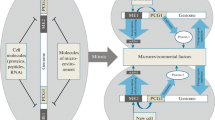
Abstract
The paper summarizes data on the existence of evolutionarily fixed species balance of interactions between peptides, RNA, and DNA that provide the stability of the development and functioning of tissues and organs in ontogenesis. Identification of key structures that disturb this balance with aging can become a basis for a specific targeted effect on reversible epigenetic mechanisms of their origin. Non-coding RNAs that, in addition to the function of ribozymes and effectors of RNA interference, are capable of being translated into peptides, which can be the most convenient targets. The effect of the latter on non-coding RNAs and involvement in the same biological processes can become a basis for a complex approach in the development of new geroprotective drugs. It was suggested that peculiarities of the expression of non-coding RNAs typical for a cell type and stage of development reflect transposon activation patterns (programmed at the species level) required for setting of tissue-specific gene networks. This is caused by the formation of non-coding RNAs from transposon transcripts that are regulators of the expression of protein-coding genes in successive cell divisions.

Similar content being viewed by others
REFERENCES
Mustafin, R.N. and Khusnutdinova, E.K., The interaction of transposons with epigenetic factors in aging, Usp. Gerontol., 2017, vol. 30, no. 4, pp. 516–528.
Khavinson, V.Kh., Solovyov, A.Yu., and Shataeva, L.K., Molecular mechanism of interaction between oligopeptides and double-stranded DNA, Bull. Exp. Biol. Med., 2006, vol. 141, no. 4, pp. 457–461.
Khavinson, V.Kh., Peptidnaya regulyatsiya stareniya (Peptide Regulation of Aging), St. Petersburg: Nauka, 2009.
Abdolmohsen, K., Panda, A., Kang, M.J., et al., Senescence-associated lncRNAs: senescence-associated long noncoding RNAs, Aging Cell, 2013, vol. 12, pp. 890–900.
Abdelmohsen, K. and Gorospe, M., Noncoding RNA control of cellular senescence, Wiley Interdiscip. Rev.: RNA, 2015, vol. 6, no. 6, pp. 615–629.
Anderson, D.M., Anderson, K.M., Chang, C.L., et al., A micropeptide encoded by a putative long noncoding RNA regulates muscle performance, Cell, 2015, vol. 160, pp. 595–606.
Aschacher T., Wolf B., Enzmann F., et al., LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines, Oncogene, 2016, vol. 35, pp. 94–104.
Aschacher, T., Wolf, B., Aschacher, O., et al., Long interspersed element-1 ribonucleoprotein particles protect telomeric ends in alternative lengthening of telomeres dependent cells, Neoplasia, 2020, vol. 22, pp. 61–75.
Barry, G., Guennewig, B., Fung, S., et al., Long non-coding RNA expression during aging in the human subependymal zone, Front. Neurol., 2015, vol. 6, p. 45.
Bravo, J.I., Nozownik, S., Danthi, P.S., and Benayoun, B.A., Transposable elements, circular RNAs and mitochondrial transcription in age-related genomic regulation, Development, 2020, vol. 147, pp. 175786.
Briggs, J.A., Wolvetang, E.J., Mattick, J.S., et al., Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution, Neuron, 2015, vol. 88, pp. 861–877.
Cabili, M.N., Trapnell, C., Goff, L., et al., Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses, Genes Dev., 2011, vol. 25, no. 18, pp. 1915–1927.
Cao, Q., Wu, J., Wang, X., and Song, C., Noncoding RNAs in vascular aging, Oxid. Med. Cell. Longevity, 2020, vol. 2020, art. ID 7914957.
Cardelli, M., The epigenetic alteration of endogenous retroelements in aging, Mech. Ageing Dev., 2018, vol. 174, pp. 30–46.
Casacuberta, E., Drosophila: retrotransposons making up telomeres, Viruses, 2017, vol. 9, p. 192.
