Abstract
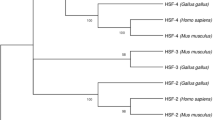
The Hsp60/Hsp10 chaperonin system is one of the most studied systems of cell emergency responses to stresses associated with changes in environmental conditions. In this regard, we have performed a bioinformatics analysis of 164 amino acid sequences of Hsp60 and 125 amino acid sequences of Hsp10 in five classes of chordata. This enabled uncovering the relationship between the identity of the amino acid composition of Hsp60/Hsp10 and the evolutionary distance between classes of chordata. In the course of the study of the chaperonin crystal structure, potentially significant amino acid motifs responsible for the oligomerization of Hsp60 and Hsp10 monomers and the association/dissociation of the Hsp60 and Hsp10 hetero-dimer have been identified. In addition, we have established that Hsp60 and Hsp10 can form amyloid fibrils due to structural features through the alternative using of the oligomerization sites of monomers as well as association/dissociation sites.







Similar content being viewed by others
REFERENCES
Corrao S., Anzalone R., Lo Iacono M., Corsello T., Di Stefano A., D’Anna S.E., Balbi B., Carone M., Sala A., Corona D., Timperio A.M., Zolla L., Farina F., Conway de Macario E., et al. 2014. Hsp10 nuclear localization and changes in lung cells response to cigarette smoke suggest novel roles for this chaperonin. Open Biol. 4 (10), pii 140125. doi 10.1098/rsob.140125
Khamis I., Chan D.W., Shirriff C.S., Campbell J.H., Heikkila J.J. 2016. Expression and localization of the Xenopus laevis small heat shock protein, HSPB6 (HSP20), in A6 kidney epithelial cells. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 201, 12–21.
Soltys B.J., Gupta R.S. 1996. Immunoelectron microscopic localization of the 60-kDa heat shock chaperonin protein (Hsp60) in mammalian cells. Exp. Cell Res. 222 (1), 16–27.
Baringou S., Rouault J.-D., Koken M., Hardivillier Y., Hurtado L., Leignel V. 2016. Diversity of cytosolic HSP70 heat shock protein from decapods and their phylogenetic placement within Arthropoda. Gene. 591 (1), 97–107. doi 10.1016/j.gene.2016.06.061
Ebrahimi M., Mohammadi P., Daryadel A., Baharvand H. 2010. Assessment of heat shock protein (HSP60, HSP72, HSP90, and HSC70) expression in cultured limbal stem cells following air lifting. Mol. Vis. 16, 1680–1688.
Moon A., Bacchini P., Bertoni F., Olvi L.G., Santini-Arawo E., Kim Y.W., Park Y.-K. 2010). Expression of heat shock proteins in osteosarcomas. Pathology (Phila.). 42 (5), 421–425.
Cheng Y.F., Sun J.R., Chen H.B., Abdelnasir A., Tang S., Kemper N., Hartung J., Bao E.D. 2014. Association of Hsp60 expression with damage to rat myocardial cells exposed to heat stress in vivo and in vitro. Genet. Mol. Res. GMR. 13 (4), 9371–9381.
Seveso D., Montano S., Strona G., Orlandi I., Galli P., Vai M. 2016. Hsp60 expression profiles in the reef-building coral Seriatopora caliendrum subjected to heat and cold shock regimes. Mar. Environ. Res. 119, 1–11.
Donovan M.R., Marr M.T. 2016. dFOXO activates large and small heat shock protein genes in response to oxidative stress to maintain proteostasis in Drosophila. J. Biol. Chem. 291 (36), 19 042–19 050. doi 10.1074/ jbc.M116.723049
Singh M.P., Ravi Ram K., Mishra M., Shrivastava M., Saxena D.K., Chowdhuri D.K. 2010. Effects of co-exposure of benzene, toluene and xylene to Drosophila melanogaster: Alteration in hsp70, hsp60, hsp83, hsp26, ROS generation and oxidative stress markers. Chemosphere. 79 (5), 577–587.
Azarnia Tehran D., Pirazzini M., Leka O., Mattarei A., Lista F., Binz T., Rossetto O., Montecucco C. 2016. Hsp90 is involved in the entry of clostridial neurotoxins into the cytosol of nerve terminals. Cell. Microbiol. 19 (2). doi 10.1111/cmi.12647
Inda C., Bolaender A., Wang T., Gandu S. R., Koren Iii J. 2016. Stressing out Hsp90 in neurotoxic proteinopathies. Curr. Top. Med. Chem. 16 (25), 2829–2838.
Zhao P., Zhang K., Guo G., Sun X., Chai H., Zhang W., Xing M. 2016. Heat shock protein alteration in the gastrointestinal tract tissues of chickens exposed to arsenic trioxide. Biol. Trace Elem. Res. 170 (1), 224–236.
