Abstract
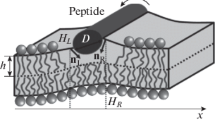
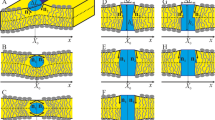
A number of processes in living cells are accompanied by significant changes of the geometric curvature of lipid membranes. In turn, heterogeneity of the lateral curvature can lead to spatial redistribution of membrane components, most important of which are transmembrane proteins and liquid-ordered lipid-protein domains. These components have a so-called hydrophobic mismatch: the length of the transmembrane domain of the protein, or the thickness of the bilayer of the domain differ from the thickness of the surrounding membrane. In this work we consider redistribution of membrane components with hydrophobic mismatch in membranes with non-uniform geometric curvature. Dependence of the components’ energy on the curvature is calculated in terms of theory of elasticity of liquid crystals adapted to lipid membranes. According to the calculations, transmembrane proteins prefer regions of the membrane with zero curvature. Liquid-ordered domains having a size of a few nm distribute mainly into regions of the membrane with small negative curvature appearing in the cell plasma membrane in the process of endocytosis. The distribution of domains of a large radius is determined by a decrease of their perimeter upon bending; these domains distribute into membrane regions with relatively large curvature.
Similar content being viewed by others
References
Capraro B.R., Yoon Y., Cho W., Baumgart T. 2010. Curvature sensing by the epsin N-terminal homology (ENTH) domain measured on cylindrical lipid membrane tethers. J. Am. Chem. Soc. 132, 1200–1201.
Kamal M.M., Mills D., Grzybek M., Howard J. 2009. Measurement of the membrane curvature preference of phospholipids reveals only weak coupling between lipid shape and leaflet curvature. Proc. Natl. Acad. Sci. USA. 106, 22245–22250.
Quemeneur F., Sigurdsson J.K., Renner M., Atzberger P.J., Bassereau P., Lacoste D. 2014. Membrane shaping proteins and implications for their mobility in lipid membranes. Proc. Natl. Acad. Sci. USA. 111, 5083–5087.
Bashkirov P.V., Chekashkina K.V., Akimov S.A., Kuzmin P.I., Frolov V.A. 2011. Variation of lipid membrane composition caused by strong bending. Biol. Membrany (Rus.). 28, 145–152.
Galimzyanov T.R., Akimov S.A. 2011. Phase separation in lipid membranes induced by the elastic properties of components. JETP Letters. 93, 463–469.
Karpunin D.V., Akimov S.A., Frolov V.A. 2005. Pore formation in lipid membranes containing lysolipids and cholesterol. Biol. Membrany (Rus.). 22, 429–432.
Simons K., Ikonen E. 1997. Functional rafts in cell membranes. Nature. 387, 569–572.
Edidin M. 2001. Shrinking patches and slippery rafts: scales of domains in the plasma membrane. Trends Cell. Biol. 11, 492–496.
Ayuyan A.G., Cohen F.S. 2008. Raft composition at physiological temperature and pH in the absence of detergents. Biophys. J. 94, 2654–2666.
Heftberger P., Kollmitzer B., Rieder A.A., Amenitsch H., Pabst G. 2015. In situ determination of structure and fluctuations of coexisting fluid membrane domains. Biophys. J. 108, 854–862.
Garcia-Saez A.J., Chiantia S., Schwille P. 2007. Effect of line tension on the lateral organization of lipid membranes. J. Biol. Chem. 282, 33537–33544.
Rinia H.A., Snel M.M.E., van der Eerden J.P.J.M., de Kruijff B. 2001. Imaging domains in model membranes with atomic force microscopy. FEBS Lett. 501, 194–199.
Weiss T.M., van der Wel P.C.A., Killian J.A., Koeppe R.E., Huang H.W. 2003. Hydrophobic mismatch between helices and lipid bilayers. Biophys. J. 84, 379–385.
Gandhavadi M., Allende D., Vidal A., Simon S.A., McIntosh T.J. 2002. Structure, composition, and peptide binding properties of detergent soluble bilayers and detergent resistant rafts. Biophys. J. 82, 1469–1482.
