Abstract
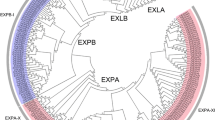
Expansins are ancient proteins involved in cell wall loosening during diverse biological processes in plants. They may participate inplant developmental processes and in responses to multiple external stresses. Triticum urartu is a diploid wheat, which is the A genome donor in hexaploid wheat (AABBDD). Wheat is an important agricultural crop. In this study, a total of 39 expansin (EXP) genes in T. urartu were identified and categorised into three subfamilies according to their phylogenetic relationship, consisting of 19 EXPA genes, 18 EXPB genes and 2 EXLA genes, but no EXLB genes. The structures of T. urartu EXP (TuEXP) genes were conserved. Expression analysis and transcript profiling revealed differential expression of TuEXPs in different tissues and developmental stages. Expression patterns of TuEXPs genes were also investigated in different tissues under different phytohormone treatments at the three leafstage. The expression of most of the analyzed genes were either up-or down-regulated, which could help research on expansin-related mechanisms.






Similar content being viewed by others
REFERENCES
Cosgrove, D.J., Li, L.C., Cho, H.T., et al., The growing world of expansins, Plant Cell Physiol., 2002, vol. 43, pp. 1436—1444.
McQueen-Mason, S., Durachko, D.M., and Cosgrove, D.J., Two endogenous proteins that induce cell wall extension in plants, Plant Cell, 1992, vol. 4, pp. 1425—1433.
Cosgrove, D.J., Plant expansins: diversity and interactions with plant cell walls, Curr. Opin. Plant Biol., 2015, vol. 25, pp. 162—172.
Wang, T., Park, Y.B., Caporini, M.A., et al., Sensitivity-enhanced solid-state NMR detection of expansin’s target in plant cell walls, Proc. Natl. Acad. Sci. U.S.A., 2013, vol. 110, no. 41, pp. 16444—16449.
Sampedro, J. and Cosgrove, D.J., The expansin superfamily, Genome Biol., 2005, vol. 6, no. 12, p. 242.
Georgelis, N., Yennawar, N.H., and Cosgrove, D.J., Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin, Proc. Natl. Acad. Sci. U.S.A., 2012, vol. 109, no. 37, pp. 14830—14835.
Zhang, W., Yan, H., Chen, W., et al., Genome-wide identification and characterization of maize expansin genes expressed in endosperm, Mol. Genet. Genomics, 2014, vol. 289, no. 6, pp.1061—1074.
Zhang, J.F, Xu, Y.Q, Dong, J.M, et al., Genome-wide identification of wheat (Triticum aestivum) expansins and expansin expression analysis in cold-tolerant and cold-sensitive wheat cultivars, PLoS One, 2018, vol. 13. e0195138
Carey, R.E. and Cosgrove, D.J., Portrait of the expansin superfamily in Physcomitrella patens: comparisons with angiosperm expansins, Ann. Bot., 2007, vol. 99, no. 6, pp.1131—1141.
Rose, J.K.C, Cosgrove, D.J, Albersheim, P., et al., Dectection of expansin proteins and activity during tomato fruit ontogeny, Plant Physiol., 2000, vol. 123, no. 4, pp. 1583—1592.
Cho, D.J. and Cosgrove, D.J., Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana,Proc. Natl. Acad. Sci. U.S.A., 2000, vol. 97, no. 17, pp. 9783—9788.
Guo, W., Zhao, J., Li, X., Qin, L., et al., A soybean beta-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses, Plant J., 2011, vol. 66, no. 3, pp. 541—552.
Yu, Z.M., Kang B., He, X. W., et al., Root hair-specific expansins modulate root hair elongation in rice, Plant J., 2011, vol. 66, no. 5, pp. 725—734.
Kuluev, B.R., Safiullina, M.G., Kniazev, A.V., and Chemeris, A.V., Effect of ectopic expression of NtEXPA5 gene on cell size and growth of organs of transgenic tobacco plants, Ontogenez, 2013, vol. 44, pp. 34—41.
Harrison, E.P., McQueen-Mason, S.J., and Manning, K., Expression of six expansin genes in relation to extension activity in developing strawberry fruit, J. Exp. Bot., 2001, vol. 52, no. 360, pp. 1437—1446.
Ishimaru, M., Smith, D.L., Gross, K.C., and Kobayashi, S., Expression of three expansin genes during development and maturation of Kyoho grape berries, J. Plant Physiol., 2007, vol. 164, no. 12, pp. 1675—1682.