Cooper, C., Vincett, D., Yan, Y., et al., Steroid receptor RNA activator bi-faceted genetic system: heads or tails, Biochimie, 2011, vol. 93, pp. 1973–1980.
Dong, X., Chen, K., Cuevas-Diaz Duran, R., et al., Comprehensive identification of long non-coding RNAs in purified cell types from the brain reveals functional lncRNA in OPC fate determination, PLoS Genet., 2015, vol. 11, pp. 1–26.
Ferron, S.R., Mira, H., Franco, S., et al., Telomere shortening and chromosomal instability abrogates proliferation of adult but not embryonic neural stem cells, Development, 2004, vol. 131, pp. 4059–4070.
Ferron, S.R., Marques-Torrejon, M.A., Mira, H., et al., Telomere shortening in neural stem cells disrupts neuronal differentiation and neurogenesis, J. Neurosci., 2009, vol. 29, pp. 14394–14407.
Fico, A., Fiorenzano, A., Pascale, E., et al., Long non-coding RNA in stem cell pluripotency and lineage commitement: functions and evolutionary conservation, Cell. Mol. Life Sci., 2019, vol. 76, pp. 1459–1471.
Gerdes, P., Richardson, S.R., Mager, D.L., and Faulkner, G.J., Transposable elements in the mammalian embryo: pioneers surviving through stealth and service, Genome Biol., 2016, vol. 17, pp. 100–116.
Grammatikakis, I., Panda, A.C., Abdelmohsen, K., and Gorospe, M., Long noncoding RNAs (lncRNAs) and the molecular hallmarks of aging, Aging (Albany, NY), 2014, vol. 6, pp. 992–1009.
Guzman, H., Sanders, K., Idica, A., et al., miR-128 inhibits telomerase activity by targeting TERT mRNA, Oncotarget, 2018, vol. 9, pp. 13244–13253.
Hadjiargyrou, M. and Delihas, N., The intertwining of transposable elements and non-coding RNAs, Int. J. Mol. Sci., 2013, vol. 14, no. 7, pp. 13307–13328.
Honson, D.D. and Macfarlan, T.S., A lncRNA-like role for LINE1s in development, Dev. Cell, 2018, vol. 46, pp. 132–134.
Ingolia, N.T., Lareau, L.F., and Weissman, J.S., Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes, Cell, 2011, vol. 147, pp. 789–802.
Johnson, R. and Guigo, R., The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs, RNA, 2014, vol. 20, pp. 959–976.
Kapusta, A., Kronenberg, Z., Lynch, V.J., et al., Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs, PLoS Genet., 2013, vol. 9, p. e1003470.
Kelley, D. and Rinn, J., Transposable elements reveal a stem cell-specific class of long noncoding RNAs, Genome Biol., 2012, vol. 13, no. 11, p. R107.
Kondo, T., Plaza, S., Zanet, J., et al., Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis, Science, 2010, vol. 329, pp. 336–339.
Kordyukova, M., Olovnikov, I., and Kalmykova, A., Transposon control mechanisms in telomere biology, Curr. Opin. Genet. Dev., 2018, vol. 49, pp. 56–62.
Kour, S. and Rath, P.C., Age-dependent differential expression profile of a novel intergenic long noncoding RNA in rat brain, Int. J. Dev. Neurosci., 2015, vol. 47, pp. 286–297.
Kour, S. and Rath, P.C., Long noncoding RNAs in aging and age-related diseases, Ageing Res. Rev., 2016, vol. 26, pp. 1–21.
Ladoukakis, E., Pereira, V., Magny, E.G., et al., Hundreds of putatively functional small open reading frames in Drosophila, Genome Biol., 2011, vol. 12, p. 118.
Lambowitz, A.M. and Belfort, M., Mobile bacterial group II introns at the crux of eukaryotic evolution, Microbiol. Spectr., 2015, vol. 3, MDNA3-0050-2014.
