimicrob Agents Chemother. 2011, 55 (6): 2528-2531. 10.1128/AAC.01314-10.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2334/12/373/prepub
Acknowledgments
This work was supported by the National Natural Science Foundation of China [NO. 30972592] and Chinese High Tech Research & Development (863) Program [NO. 2011AA020104].
We thank platform Genotyping of Pathogens and Public Health (Institut Pasteur) for coding our MLST alleles and profiles.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
All authors have no reported conflicts to disclose.
Authors’ contributions
JJL was involved in all processes related to study design, collecting samples and reviewing medical records, microbiological and molecular studies, analyses of data, and wrote this paper. ZKS and MD contributed to sample collection and microbiological and molecular studies. SB, FSH, HFM, and ZKJ contributed to this study by sample collection. LJL contributed to this study by reviewing and making comments on all drafts of this paper. JFS reviewed and revised this paper, and gave final approval to submit for publication. All authors have read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Li, JJ., Sheng, ZK., Deng, M. et al. Epidemic of Klebsiella pneumoniae ST11 Clone Coproducing KPC-2 and 16S rRNA Methylase RmtB in a Chinese University Hospital. BMC Infect Dis 12, 373 (2012). https://doi.org/10.1186/1471-2334-12-373
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2334-12-373