Abstract
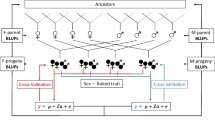
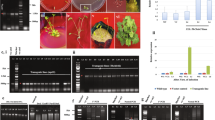
As an ornamental organ, petal plays an important role in attracting insect pollination, it’s development attracts the attention of many researchers. In this study, a sunflower mutant line with ligulate-like tubular petal (lpm) was screened out, and its disc tubular florets were characterized with elongated ligulate-like petals, longer filaments size, fewer anthers and lower seed setting rate, compared with wide type (WT) plants. From the suppression subtractive hybridization library constructed using inflorescence of lpm and WT plants, the HaYABBY was identified with expressing differentially. The cDNA and structural sequences of HaYABBY gene were 997 and 4339 bp, respectively, encoding a peptide with 219 amino acids. The analysis of HaYABBY expression showed that it presented higher transcription level in flower than root, stem and leaf of sunflower. The expression level of YABBY in disc florets of lpm was significantly higher than those of WT, especially at early floral developmental stage. The results suggested that HaYABBY could impact the floral development at early stage, particularly on petal elongation and floral symmetry morphogenesis in sunflower. Furthermore, the results implied that HaYABBY would be involved in the regulation of reproductive system development and affect their fertility. Our findings will provide some insight to understand the regulating network of floral development in sunflower, and help to cultivate new sunflower varieties with high ornamental value.






Similar content being viewed by others
REFERENCES
Funk, V.A., Bayer, R.J., Keeley, S., et al., Everywhere but Antarctica: using a supertree to understand the diversity and distribution of the Compositae, Biol. Skr., 2005, pp. 343—373.
Takeda, S., Matsumoto, N., and Okada, K., RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana, Development, 2004, vol. 131, no. 2, pp. 425—434. https://doi.org/10.1242/dev.00938
Varaud, E., Brioudes, F., Szécsi, J., et al., AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp, Plant Cell, 2011, vol. 23, no. 3, pp. 973—983. https://doi.org/10.1105/tpc.110.081653
Powell, A.E. and Lenhard, M., Control of organ size in plants, Curr. Biol., 2012, vol. 22, no. 9, pp. R360—R367. https://doi.org/10.1016/j.cub.2012.02.010
Claisse, G., Charrier, B., and Kreis, M., The Arabidopsis thaliana GSK3/Shaggy like kinase AtSK3-2 modulates floral cell expansion, Plant Mol. Biol., 2007, vol. 64, nos. 1—2, pp. 113—124. https://doi.org/10.1007/s11103-007-9138-y
Tzeng, T.Y., Kong, L.R., Chen, C.H., et al., Overexpression of the lily p70(s6k) gene in Arabidopsis affects elongation of flower organs and indicates TOR-dependent regulation of AP3, PI and SUP translation, Plant Cell Physiol., 2009, vol. 50, no. 9, pp. 1695—1709. https://doi.org/10.1093/pcp/pcp114
Li, S., Liu, Y., Zheng, L., et al., The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana, New Phytol., 2012, vol. 194, no. 3, pp. 690—703. https://doi.org/10.4161/psb.21620
Szécsi, J., Joly, C., Bordji, K., et al., BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size, EMBO J., 2006, vol. 25, no. 16, p. 3912. https://doi.org/10.1038/sj.emboj.7601270
Li, N., Huang, B., Tang, N., et al., The MADS-box gene SlMBP21 regulates sepal size mediated by ethylene and auxin in tomato, Plant Cell Physiol., 2017, vol. 58, no. 12, pp. 2241—2256. https://doi.org/10.1093/pcp/pcx158
