Abstract
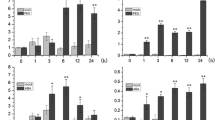
NAC is one of the most abundant plant-specific groups of transcription factors, which play important roles in plant growth and development regulation, as well as in biotic and abiotic stress responses. In the present study, a stress-responsive NAC gene, AhNAC4, was isolated from a cDNA library of peanut (Arachis hypogaea L.) immature seeds, and characterized for its role in drought tolerance. AhNAC4 shared high amino acid similarity with NAC proteins belonging to the ATAF subfamily. The expression analysis indicated that AhNAC4 was highly induced by drought, salinity and ABA treatments. Transient expression analysis showed the AhNAC4-GFP fusion protein was exclusively localized in the nucleus of onion epidermal cells. Transactivation assays in yeast cells demonstrated that AhNAC4 functioned as a transcription activator and its C-terminus contained the activation domain. Overexpression of AhNAC4 confers enhanced drought tolerance in transgenic tobacco plants. The improved drought tolerance was associated with more stomatal closure and higher water use efficiency. Collectively, our results indicated that AhNAC4 functions as an important regulator in response to drought stress.
Similar content being viewed by others
Abbreviations
- meJA:
-
methyl jasmonate
- TF:
-
transcription factor
- WT:
-
wild type
References
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., Matsubara, K., Osato, N., Kawai, J., Carninci, P., Hayashizaki, Y., Suzuki, K., Kojima, K., Takahara, Y., Yamamoto, K., et al., Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana, DNA Res., 2003, vol. 10, pp. 239–247.
Fang, Y., You, J., Xie, K., Xie, W., and Xiong, L., Systematic sequence analysis and identification of tissuespecific or stress-responsive genes of NAC transcription factor family in rice, Mol. Genet. Genomics, 2008, vol. 280, pp. 547–563.
Mochida, K., Yoshida, T., Sakurai, T., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L.S., LegumeTFDB: an integrative database of Glycine max, Lotus japonicus and Medicago truncatula transcription factors, Bioinformatics, 2010, vol. 26, pp. 290–291.
Shen, H., Yin, Y.B., Chen, F., Xu, Y., and Dixon, R.A., A bioinformatic analysis of NAC genes for plant cell wall development in relation to ligocellulosic bioenergy production, Bioenerg. Res., 2009, vol. 2, pp. 217–232.
Nuruzzaman, M., Sharoni, A.M., and Kikuchi, S., Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants, Front. Microbiol., 2013, vol. 4:248.
Tran, L.S., Nakashima, K., Sakuma, Y., Simpson, S.D., Fujita, Y., Maruyama, K., Fujita, M., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K., Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter, Plant Cell, 2004, vol. 16, pp. 2481–2498.
Wang, X., Basnayake, B.M., Zhang, H., Li, G., Li, W., Virk, N., Mengiste, T., and Song, F., The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulation of defense responses against necrotrophic fungal and bacterial pathogens, Mol. Plant–Microbe Interact., 2009, vol. 22, pp. 1227–1238.
Wu, Y., Deng, Z., Lai, J., Zhang, Y., Yang, C., Yin, B., Zhao, Q., Zhang, L., Li, Y., Yang, C., and Xie, Q., Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses, Cell Res., 2009, vol. 19, pp. 1279–1290.
Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., and Xiong, L., Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice, Proc. Natl. Acad. Sci. USA, 2006, vol. 103, pp. 12987–12992.
Ohnishi, T., Sugahara, S., Yamada, T., Kikuchi, K., Yoshiba, Y., Hirano, H.Y., and Tsutsumi, N., OsNAC6, a member of the NAC gene family, is induced by various stresses in rice, Genes Genet. Syst., 2005, vol. 80, pp. 135–139.
Nakashima, K., Tran, L.S., Nguyen, D.V., Fujita, M., Maruyama, K., Todaka, D., Ito, Y., Hayashi, N., Shinozaki, K., and Yamaguchi-Shinozaki, K., Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice, Plant J., 2007, vol. 51, pp. 617–630.
Jeong, J.S., Kim, Y.S., Baek, K.H., Jung, H., Ha, S.H., Choi, Y.D., Kim, M.K., Reuzeau, C., and Kim, J.K., Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions, Plant Physiol., 2010, vol. 153, pp. 185–197.
Liu, Z., Feng, S., Pandey, M.K., Chen, X., Culbreath, A.K., Varshney, R.K., and Guo, B., Identification of expressed resistance gene analogs from peanut (Arachis hypogaea L.) expressed sequence tags, J. Integr. Plant Biol., 2013, vol. 55, pp. 453–461.
