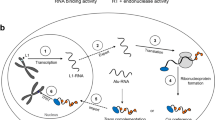
Abstract
LINE-1 retrotransposon is the most common mobile genetic element in the genomes of various mammals, including humans. Its genes are represented by the greatest number of copies. For a long time, it has been considered that the presence of LINE-1 in genome reflects the limited ability of cells to eliminate it, and the retrotransposon activity is negative owing to the insertional mutagenesis. In recent years, the increased expression of LINE-1 retrotransposon and the activity of their encoded proteins observed in mammalian cells at different stages of development and, first of all, in early embryogenesis have been discussed in the literature. Is early embryogenesis the stage of development when the organism is more susceptible to the activity of retrotransposons, or does LINE-1 play some positive role in early embryonic development? This review is aimed at classifying the available data on the epigenetic regulation and the role of LINE-1 retrotransposon in embryogenesis of mammals. The link between the mechanisms of regulation of LINE-1 expression and the waves of epigenetic reprogramming is tracked in germ cells, during fertilization, and in blastocyst, as well as during the differentiation of embryonic and extraembryonic tissues.
Similar content being viewed by others
References
Ostertag, E.M. and Kazazian, H.H., Biology of mammalian L1 retrotransposons, Annu. Rev. Genet., 2001, vol. 35, pp. 501–538. doi 10.1146/annurev.genet.35.102401.091032
Mukherjee, S., Mukhopadhyay, A., Banerjee, D., et al., Molecular pathology of haemophilia B: identification of five novel mutations including a LINE 1 insertion in Indian patients, Haemophilia, 2004, vol. 10, no. 3, pp. 259–263. doi 10.1111/j.1365-2516.2004. 00895.x
Brooks, M.B., Gu, W., Barnas, J.L., et al., A Line 1 insertion in the Factor IX gene segregates with mild hemophilia B in dogs, Mamm. Genome, 2003, vol. 14, no. 11, pp. 788–795. doi 10.1007/s00335-003-2290-z
Muhle, C., Zenker, M., Chuzhanova, N., and Schneider, H., Recurrent inversion with concomitant deletion and insertion events in the coagulation factor VIII gene suggests a new mechanism for X-chromosomal rearrangements causing hemophilia A, Hum. Mutat., 2007, vol. 28, no. 10, p. 1045. doi 10.1002/humu.9506
Feschotte, C., Transposable elements and the evolution of regulatory networks, Nat. Rev. Genet., 2008, vol. 9, no. 5, pp. 397–405. doi 10.1038/nrg2337
Goodier, J.L. and Kazazian, H.H., Retrotransposons revisited: the restraint and rehabilitation of parasites, Cell, 2008, vol. 135, no. 1, pp. 23–35. doi 10.1016/j.cell.2008.09.022
Faulkner, G.J., Kimura, Y., Daub, C.O., et al., The regulated retrotransposon transcriptome of mammalian cells, Nat. Genet., 2009, vol. 41, no. 5, pp. 563–571. doi 10.1038/ng.368
Jachowicz, J.W. and Torres-Padilla, M.E., LINEs in mice: features, families, and potential roles in early development, Chromosoma, 2015, vol. 125, no. 1, pp. 29–39. doi 10.1007/s00412-015-0520-2
Beck, C.R., Collier, P., Macfarlane, C., et al., LINE-1 retrotransposition activity in human genomes, Cell, 2010, vol. 141, no. 7, pp. 1159–1170. doi 10.1016/j.cell. 2010.05.021
Iskow, R.C., McCabe, M.T., Mills, R.E., et al., Natural mutagenesis of human genomes by endogenous retrotransposons, Cell, 2010, vol. 141, no. 7, pp. 1253–1261. doi 10.1016/j.cell.2010.05.020
Aschacher, T., Wolf, B., Enzmann, F., et al., LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines, Oncogene, 2015, vol. 35, no. 1, pp. 94–104. doi 10.1038/onc.2015.65
Guo, F., Yan, L., Guo, H., et al., The transcriptome and DNA methylome landscapes of human primordial germ cells, Cell, 2015, vol. 161, no. 6, pp. 1437–1452. doi 10.1016/j.cell.2015.05.015