Griffith, M.E., Da, S.C.A., Smyth, D.R., PETAL LOSS gene regulates initiation and orientation of second whorl organs in the Arabidopsis flower, Development, 1999, vol. 126, no. 24, pp. 5635—5644. https://doi.org/10.1083/jcb.42.2.377
Disch, S., Anastasiou, E., Sharma, V.K., et al., The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner, Curr. Biol., 2006, vol. 16, no. 3, p. 272. https://doi.org/10.1016/j.cub.2005.12.026
Shul’ga, O.A., Shennikova, A.V., Angenent, G.S., and Skriabin, K.G., MADS-box genes controlling inflorescence morphogenesis in sunflower, Ontogenez, 2008, vol. 39, no. 1, pp. 2—5. https://doi.org/10.1134/s1062360408010025
Chapman, M.A., Tang, S., Draeger, D., et al., Genetic analysis of floral symmetry in Van Gogh’s sunflowers reveals independent recruitment of CYCLOIDEA genes in the Asteraceae, PLoS Genet., 2012, vol. 8, no. 3. e1002628. https://doi.org/10.1371/journal.pgen.1002628
Fambrini, M., Salvini, M., Basile, A., and Pugliesi, C., Transposon-dependent induction of Vincent van Gogh’s sunflowers: exceptions revealed, Genesis, 2014, vol. 52, no. 4, p. 315. https://doi.org/10.1002/dvg.22743
Tanaka, W. and Hirano, H.Y., The YABBY gene TONGARI-BOUSHI1 is involved in lateral organ development and maintenance of meristem organization in the rice spikelet, Plant Cell, 2012, vol. 24, no. 1, pp. 80—95. https://doi.org/10.2307/41433951
Golz, J.F. and Hudson, A., Plant development: YABBYs claw to the fore, Curr. Biol., 1999, vol. 9, no. 22, pp. R861—R863. https://doi.org/10.1016/s0960-9822(00)80047-0
Bowman, J.L., The YABBY gene family and abaxial cell fate, Curr. Opin. Plant Biol., 2000, vol. 3, no. 1, pp. 17—22. https://doi.org/10.1016/s1369-5266(99)00035-7
Floyd, S.K. and Bowman, J.L., The ancestral developmental tool kit of land plants, Int. J. Plant Sci., 2007, vol. 168, no. 1, pp. 1—35. https://doi.org/10.1086/509079
Nishiyama, T., Fujita, T., Seki, M., et al., Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implication for land plant evolution, Proc. Natl. Acad. Sci. U.S.A., 2003, vol. 100, no. 13, p. 8007. https://doi.org/10.1073/pnas.0932694100
Bowman, J.L. and Smyth, D.R., CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains, Development, 1999, vol. 126, no. 11, pp. 2387—2396. https://doi.org/10.1007/s004290050253
Yamada, T., Ito, M., and Kato, M., YABBY2—homologue expression in lateral organs of Amborella trichopoda (Amborellaceae), Int. J. Plant Sci., 2004, vol. 165, no. 6, pp. 917—924. https://doi.org/10.1086/423793
Lee, J.Y., Baum, S.F., Oh, S.H., et al., Recruitment of CRABS CLAW to promote nectary development within the eudicot clade, Development, 2005, vol. 132, no. 22, pp. 5021—5032. https://doi.org/10.1242/dev.02067
Eckardt, N.A., YABBY genes and the development and origin of seed plant leaves, Plant Cell, 2010, vol. 22, no. 7, pp. 2103—2103. https://doi.org/10.2307/20780551
Villanueva, J.M., Broadhvest, J., Hauser, B.A., et al., INNER NO OUTER regulates abaxial–adaxial patterning in Arabidopsis ovules, Genes Dev., 1999, vol. 13, no. 23, pp. 3160—3169. https://doi.org/10.1101/gad.13.23.3160
Stahle, M.I., Kuehlich, J., Staron, L., et al., YABBYs and the transcriptional corepressors LEUNIG and LEUNIG_HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis, Plant Cell, 2009, vol. 21, no. 10, pp. 3105—3118. https://doi.org/10.1105/tpc.109.070458
Sarojam, R., Sappl, P.G., Goldshmidt, A., et al., Differentiating Arabidopsis shoots from leaves by combined YABBY activities, Plant Cell, 2010, vol. 22, no. 7, pp. 2113—2130. https://doi.org/10.1105/tpc.110.075853