Han, Y., Li, A., Li, F., et al., Characterization of a wheat (Triticum aestivum L.) expansin gene, TaEXPB23, involved in the abiotic stress response and phytohormone regulation. Plant Physiol. Biochem., 2012, vol. 54, pp. 49—58.
Lu, P., Kang, M., Jiang, X., et al., RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis,Planta, 2013, vol. 237, no. 6, pp. 1547—1559.
Xu, J., Tian, J, Belanger, F.C., and Huang, B.R., Identification and characterization of an expansin gene AsEXP1 associated with heat tolerance in C3 Agrostis grass species, J. Exp. Bot., 2007, vol. 58, no. 58, pp. 3789—3796.
Marowa, P., Ding, A., and Kong, Y., expansins: roles in plant growth and potential applications in crop improvement, Plant Cell Rep., 2016, vol. 35, no. 5, pp. 949—965.
Park, C.H., Kim, T.W., Son, S.H., et al., Bгassinosteroids control AtEXPA5 gene expression in Arabidopsis thaliana, Phytochemistry, 2010, vol. 71, пo. 4, pp. 380—387.
Lu, Y., Liu, L., Wang, X., et al., Genome-wide identification and expression analysis of the expansin gene family in tomato, Mol. Genet. Genomics, 2015, vol. 291, no. 2, pp. 597—608.
Lee, Y., Choi, D., and Kende, H., Expansins: ever-expanding numbers and functions, Curr. Opin. Plant Biol., 2001, vol. 4, no. 6, pp. 527—532.
Lee, Y. and Kende, H. Expression of alpha-expansin and expansin-like genes in deepwater rice, Plant Physiol., 2002, vol. 130, no. 3, pp. 1396—1405.
Wu, X., Song, C., Wang, B., and Cheng, J., Hidden Markov model used in protein sequence analysis, J. Biomed. Eng., 2002, vol. 19, no. 3, pp. 455—458.
Li, N.N., Pu, Y.Y., Gong, Y.C., et al., Genomic location and expression analysis of expansin gene family reveals the evolutionary and functional signficance in Triticum aestivum,Gene Genomics, 2016, vol. 38, no. 11, pp. 1021—1030.
Tamura, K., Stecher, G., Peterson, D., et al., MEGA6: Molecular Evolutionary Genetics Analysis version 6.0, Mol. Biol. Evol., 2013, vol. 30, no. 12, pp. 2725—2729.
Sperandeo, M.P., Annunziata, P., Ammendola, V., et al., Lysinuric protein intolerance: identification and functional analysis of mutations of the SLC7A7 gene, Hum. Mutat., 2005, vol. 25, no. 4, p. 410.
Petersen, T.N., Brunak, S., von Heijne, G., and Nielsen, H., SignalP 4.0: discriminating signal peptides from transmembrane regions, Nat. Methods, 2011, vol. 8, no. 10, pp. 785—786.
Guo, A.Y., Zhu, Q.H., Chen, X., and Luo, J.C., GSDS: a gene structure display server, Yi chuan, 2007, vol. 29, pp. 1023—1026.
Lin, Y.X., Jiang, H.Y., Chu, Z.X., et al., Genome-wide identification, classification and analysis of heat shock transcription factor family in maize, BMC Genomics, 2011, vol. 12, no. 76, pp. 1—14.
John, C.M., Preetam, H.S., and William, B.L. Trichomonas vaginalis: analysis of codon usage, Exp. Parasitol., 1997, vol. 87, no. 1, pp. 73–74.
Deng, W., Wang, Y., Liu, Z., et al., HemI: a toolkit for illustrating heatmaps, PLoS One, 2014, vol. 9. e111988
Fredslund, J. and Lange, M., Primique: automatic design of specific PCR primers for each sequence in a family, BMC Bioinf., 2007, vol. 8, p. 369.
Sampedro, J., Lee, Y., Carey, R.E., et al., Use of genomic history to improve phylogeny and understanding of births and deaths in a gene family, Plant J., 2005, vol. 44, no. 3, pp. 409—419.
Plotkin, J.B. and Kudla, G., Synonymous but not the same: the cause and consequences of codon bias, Nat. Rev. Genet., 2011, vol. 12, no. 1, pp. 32—42.
Shewry, P.R., Wheat, J. Exp. Bot., 2009, vol. 60, no. 6, pp. 1537—1553.