Mathias, S.L., Scott, A.F., Kazazian, H.H., et al., Reverse transcriptase encoded by a human transposable element, Science, 1991, vol. 254, no. 5039, pp. 1808–1810. doi 10.1126/science.1722352
Fedorov, A., Regulation of mammalian LINE1 retrotransposon transcription, Cell Tiss. Biol., 2009, vol. 3, no. 1, p 1. doi 10.1134/S1990519X09010015
Martin, S.L., Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1, RNA Biol., 2010, vol. 7, no. 6, pp. 706–711. doi 10.4161/rna.7.6.13766
An, W., Dai, L., Niewiadomska, A.M., et al., Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs, Mob. DNA, 2011, vol. 2, no. 1, p. 2. doi 10.1186/1759-8753-2-2
Dai, L., Huang, Q., and Boeke, J.D., Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition, BMC Biochem., 2011, vol. 12, p. 18. doi 10.1186/1471-2091-12-18
Dewannieux, M. and Heidmann, T., LINEs, SINEs and processed pseudogenes: parasitic strategies for genome modeling, Cytogenet. Genome Res., 2005, vol. 110, nos. 1–4, pp. 35–48. doi 10.1159/000084936
Doucet, A.J., Hulme, A.E., Sahinovic, E., et al., Characterization of LINE-1 ribonucleoprotein particles, PLoS Genet., 2010, vol. 6, no. 10. doi 10.1371/journal. pgen.1001150
Aravin, A.A., Sachidanandam, R., Bourc’his, D., et al., A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice, Mol. Cell, 2008, vol. 31, no. 6, pp. 785–799. doi 10.1016/j.molcel.2008.09.003
Aravin, A.A., van der Heijden, G.W., Castaneda, J. et al., Cytoplasmic compartmentalization of the fetal piRNA pathway in mice, PLoS Genet., 2009, vol. 5, no. 12. e1000764. doi 10.1371/journal.pgen.1000764
Smith, Z.D., Chan, M.M., Mikkelsen, T.S., et al., A unique regulatory phase of DNA methylation in the early mammalian embryo, Nature, 2012, vol. 484, no. 7394, pp. 339–344. doi 10.1038/nature10960
Malki, S., van der Heijden, G.W., O’Donnell, K.A., et al., A role for retrotransposon LINE-1 in fetal oocyte attrition in mice, Dev. Cell, 2014, vol. 29, no. 5, pp. 521–533. doi 10.1016/j.devcel.2014.04.027
Luo, Y.B., Zhang, L., Lin, Z.L., et al., Distinct subcellular localization and potential role of LINE1-ORF1P in meiotic oocytes, Histochem. Cell Biol., 2015, vol. 145, no. 1, pp. 93–104. doi 10.1007/s00418-015-1369-4
Aravin, A.A., Sachidanandam, R., Girard, A., et al., Developmentally regulated piRNA clusters implicate MILI in transposon control, Science, 2007, vol. 316, no. 5825, pp. 744–747. doi 10.1126/science.1142612
Heyn, H., Ferreira, H.J., Bassas, L., et al., Epigenetic disruption of the PIWI pathway in human spermatogenic disorders, PLoS One, 2012, vol. 7, no. 10. e47892. doi 10.1371/journal.pone.0047892
Gasior, S.L., Wakeman, T.P., Xu, B., and Deininger, P.L., The human LINE-1 retrotransposon creates DNA dou ble-strand breaks, J. Mol. Biol., 2006, vol. 357, no. 5, pp. 1383–1393. doi 10.1016/j.jmb.2006.01.089
Vitullo, P., Sciamanna, I., Baiocchi, M., et al., LINE-1 retrotransposon copies are amplified during murine early embryo development, Mol. Reprod. Dev., 2012, vol. 79, no. 2, p. 118–127. doi 10.1002/mrd.22003
Tian, M., Bao, H., Martin, F.L., et al., Association of DNA methylation and mitochondrial DNA copy number with human semen quality, Biol. Reprod., 2014, vol. 91, no. 4, p. 101. doi 10.1095/biolreprod.114.122465
Houshdaran, S., Cortessis, V.K., Siegmund, K., et al., Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm, PLoS One, 2007, vol. 2, no. 12. e1289. doi 10.1371/journal.pone.0001289
Evsikov, A.V., de Vries, W.N., Peaston, A.E., et al., Systems biology of the 2-cell mouse embryo, Cytogenet. Genome Res., 2004, vol. 105, nos. 2–4, pp. 240–250. doi 10.1159/000078195
Peaston, A.E., Evsikov, A.V., Graber, J.H., et al., Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos, Dev. Cell, 2004, vol. 7, no. 4, pp. 597–606. doi 10.1016/j.devcel.2004.09.004