Golz, J.F., Roccaro, M.R., and Hudson, A., GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves, Development, 2004, vol. 131, no. 15, pp. 3661—3670. https://doi.org/10.1242/dev.01221
Navarro, C., Efremova, N., Golz, J.F., et al., Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development, Development (Cambridge, England), 2004, vol. 131, no. 15, pp. 3649—3659. https://doi.org/10.1242/dev.01205
Kim, M., Pham, T., Hamidi, A., et al., Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves, Development, 2003, vol. 130, no. 18, pp. 4405—4415. https://doi.org/10.1242/dev.00655
Siegfried, K.R., Eshed, Y., Baum, S.F., et al., Members of the YABBY gene family specify abaxial cell fate in Arabidopsis, Development (Cambridge, England), 1999, vol. 126, no. 18, pp. 4117—4128. https://doi.org/10.1021/ie020097t
Sawa, S., Watanabe, K., Goto, K., et al., FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains, Genes Dev., 1999, vol. 13, no. 9, pp. 1079—1088. https://doi.org/10.1101/gad.13.9.1079
Juarez, M.T., Kui, J.S., Thomas, J., and Timmermans, M.C., microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity, Nature, 2004, vol.428, no. 6978, pp. 84—88. https://doi.org/10.1038/nature02363
Juarez, M.T., Twigg, R.W., and Timmermans, M.C.P., Specification of adaxial cell fate during maize leaf development, Development, 2004, vol. 131, no. 18, pp. 4533—4544. https://doi.org/10.1242/dev.01328
Zhao, W., Su, H.Y., Song, J., et al., Ectopic expression of TaYAB1, a member of YABBY gene family in wheat, causes the partial abaxialization of the adaxial epidermises of leaves and arrests the development of shoot apical meristem in Arabidopsis, Plant Sci., 2006, vol. 170, no. 2, pp. 364—371. https://doi.org/10.1016/j.plantsci.2005.09.008
Dai, M., Hu, Y., Zhao, Y., et al., A WUSCHEL-like homeobox gene represses a YABBY gene expression required for rice leaf development, Plant Physiol., 2007, vol. 144, no. 1, pp. 380—390. https://doi.org/10.1104/pp.107.095737
Toriba, T., Harada, K., Takamura, A., et al., Molecular characterization the YABBY gene family in Oryza sativa and expression analysis of OsYABBY1, Mol. Genet. Genomics, 2007, vol. 277, no. 5, pp. 457—468. https://doi.org/10.1007/s00438-006-0202-0
Yamaguchi, T., Nagasawa, N., Kawasaki, S., et al., The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa, Plant Cell, 2004, vol. 16, no. 2, pp. 500—509. https://doi.org/10.1105/tpc.018044
Ohmori, Y., Abiko, M., Horibata, A., and Hirano, H.Y., A transposon, Ping, is integrated into intron 4 of the DROOPING LEAF gene of rice, weakly reducing its expression and causing a mild drooping leaf phenotype, Plant Cell Physiol., 2008, vol. 49, no. 8, pp. 1176—1184. https://doi.org/10.1093/pcp/pcn093
Ohmori, Y., Toriba, T., Nakamura, H., et al., Temporal and spatial regulation of DROOPING LEAF gene expression that promotes midrib formation in rice, Plant J., 2011, vol. 65, no. 1, pp. 77–86. https://doi.org/10.1111/j.1365-313X.2010.04404.x
Tanaka, W., Toriba, T., Ohomori, Y., and Hirano, H.Y., Formation of two florets within a single spikelet in the rice tongari-boushi1 mutant, Plant Signal Behav., 2012, vol. 7, no. 7, pp. 793—795. https://doi.org/10.4161/psb.20522
Jang, S., Hur, J., Kim, S.J., et al., Ectopic expression of OsYAB1 causes extra stamens and carpels in rice, Plant Mol. Biol., 2004, vol. 56, no. 1, pp. 133—143. https://doi.org/10.1007/s11103-004-2648-y