Govind, G., Harshavardhan, V.T., Patricia, J.K., Dhanalakshmi, R., Senthil Kumar, M., Sreenivasulu, N., and Udayakumar, M., Identification and functional validation of a unique set of drought induced genes preferentially expressed in response to gradual water stress in peanut, Mol. Genet. Genomics, 2009, vol. 281, pp. 591–605.
Liu, X., Zhang, B.Y., Hong, L., Su, L.C., Liang, X.Q., Li, X.Y., and Li, L., Molecular characterization of Arachis hypogaea NAC 2 (AhNAC2) reveals it as a NAC-like protein in peanut, Biotechnol. Biotechnol. Equip., 2010, vol. 24, pp. 2066–2070.
Liu, X., Liu, S., Wu, J.L., Zhang, B.Y., Li, X.Y., Yan, Y.C., and Li, L., Overexpression of Arachis hypogaea NAC3 in tobacco enhances dehydration and drought tolerance by increasing superoxide scavenging, Plant Physiol. Biochem., 2013, vol. 70, pp. 354–359.
Shao, Q.S., Shu, S., Du, J., Xing, W.W., and Guo, S.R., Effects of NaCl stress on nitrogen metabolism of cucumber seedlings, Russ. J. Plant Physiol., 2015, vol. 62, pp. 595–603.
Bi, Y.P., Liu, W., Xia, H., Su, L., Zhao, C.Z., Wan, S.B., and Wang, X.J., EST sequencing and gene expression profiling of cultivated peanut (Arachis hypogaea L.), Genome, 2010, vol. 53, pp. 832–839.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S., MEGA6: Molecular Evolutionary Genetics Analysis version 6, Mol. Biol. Evol., 2013, vol. 30, pp. 2725–2729.
Jiang, S., Sun, Y., and Wang, S., Selection of reference genes in peanut seed by real-time quantitative polymerase chain reaction, Int. J. Food Sci. Technol., 2011, vol. 46, pp. 2191–2196.
Livak, K.J. and Schmittgen, T.D., Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method, Methods, 2001, vol. 25, pp. 402–408.
Horsch, R.B., Fry, J.E., Hoffmann, N.L., Eichholtz, D., Rogers, S.G., and Fraley, R.T., A simple and general method for transferring genes into plants, Science, 1985, vol. 227, pp. 1229–1231.
Yu, H., Chen, X., Hong, Y.Y., Wang, Y., Xu, P., Ke, S.D., Liu, H.Y., Zhu, J.K., Oliver, D.J., and Xiang, C.B., Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density, Plant Cell, 2008, vol. 20, pp. 1134–1151.
Finn, R.D., Coggill, P., Eberhardt, R.Y., Eddy, S.R., Mistry, J., Mitchell, A.L., Potter, S.C., Punta, M., Qureshi, M., Sangrador-Vegas, A., Salazar, G.A., Tate, J., and Bateman, A., The Pfam protein families database: towards a more sustainable future, Nucleic Acids Res., 2016, vol. 44, pp. D279–285.
Hua, S.J. and Sun, Z.R., Support vector machine approach for protein subcellular localization prediction, Bioinformatics, 2001, vol. 17, pp. 721–728.
Olsen, A.N., Ernst, H.A., Leggio, L.L., and Skriver, K., NAC transcription factors: structurally distinct, functionally diverse, Trends Plant Sci., 2005, vol. 10, pp. 79–87.
Pinheiro, G.L., Marques, C.S., Costa, M.D., Reis, P.A., Alves, M.S., Carvalho, C.M., Fietto, L.G., and Fontes, E.P., Complete inventory of soybean NAC transcription factors: sequence conservation and expression analysis uncover their distinct roles in stress response, Gene, 2009, vol. 444, pp. 10–23.
Meng, Q., Zhang, C., Gai, J., and Yu, D., Molecular cloning, sequence characterization and tissue-specific expression of six NAC-like genes in soybean (Glycine max (L.) Merr.), J. Plant Physiol., 2007, vol. 164, pp. 1002–1012.
Tran, L.P., Quach, T.N., Guttikonda, S.K., Aldrich, D.L., Kumar, R., Neelakandan, A., Vallivodan, B., and Nguyen, H.T., Molecular characterization of stress-inducible GmNAC genes in soybean, Mol. Genet. Genomics, 2009, vol. 281, pp. 647–664.
Fang, Y., Liao, K., Du, H., Xu, Y., Song, H., Li, X., and Xiong, L., A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice, J. Exp. Bot., 2015, vol. 66, pp. 6803–6817.
Author information
Authors and Affiliations
Corresponding author
Additional information
The article is published in the original.
Rights and permissions
About this article
Cite this article
Tang, G.Y., Shao, F.X., Xu, P.L. et al. Overexpression of a peanut NAC gene, AhNAC4, confers enhanced drought tolerance in tobacco. Russ J Plant Physiol 64, 525–535 (2017). https://doi.org/10.1134/S1021443717040161
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1021443717040161