Lauressergues, D., Couzigou, J.M., Clemente, H.S., et al., Primary transcripts of microRNAs encode regulatory peptides, Nature, 2015, vol. 520, no. 7545, pp. 90–93.
Lee, H.E., Huh, J.W., and Kim, H.S., Bioinformatics analysis of evolution and human disease related transposable element-derived microRNAs, Life (Basel), 2020, vol. 10, p. 95.
Li, L.J., Leng, R.X., and Fan, Y.G., Translation of noncoding RNAs: focus on lncRNAs, pri-miRNAs, and circRNAs, Exp. Cell Res., 2017, vol. 361, pp. 1–8.
Li, X., Zhang, J., Yang, Y., et al., MicroRNA-340-5p increases telomere length by targeting telomere protein POT1 to improve Alzheimer’s disease in mice, Cell Biol. Int., 2021, vol. 45, no. 6, pp. 1306–1315. https://doi.org/10.1002/cbin.11576
Lou, Z., Zhu, J., Li, X., et al., LncRNA Sirt1-AS upregulates Sirt1 to attenuate aging related deep venous thrombosis, Aging (Albany, NY), 2021, vol. 13, pp. 6918–6935.
Lu, X., Sachs, F., Ramsay, L., et al., The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity, Nat. Struct. Mol. Biol., 2014, vol. 21, pp. 423–425.
Lu, S., Zhang, J., Lian, X., et al., A hidden human proteome encoded by ‘non-coding’ genes, Nucleic Acids Res., 2019, vol. 47, no. 15, pp. 8111–8125. https://doi.org/10.1093/nar/gkz646
Ma, J.J., Ju, X., Xu, R.J., et al., Telomerase reverse transcriptase and p53 regulate mammalian peripheral nervous system and CNS axon regeneration downstream of c-Myc, J. Neurosci., 2019, vol. 39, pp. 9107–9118.
Magny, E.G., Pueyo, J.I., Pearl, F.M., et al., Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames, Science, 2013, vol. 341, pp. 1116–1120.
Matsumoto, A., Pasut, A., Matsumoto, M., et al., mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide, Nature, 2017, vol. 541, pp. 228–232.
Maxwell, P.H., What might retrotransposons teach us about aging, Curr. Genet., 2016, vol. 62, pp. 277–282.
Mercer, T.R., Dinger, M.E., Sunkin, S.M., et al., Specific expression of long noncoding RNAs in the mouse brain, Proc. Natl. Acad. Sci. U.S.A., 2008, vol. 105, pp. 716–721.
Mercer, T.R. and Mattick, J.S., Structure and function of long noncoding RNAs in epigenetic regulation, Nat. Struct. Mol. Biol., 2013, vol. 20, pp. 300–307.
Mueller, C., Aschacher, T., Wolf, B., and Bergmann, M., A role of LINE-1 in telomere regulation, Front. Biosci., 2018, vol. 23, pp. 1310–1319.
Mus, E., Hof, P.R., and Tiedge, H., Dendritic BC200 RNA in aging and in Alzheimer’s disease, Proc. Natl. Acad. Sci. U.S.A., 2007, vol. 104, no. 25, pp. 10679–10684.
Nelson, B.R., Makarewich, C.A., Anderson, D.M., et al., A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle, Science, 2016, vol. 351, pp. 271–275.
Nichuguti, N. and Fujiwara, H., Essential factors involved in the precise targeting and insertion of telomere-specific non-LTR retrotransposon, SART1Bm, Sci. Rep., 2020, vol. 10, p. 8963.
Ng, S.Y., Bogu, G.K., Soh, B.S., and Stanton, L.W., The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis, Mol. Cell, 2013, vol. 51, pp. 349–359.
Park, J. and Belden, W.J., Long non-coding RNAs have age-dependent diurnal expression that coincides with age-related changes in genome-wide facultative heterochromatin, BMC Genomics, 2018, vol. 19, p. 777.
