Abstract
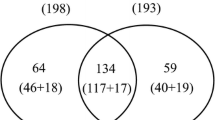
To explore proteomic characters of Kunitz-type trypsin inhibitors (KTIs) deleted soybean (Glycine max (L.) Merr.), seeds without KTIs and its female parent with KTIs were analyzed for differentially expressed proteins and specially expressed proteins using a label-free quantitative approach. A total of 1093 proteins were identified, with 139 up-regulated proteins among 169 differentially expressed proteins in both. Functional classification revealed that differentially expressed proteins were involved in almost functional categories, and carbohydrate metabolic process was the most abundant by enrichment analysis. Pathway analysis indicated that the differentially expressed proteins were mainly enriched into ribosome pathway, then carbohydrate metabolism. KTI3 (trypsin inhibitors A) interacted with β-conglycinin was independent from a complex protein network by proteins interactions analysis. Expression of KTI3 decreased in the seed of KTIs deleted soybean, which made for protein biosynthetic processes, metabolic processes and resistance processes. The specially expressed proteins analysis provided new information for the proteomic metabolism of KTIs deleted soybean resource during seed development, but it need further investigate for unknown information of KTIs deleted soybean.



Similar content being viewed by others
REFERENCES
Batista, I.F.C., Oliva, M.L.V., Araujo, M.S., Sampaio, M.U., Richardson, M., Fritz, H., and Sampaio, C.A.M., Primary structure of a Kunitz-type trypsin inhibitor from Enterolobium contortisiliquum seeds, Phytochemistry, 1996, vol. 41, pp. 1017–1022.
Natarajan, S., Xu, C., Bae, H., and Bailey, B.A., Proteomic and genomic characterization of Kunitz trypsin inhibitors in wild and cultivated soybean genotypes, J. Plant Physiol., 2007, vol. 164, pp. 756–763.
Kaur, S., Sharma, S., Dar, B.N., and Singh, B., Optimization of process for reduction of antinutritional factors in edible cereal brans, Food Sci. Technol. Int., 2012, vol. 18, pp. 445–454.
Chen, Y., Xu, Z., Zhang, C., Kong, X., and Hua, Y., Heat-induced inactivation mechanisms of Kunitz trypsin inhibitor and Bowman-Birk inhibitor in soymilk processing, Food Chem., 2014, vol. 154, pp. 108–116.
Görg, A., Obermaier, C., Boguth, G., Harder, A., Scheibe, B., Wildgruber, R., and Weiss, W., The current state of two-dimensional electrophoresis with immobilized pH gradients, Electrophoresis, 2000, vol. 21, pp. 1037–1053.
Finnie, C., Melchior, S., Roepstorff, P., and Svensson, B., Proteome analysis of grain filling and seed maturation in barley, Plant Physiol., 2002, vol. 129, pp. 1308–1319.
Tasleem-Tahir, A., Nadaud, I., Girousse, C., Martre, P., Marion, D., and Branlard, G., Proteomic analysis of peripheral layers during wheat (Triticum aestivum L.) grain development, Proteomics, 2011, vol. 11, pp. 371–379.
Song, X., Ni, Z., Yao, Y., Xie, C., Li, Z., Wu, H., Zhang, Y., and Sun, Q., Wheat (Triticum aestivum L.) root proteome and differentially expressed root proteins between hybrid and parents, Proteomics, 2007, vol. 7, pp. 3538–3557.
Zhu, W., Smith, J.W., and Huang, C.M., Mass spectrometry-based label-free quantitative proteomics, J. Biomed. Biotechnol., 2010, vol. 1, pp. 1–6.
Marcon, C., Lamkemeyer, T., Malik, W.A., Ungrue, D., Piepho, H.P., and Hochholdinger, F., Heterosis-associated proteome analyses of maize (Zea mays L.) seminal roots by quantitative label-free LC-MS, J. Proteomics, 2013, vol. 93, pp. 295–302.
Lee, J. and Koh, H.J., A label-free quantitative shotgun proteomics analysis of rice grain development, Proteome Sci., 2011, vol. 9, pp. 1–10.
Wang, X., Oh, M., Sakata, K., and Komatsu, S., Ge-l‑free/label-free proteomic analysis of root tip of soybean over time under flooding and drought stresses, J. Proteomics, 2016, vol. 130, pp. 42–55.
Kaufmann, K., Smaczniak, C., De Vries, S., Angenent, G.C., and Karlova, R., Proteomics insights into plant signaling and development, Proteomics, 2011, vol. 11, pp. 744–755.
Mirzaei, M., Soltani, N., Sarhadi, E., Pascovici, D., Keighley, T., Salekdeh, G.H., Haynes, P.A., and Atwell, B.J., Shotgun proteomic analysis of long-distance drought signaling in rice roots, J. Proteome Res., 2012, vol. 11, pp. 348–358.