Peaston, A.E., Knowles, B.B., and Hutchison, K.W., Genome plasticity in the mouse oocyte and early embryo, Biochem. Soc. Trans., 2007, vol. 35, no. 3, pp. 618–622. doi 10.1042/BST0350618
Fadloun, A., Le Gras, S., Jost, B., et al., Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA, Nat. Struct. Mol. Biol., 2013, vol. 20, no. 3, pp. 332–338. doi 10.1038/nsmb.2495
Brouha, B., Meischl, C., Ostertag, E., et al., Evidence consistent with human L1 retrotransposition in maternal meiosis I, Am. J. Hum. Genet., 2002, vol. 71, no. 2, pp. 327–336. doi 10.1086/341722
Garcia-Perez, J.L., Marchetto, M.C., Muotri, A.R., et al., LINE-1 retrotransposition in human embryonic stem cells, Hum. Mol. Genet., 2007, vol. 16, no. 13, pp. 1569–1577. doi 10.1093/hmg/ddm105
Kano, H., Godoy, I., Courtney, C., et al., L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism, Genes Dev., 2009, vol. 23, no. 11, pp. 1303–1312. doi 10.1101/gad.1803909
Ostertag, E.M., DeBerardinis, R.J., Goodier, J.L., et al., A mouse model of human L1 retrotransposition, Nat. Genet., 2002, vol. 32, no. 4, pp. 655–660. doi 10.1038/ng1022
van den Hurk, J.A., Meij, I.C., Seleme, M.C., et al., L1 retrotransposition can occur early in human embryonic development, Hum. Mol. Genet., 2007, vol. 16, no. 13, pp. 1587–1592. doi 10.1093/hmg/ddm108
van den Hurk, J.A., van de Pol, D.J., Wissinger, B., et al., Novel types of mutation in the choroideremia (CHM) gene: a full-length L1 insertion and an intronic mutation activating a cryptic exon, Hum. Genet., 2003, vol. 113, no. 3, pp. 268–275. doi 10.1007/s00439-003- 0970-0
Li, J., Kannan, M., Trivett, A.L., et al., An antisense promoter in mouse L1 retrotransposon open reading frame-1 initiates expression of diverse fusion transcripts and limits retrotransposition, Nucleic Acids Res., 2014, vol. 42, no. 7, pp. 4546–4562. doi 10.1093/nar/gku091
Beraldi, R., Pittoggi, C., Sciamanna, I., et al., Expression of LINE-1 retroposons is essential for murine preimplantation development, Mol. Reprod. Dev., 2006, vol. 73, no. 3, pp. 279–287. doi 10.1002/mrd.20423
Kigami, D., Minami, N., Takayama, H., and Imai, H., MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos, Biol. Reprod., 2003, vol. 68, no. 2, pp. 651–654. doi 10.1095/biolreprod.102. 007906
Mangiacasale, R., Pittoggi, C., Sciamanna, I., et al., Exposure of normal and transformed cells to nevirapine, a reverse transcriptase inhibitor, reduces cell growth and promotes differentiation, Oncogene, 2003, vol. 22, no. 18, pp. 2750–2761. doi 10.1038/sj.onc. 1206354
Sciamanna, I., Landriscina, M., Pittoggi, C., et al., Inhibition of endogenous reverse transcriptase antagonizes human tumor growth, Oncogene, 2005, vol. 24, no. 24, pp. 3923–3931. doi 10.1038/sj.onc.1208562
Hall, L.L., Carone, D.M., Gomez, A.V., et al., Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes, Cell, 2014, vol. 156, no. 5, pp. 907–919. doi 10.1016/j.cell.2014.01.042
Lyon, M.F., X-chromosome inactivation: a repeat hypothesis, Cytogenet. Cell Genet., 1998, vol. 80, nos. 1–4, pp. 133–137. doi 10.1159/000014969
Lyon, M.F., LINE-1 elements and X chromosome inactivation: a function for “junk” DNA?, Proc. Natl. Acad. Sci. U.S.A., 2000, vol. 97, no. 12, pp. 6248–6249. doi 10.1073/pnas.97.12.6248
Waterston, R.H., Lindblad-Toh, K., Birney, E., et al., Initial sequencing and comparative analysis of the mouse genome, Nature, 2002, vol. 420, nos. 6915, pp. 520–562. doi 10.1038/nature01262
Boyle, A.L., Ballard, S.G., and Ward, D.C., Differential distribution of long and short interspersed element sequences in the mouse genome: chromosome karyotyping by fluorescence in situ hybridization, Proc. Natl. Acad. Sci. U.S.A., 1990, vol. 87, no. 19, pp. 7757–7761.