Pavlicev, M., Hiratsuka, K., Swaggart, K., et al., Detecting endogenous retrovirus-driven tissue-specific gene transcription, Genome Biol. Evol., 2015, vol. 7, pp. 1082–1097.
Pereira Fernandes, D.P., Bitar, M., Jacobs, F.M., and Barry, G., Long non-coding RNAs in neuronal aging, Noncoding RNA, 2018, vol. 4, p. E12.
Popa, A., Labrigand, P., Barbry, R., and Waldmann, R., Pateamine A-sensitive ribosome profiling reveals the scope of translation in mouse embryonic stem cells, BMC Genomics, 2016, vol. 17, p. 52.
Qian, W., Cai, X., and Qian, Q., Sirt1 antisense long non-coding RNA attenuates pulmonary fibrosis through sirt1-mediated epithelial-mesenchymal transition, Aging (Albany, NY), 2020, vol. 12, pp. 4322–4336.
Rodriguez-Martin, B., Alvarez, E.G., Baez-Ortega, A., et al., Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition, Nat. Genet., 2020, vol. 52, pp. 306–319.
Ruiz-Orera, J., Messeguer, X., Subirana, J.A., and Alba, M.M., Long non-coding RNAs as a source of new peptide, eLife, 2014, vol. 3, p. e03523.
Schratz, K.E., Extrahematopoietic manifestations of the short telomere syndromes, Hematol. Am. Soc. Educ. Progr., 2020, vol. 2020, pp. 115–122.
Scheidler, C.M., Kick, L.M., and Schneider, S., Ribosomal peptides and small proteins on the rise, ChemBioChem, 2019, vol. 20, pp. 1479–1486.
Schroeder, E.A., Raimundo, N., and Shadel, G.S., Epigenetic silencing mediates mitochondria stress-induced longevity, Cell Metab., 2013, vol. 17, pp. 954–964.
Tiwari, B., Jones, A.E., Caillet, C.J., et al., P53 directly repress human LINE1 transposons, Genes. Dev., 2020, vol. 34, pp. 1439–1451.
Trapathi, V., Shen, Z., Chakraborty, A., et al., Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB, PLoS Genet., 2013, vol. 9, p. e1003368.
Trembinski, D.J., Bink, D.I., Theodorou, K., et al., Aging-regulated anti-apoptotic long non-coding RNA Sarrah augments recovery from acute myocardial infarction, Nat. Commun., 2020, vol. 11, p. 2039.
Trizzino, M., Kapusta, A., and Brown, C.D., Transposable elements generate regulatory novelty in a tissue-specific fashion, BMC Genomics, 2018, vol. 19, p. 468.
Wang, H., Iacoangeli, A., Popps, S., et al., Dendritic BC1 RNA: functional role in regulation of translation initiation, J. Neurosci., 2002, vol. 22, no. 23, pp. 10 232–10 241.
Wang, T., Zeng, J., Lowe, C.B., et al., Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53, Proc. Natl. Acad. Sci. U.S.A., 2007, vol. 104, pp. 18 613–18 618.
Zhang, J., Mujahid, H., Hou, Y., et al., Plant long ncRNAs: a new frontier for gene regulatory control, Am. J. Plant Sci., 2013, vol. 4, no. 5, art. ID 32 139.
Zhao, Y., Yuan, J., and Chen, R., NONCODEv4: Annotation of noncoding RNAs with emphasis on long noncoding RNAs, in Long Non-Coding RNAs: Methods and Protocols, Methods Mol. Biol. Ser., vol. 1402, New York: Springer-Verlag, 2016, pp. 243–254.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author declares that he has no conflicts of interest. This article does not contain any studies involving animals or human participants performed by the author.
Additional information
Translated by A. Barkhash
Rights and permissions
About this article
Cite this article
Mustafin, R.N. Relationship of Peptides and Long Non-Coding RNAs with Aging. Adv Gerontol 11, 351–361 (2021). https://doi.org/10.1134/S2079057021040081
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S2079057021040081