Scheiffele P., Roth M.G., Simons K. 1997. Interaction of influenza virus haemagglutinin with sphingolipidcholesterol membrane domains via its transmembrane domain. EMBO J. 16, 5501–5508.
Scheiffele P., Rietveld A., Simons K. 1999. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J. Biol. Chem. 274, 2038–2044.
Takeda M., Leser G.P., Russell C.J., Lamb R.A. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA. 100, 14610–14617.
Galimzyanov T.R., Molotkovsky R.J., Kuzmin P.I., Akimov S.A. 2011. Stabilization of bilayer structure of raft due to elastic deformations of membrane. Biol. Membrany (Rus.). 28, 307–314.
Galimzyanov T.R., Molotkovsky R.J., Bozdaganyan M.E., Cohen F.S., Pohl P., Akimov S.A. 2015. Elastic membrane deformations govern interleaflet coupling of lipid-ordered domains. Phys. Rev. Lett. 115, 088101-1–88101-5.
Risselada H.J., Marrink S.J. 2008. The molecular face of lipid rafts in model membranes. Proc. Natl. Acad. Sci. USA. 105, 17367–17372.
Perlmutter J.D., Sachs J.N. 2011. Interleaflet interaction and asymmetry in phase separated lipid bilayers: molecular dynamics simulations. J. Am. Chem. Soc. 133, 6563–6577.
Galimzyanov T.R., Molotkovsky R.J., Kheyfets B.B., Akimov S.A. 2013. Energy of the interaction between membrane lipid domains calculated from splay and tilt deformations. JETP Letters. 96, 681–686.
Hamm M., Kozlov M.M. 2000. Elastic energy of tilt and bending of fluid membranes. Eur. Phys. J. E. 3, 323–335.
Rawicz W., Olbrich K.C., McIntosh T., Needham D., Evans E. 2000. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 79, 328–339.
Hamm M., Kozlov M.M. 1998. Tilt model of inverted amphiphilic mesophases. Eur. Phys. J. B. 6, 519–528.
Pan J., Tristram-Nagle S., Nagle J.F. 2009. Effect of cholesterol on structural and mechanical properties of membranes depends on lipid chain saturation. Phys. Rev. E 80, 021931.
Baumgart T., Das S., Webb W.W., Jenkins J.T. 2005. Membrane elasticity in giant vesicles with fluid phase coexistence. Biophys. J. 89, 1067–1080.
van Meer G., Voelker D.R., Feigenson G.W. 2008. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124.
Baumgart T., Hess S.T., Webb W.W. 2003. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 425, 821–824.
Akimov S.A., Hlaponin E.A., Bashkirov P.V., Boldyrev I.A., Mikhalyov I.I., Telford W.G., Molotkovskaya I.M. 2009. Ganglioside GM1 increases line tension at raft boundary in model membranes. Biol. Membrany (Rus.). 26, 234–239.
Tang Q., Edidin M. 2003. Lowering the barriers to random walks on the cell surface. Biophys. J. 84, 400–407.
Sasaki S., Yui N., Noda Y. 2014. Actin directly interacts with different membrane channel proteins and influences channel activities: AQP2 as a model. Biochim. Biophys. Acta. 1838, 514–520.
Watanabe S., Trimbuch T., Camacho-Perez M., Rost B.R., Brokowski B., Sohl-Kielczynski B., Felies A., Davis M.W., Rosenmund C., Jorgensen E.M. 2014. Clathrin regenerates synaptic vesicles from endosomes. Nature. 515, 228–233.
Wahl S., Katiyar R., Schmitz F. 2013. A local, periactive zone endocytic machinery at photoreceptor synapses in close vicinity to synaptic ribbons. J. Neurosci. 33, 10278–10300.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © D.S. Osipenko, T.R. Galimzyanov, S.A. Akimov, 2016, published in Biologicheskie Membrany, 2016, Vol. 33, No. 3, pp. 176–186.
Rights and permissions
About this article
Cite this article
Osipenko, D.S., Galimzyanov, T.R. & Akimov, S.A. Lateral redistribution of transmembrane proteins and liquid-ordered domains in lipid membranes with inhomogeneous curvature. Biochem. Moscow Suppl. Ser. A 10, 259–268 (2016). https://doi.org/10.1134/S1990747816030077
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1990747816030077