Sharma, A., Sharma, N., Bhalla, P., and Singh, M., Comparative and evolutionary analysis of grass pollen allergens using Brachypodium distachyon as a model system, PLoS One, 2017, vol. 12. e0169686
Zhu, Y., Wu, N., Song, W., et al., Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies, BMC Plant Biol., 2014, vol. 14, no. 1, p. 93.
Dal Santo, S., Vannozzi, A., Tornielli, G.B., et al., Genome-wide analysis of the expansin gene superfamily reveals grapevine-specific structural and functional characteristics, PLoS One, 2013, vol. 8. e62206
Lu, Y., Liu, L., Wang, X., et al., Genome-wide identification and expression analysis of the expansin gene family in tomato, Mol. Genet. Genomics, 2015, vol. 291, no. 2, pp. 597—608.
O’Neill, Y., The composition and structure of plant primary walls, Plant Cell Wall, 2003, vol. 8, pp. 1—54.
Li, Y., Darley, C.P., Ongaro, V., et al., Plant expansins are a complex multigene family with an ancient evolutionary origin, Plant Physiol., 2002, vol. 128, no. 3, pp. 854—864.
Seader, V.H., Thornsberry, J.M., and Carey, R.E., Utility of the Amborella trichopoda expansin superfamily in elucidating the history of angiosperm expansins, J. Plant Res., 2016, vol. 129, no. 2, pp. 199—207.
Peng, X.J, Zhao, Y., Cao, J.G., et al., CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS One, 2012, vol. 7. e40120
Zhao, Y., Zhou, Y.Q., Jiang, H.Y., et al., Systematic analysis of sequences and expression patterns of drought-responsive member of HD-Zip gene family in maize, PLoS One, 2011, vol. 6. e28488
Li, X., Zhao, J., Walk, T.C., and Liao, H., Characterization of soybean β-expansin genes and their expression responses to symbiosis, nutrient deficiency, and hormone treatment, Appl. Microbiol. Biotechnol., 2014, vol. 98, no. 6, pp. 2805—2817.
Xue, T., Wang, D., Zhang, S., et al., Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis,BMC Genomics, 2008, vol. 9, no. 550, pp. 1—21.
Cho, H.T., and Cosgrove, D.J., Regulation of root hair initiation and expansin gene expression in Arabidopsis,Plant Cell, 2002, vol. 14, no. 12, pp. 3237—3253.
Wu, Y., Thorne, E.T., Sharp, R.E., and Cosgrove, D.J., Modification of expansin transcript levels in the maize primary root at low water potentials, Plant Physiol., 2001, vol. 126, no. 4, pp. 1471—1479.
Cho, H.T. and Kende, H., Expression of expansin genes is correlated with growth in deepwater rice, Plant Cell, 1997, vol. 9, no. 9, pp. 1661—1671.
Downes, B.P., Steinbaker, C.R., and Crowell, D.N., Expression and processing of a hormonally regulated beta-expansin from soybean, Plant Physiol., 2001, vol. 126, no. 1, pp. 244—252.
Li, F., Han, Y., Feng, Y., et al., Expression of wheat expansin driven by the RD29 promoter in tobacco confers water-stress tolerance without impacting growth and development, J. Biotechnol., 2013, vol. 163, no. 3, pp. 281—291.
Li, F., Xing, S., Guo, Q., et al., Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco, J. Plant Physiol., 2011, vol. 168, no. 9, pp. 960—966.
Xu, Q., Xu, X., Shi, Y., et al., Transgenic tobacco plants overexpressing a grass PpEXP1 gene exhibit enhanced tolerance to heat stress, PLoS One, 2014, vol. 9. e100792
Zhou, J., Xie, J., Liao, H., and Wang, X., Overexpression of beta-expansin gene GmEXPB2 improves phosphorus efficiency in soybean, Physiol. Plant., 2014, vol. 150, no. 2, pp. 194—204.
Yan, A., Wu, M., Yan, L., et al., AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis,PLoS One, 2014, vol. 9. e85208
ACKONWLEDGMENTS
This research was funded by Capability improving and scientific research training project in the talent training fund of national natural science fund (J1210069). Thank the National Crop Germplasm Bank for providing seeds.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Supplementary material
Rights and permissions
About this article
Cite this article
Peng, L., Xu, Y., Feng, X. et al. Identification and Characterization of the Expansin Genes in Triticum urartu in Response to Various Phytohormones. Russ J Genet 56, 441–453 (2020). https://doi.org/10.1134/S1022795420040109
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795420040109