Salavati, A., Khatoon, A., Nanjo, Y., and Komatsu, S., Analysis of proteomic changes in roots of soybean seedlings during recovery after flooding, J. Proteomics, 2012, vol. 75, pp. 878–893.
Broadway, R.M. and Duffey, S.S., Plant proteinase inhibitors mechanism of action and effect on the growth and digestive physiology of larval Heliothis zea and Spodoptera exigua, J. Insect Physiol., 1986, vol. 32, pp. 827–833.
Griffin, N.M., Yu, J., Long, F., Oh, P., Shore, S., Li, Y., Koziol, J.A., and Schnitzer, J.E., Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis, Nature Biotechnol., 2010, vol. 28, pp. 83–89.
Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., and Tanabe, M., KEGG for integration and interpretation of large-scale molecular data sets, Nucleic Acids Res., 2012, vol. 40, pp. 109–114.
Franceschini, A., Szklarczyk, D., Frankild, S., Kuhn, M., Simonovic, M., Roth, A., Lin, J., Minguez, P., Bork, P., von Mering, C., and Jensen, L.J., STRING v9.1: protein–protein interaction networks, with increased coverage and integration, Nucleic Acids Res., 2013, vol. 41, pp. 808–815.
Cox, J. and Mann, M., MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification, Nature Biotechnol., 2008, vol. 26, pp. 1367–1372.
Qin, J., Gu, F., Liu, D., Yin, C.C., Zhao, S.J., Chen, H., Zhang, J.N., Yang, C.Y., Zhan, X., and Zhang, M.C., Proteomic analysis of elite soybean Jidou17 and its parents using iTRAQ-based quantitative approaches, P-roteome Sci., 2013, vol. 11, pp. 334–345.
Kanehisa, M., Sato, Y., and Morishima, K., BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences, J. Mol. Biol., 2016, vol. 428, pp. 726–731.
Horiguchi, G., Lijsebettens, M.V., Candela, H., Micol, J.L., and Tsukaya, H., Ribosomes and translation in plant developmental control, Plant Sci., 2012, vols. 191–192, pp. 24–34.
Sheoran, I.S., Ross, A.R.S., Olson, D.J.H., and Sawhney, V.K., Differential expression of proteins in the wild type and 7B-1 male-sterile mutant anthers of tomato (Solanum lycopersicum): a proteomic analysis, J. Proteomics, 2009, vol. 71, pp. 624–636.
Borland, A.M., Guo, H.B., Yang, X.H., and Cushman, J.C., Orchestration of carbohydrate processing for crassulacean acid metabolism, Curr. Opin. Plant Biol., 2016, vol. 31, pp. 118–124.
Gavalda, S., Braga, R., Dax, C., Vigroux, A., and Blonski, C., N-Sulfonyl hydroxamate derivatives as inhibitors of class II fructose-1,6-diphosphate aldolase, Bioorganic Med. Chem. Lett., 2006, vol. 15, pp. 5375–5377.
Bailey-Serres, J., Tom, J., and Freeling, M., Expression and distribution of cytosolic 6-phosphogluconate dehydrogenase isozymes in maize, Biochem. Genet., 1992, vol. 30, pp. 233–246.
Bianchi, D., Bertrand, O., Haupt, K., and Coello, N., Effect of gluconic acid as a secondary carbon source on non-growing L-lysine producers cells of Corynebacterium glutamicum. Purification and properties of 6-phosphogluconate dehydrogenase, Enzyme Microb. Technol., 2001, vol. 28, pp.754–759.
Kitamura, K., Genetic improvement of nutrition and food processing quality in soybean, Jpn. Agric. Res. Quart., 1995, vol. 29, pp. 1–8.
Jofuku, K.D. and Goldberg, R.B., Kunitz inhibitor genes are differentially expressed during the soybean life cycle and in transformed tobacco plant, Plant Cell, 1989, vol. 1, pp. 1079–1093.
ACKNOWLEDGMENTS
This work was supported by the grants from the National Science and Technology Support Project (project no. 2014 BAD11B01); Heilongjiang Postdoctoral Funded project (project no. LBH-Z13040).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
The article is published in the original.
Abbreviations: 2-DE—two-dimensional gel electrophoresis; GO—gene ontology; HCD—high energy collisional dissociation; KO—KEGG orthology; KTI—Kunitz-type trypsin inhibitor; MS—mass spectrometry; TIs—trypsin inhibitors.
Rights and permissions
About this article
Cite this article
Jiang, Y., Li, Y.M., Wang, S.D. et al. Proteomic Analysis of Kunitz-Type Trypsin Inhibitor Deleted Soybean. Russ J Plant Physiol 66, 469–476 (2019). https://doi.org/10.1134/S1021443719030099
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1021443719030099