Letousey, P., De Zélicourt, A., Vieira, D.S., et al., Molecular analysis of resistance mechanisms to Orobanche cumana in sunflower, Plant Pathol., 2007, vol. 56, no. 3, pp. 536—546. https://doi.org/10.1111/j.1365-3059.2007.01575.x
Mohamed Ahmed, I.A., Eltayeb, M.M., Habora, M.E.E., et al., Identification of the key genes involved in the degradation of homocholine by Pseudomonas sp. strain A9 by using suppression subtractive hybridization, Process Biochem., 2017, vol. 52, pp. 94—105. https://doi.org/10.1016/j.procbio.2016.10.009
Zhang, Z. and Zhang, Q., Molecular cloning, characterization and expression of heat shock protein 70 gene from the oyster Crassostrea hongkongensis responding to thermal stress and exposure of Cu2+ and malachite green, Gene, 2012, vol. 497, no. 2, pp. 172—180. https://doi.org/10.1016/j.gene.2012.01.058
Rao, X.Y., Huang, X.L., Zhou, Z.H., Lin, X., An improvement of the 2^(–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis, Biostat. Bioinf. Biomath., 2013, vol. 3, no. 3, pp. 71—85. https://doi.org/10.1016/S0920-5489(99)92176-1
Endress, P.K., Origins of flower morphology, J. Exp. Zool., 2001, vol. 291, no. 2, pp. 105—115. https://doi.org/10.1002/jez.1063
Whitney, H.M. and Glover, B.J., Morphology and development of floral features recognised by pollinators, Arthropod—Plant Int., 2007, vol. 1, no. 3, pp. 147—158. https://doi.org/10.1007/s11829-007-9014-3
Meredith, M., Thomas, P.J., Rudall, A.G., et al., Development of a complex floral trait: the pollinator-attracting petal spots of the beetle daisy, Gorteria diffusa (Asteraceae), Am. J. Bot., 2009, vol. 96, no. 12, p. 2184. https://doi.org/10.3732/ajb.0900079
Frediani, D. and Pinzauti, M., Effects of entomophilous pollination on sunflower seed production, Terra Pugliese, 1978, vol. 27, pp. 15—18.
Sawa, S., Ito, T., Shimura, Y., and Okada, K., FILAMENTOUS FLOWER controls the formation and development of Arabidopsis inflorescences and floral meristems, Plant Cell, 1999, vol. 11, no. 1, pp. 69—86. https://doi.org/10.1105/tpc.11.1.69
Orashakova, S., Lange, M., Lange, S., et al., The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation, Plant J., 2009, vol. 58, no. 4, pp. 682—693. https://doi.org/10.1111/j.1365-313X.2009.03807.x
Yamada, T., Yokota, S., Hirayama, Y., et al., Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms, Plant J., 2011, vol. 67, no. 1, pp. 26—36. https://doi.org/10.1111/j.1365-313x.2011.04570.x
Funding
This work is financially supported by National Natural Science Fund of China (#31171587), Innovation Team Program of Sichuan Education Department (16TD0020), and Talent Fund of China West Normal University (17YC354).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
APPENDIX
APPENDIX
Alignment of amino acids specific to HaYABBY and subfamily in different Arabidopsis thaliana. Completely conserved and highly conserved residues are colored black and grey, respectively. Dashes are gaps introduced to maximize alignment. The alignment was produced from 5 amino acid sequences from YABBY subfamily of At (Arabidopsis thaliana).The amino acids of YABBY were aligned using CLUSTAL X and analyzed with MEGA version 6.
Rights and permissions
About this article
Cite this article
Wu, Y., Lei, D., Su, Z. et al. HaYABBY Gene Is Associated with the Floral Development of Ligulate-Like Tubular Petal Mutant Plants of Sunflower. Russ J Genet 56, 1457–1468 (2020). https://doi.org/10.1134/S1022795420120145
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795420120145