Bailey, J.A., Carrel, L., Chakravarti, A., and Eichler, E.E., Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis, Proc. Natl. Acad. Sci. U.S.A., 2000, vol. 97, no. 12, pp. 6634–6639. doi 10.1073/pnas. 97.12.6634
Mikkelsen, T.S., Wakefield, M.J., Aken, B., et al., Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences, Nature, 2007, vol. 447, no. 7141, pp. 167–177. doi 10.1038/nature05805
Abrusan, G., Giordano, J., and Warburton, P.E., Analysis of transposon interruptions suggests selection for L1 elements on the X chromosome, PLoS Genet., 2008, vol. 4, no. 8. e1000172. doi 10.1371/journal.pgen. 1000172
Allen, E., Horvath, S., Tong, F., et al., High concentrations of long interspersed nuclear element sequence distinguish monoallelically expressed genes, Proc. Natl. Acad. Sci. U.S.A., 2003, vol. 100, no. 17, pp. 9940–9945. doi 10.1073/pnas.1737401100
Paco, A., Adega, F., and Chaves, R., LINE-1 retrotransposons: from ‘parasite’ sequences to functional elements, J. Appl. Genet., 2014. doi 10.1007/s13353-014-0241-x
Chow, J.C., Ciaudo, C., Fazzari, M.J., et al., LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation, Cell, 2010, vol. 141, no. 6, pp. 956–969. doi 10.1016/j.cell.2010.04.042
Rosser, J.M., and An, W., L1 expression and regulation in humans and rodents, Front. Biosci., 2012, vol. 4, pp. 2203–2225. 10.2741
Vasil’ev, S.A., Tolmacheva, E.N., Kashevarova, A.A., et al., Methylation status of LINE-1 retrotransposon in chromosomal mosaicism during early stages of human embryonic development, Mol. Biol. (Moscow), 2015, vol. 49, no. 1, pp. 144–152. doi 10.7868/S0026898414060196
Price, E.M., Cotton, A.M., Penaherrera, M.S., et al., Different measures of “genome-wide” DNA methylation exhibit unique properties in placental and somatic tissues, Epigenetics, 2012, vol. 7, no. 6, pp. 652–663. doi 10.4161/epi.20221
He, Z.M., Li, J., Hwa, Y.L., et al., Transition of LINE-1 DNA methylation status and altered expression in first and third trimester placentas, PLoS One, 2014, vol. 9, no. 5. e96994. doi 10.1371/journal.pone.0096994
Mi, S., Lee, X., Li, X., et al., Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis, Nature, 2000, vol. 403, no. 6771, pp. 785–789. doi 10.1038/35001608
Sugimoto, J., Sugimoto, M., Bernstein, H., et al., A novel human endogenous retroviral protein inhibits cell–cell fusion, Sci. Rep., 2013, vol. 3, p. 1462. doi 10.1038/srep01462
Yin, L.J., Zhang, Y., Lv, P.P., et al., Insufficient maintenance DNA methylation is associated with abnormal embryonic development, BMC Med., 2012, vol. 10, p. 26. doi 10.1186/1741-7015-10-26
Perrin, D., Ballestar, E., Fraga, M.F., et al., Specific hypermethylation of LINE-1 elements during abnormal overgrowth and differentiation of human placenta, Oncogene, 2007, vol. 26, no. 17, pp. 2518–2524. doi 10.1038/sj.onc.1210039
Tolmacheva, E.N., Kashevarova, A.A., Skryabin, N.A., and Lebedev, I.N., Epigenetic effects of trisomy 16 in human placenta, Mol. Biol. (Moscow), 2013, vol. 47, no. 3, pp. 373–381. doi 10.7868/S002689841303018X
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © S.A. Vasilyev, E.N. Tolmacheva, I.N. Lebedev, 2016, published in Genetika, 2016, Vol. 52, No. 12, pp. 1349–1357.
Rights and permissions
About this article
Cite this article
Vasilyev, S.A., Tolmacheva, E.N. & Lebedev, I.N. Epigenetic regulation and role of LINE-1 retrotransposon in embryogenesis. Russ J Genet 52, 1219–1226 (2016). https://doi.org/10.1134/S1022795416120152
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1022795416120152