Gammazza A.M., Bavisotto C.C., Barone R., de Macario E.C., Macario A.J.L. 2016. Alzheimer’s disease and molecular chaperones: Current knowledge and the future of chaperonotherapy. Curr. Pharm. Des. 22 (26), 4040–4049.
Zhao C., Li H., Zhao X.-J., Liu Z.-X., Zhou P., Liu Y., Feng M.-J. 2016. Heat shock protein 60 affects behavioral improvement in a rat model of Parkinson’s disease grafted with human umbilical cord mesenchymal stem cell-derived dopaminergic-like neurons. Neurochem. Res. 41 (6), 1238–1249.
Huckriede A., Heikema A., Sjollema K., Briones P., Agsteribbe E. 1995. Morphology of the mitochondria in heat shock protein 60 deficient fibroblasts from mitochondrial myopathy patients. Effects of stress conditions. Virchows Arch. Int. J. Pathol. 427 (2), 159–165.
Sugimoto M., Furuoka H., Sugimoto Y. 2003. Deletion of one of the duplicated Hsp70 genes causes hereditary myopathy of diaphragmatic muscles in Holstein-Friesian cattle. Anim. Genet. 34 (3), 191–197.
Nisemblat S., Yaniv O., Parnas A., Frolow F., Azem A. 2015. Crystal structure of the human mitochondrial chaperonin symmetrical football complex. Proc. Natl. Acad. Sci. U. S. A. 112 (19), 6044–6049.
Okamoto T., Ishida R., Yamamoto H., Tanabe-Ishida M., Haga A., Takahashi H., Takahashi K., Goto D., Grave E., Itoh H. 2015. Functional structure and physiological functions of mammalian wild-type HSP60. Arch. Biochem. Biophys. 586, 10–19.
Hayer-Hartl M., Bracher A., Hartl F.U. 2016. The GroEL-GroES chaperonin machine: A nano-cage for protein folding. Trends Biochem. Sci. 41 (1), 62–76.
Karlin S., Brocchieri L. 2000. Heat shock protein 60 sequence comparisons: Duplications, lateral transfer, and mitochondrial evolution. Proc. Natl. Acad. Sci. U. S. A. 97 (21), 11 348–11 353.
Sameshima T., Ueno T., Iizuka R., Ishii N., Terada N., Okabe K., Funatsu T. 2008. Football- and bullet-shaped GroEL-GroES complexes coexist during the reaction cycle. J. Biol. Chem. 283 (35), 23 765–23 773.
Mandal K., Foteinos G., Jahangiri M., Xu Q. 2005. Role of antiheat shock protein 60 autoantibodies in atherosclerosis. Lupus. 14 (9), 742–746.
Grundtman C., Kreutmayer S.B., Almanzar G., Wick M.C., Wick G. 2011. Heat shock protein 60 and immune inflammatory responses in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 31 (5), 960–968.
Pfister G., Stroh C.M., Perschinka H., Kind M., Knoflach M., Hinterdorfer P., Wick G. 2005. Detection of HSP60 on the membrane surface of stressed human endothelial cells by atomic force and confocal microscopy. J. Cell. Sci. 118 (8), 1587–1594.
Zhu H., Fang X., Zhang D., Wu W., Shao M., Wang L., Gu J. 2016. Membrane-bound heat shock proteins facilitate the uptake of dying cells and cross-presentation of cellular antigen. Apoptosis Int. J. Program. Cell Death. 21 (1), 96–109.
Fruchart J.-C., Nierman M.C., Stroes E.S.G., Kastelein J.J.P., Duriez P. 2004. New risk factors for atherosclerosis and patient risk assessment. Circulation. 109 (23, Suppl. 1), III-15–III-19.
Lanter B.B., Sauer K., Davies D.G. 2014. Bacteria present in carotid arterial plaques are found as biofilm deposits which may contribute to enhanced risk of plaque rupture. mBio. 5 (3), e01206-14.
Ameriso S.F., Fridman E.A., Leiguarda R.C., Sevlever G.E. 2001. Detection of Helicobacter pylori in human carotid atherosclerotic plaques. Stroke. 32 (2), 385–391.
Izadi M., Fazel M., Sharubandi S.H., Saadat S.H., Farahani M.M., Nasseri M.H., Dabiri H., SafiAryan R., Esfahani A.A., Ahmadi A., Jonaidi Jafari N., Ranjbar R., Jamali-Moghaddam S.-R., Kazemi-Saleh D., Kalantar-Motamed M.H., Taheri S. 2012. Helicobacter species in the atherosclerotic plaques of patients with coronary artery disease. Cardiovasc. Pathol. 21 (4), 307–311.
Belland R.J., Ouellette S.P., Gieffers J., Byrne G.I. 2004. Chlamydia pneumoniae and atherosclerosis. Cell. Microbiol. 6 (2), 117–127.
Farsak B., Yildirir A., Akyön Y., Pinar A., Öç M., Böke E., Kes S., Tokgözoğlu L. 2000. Detection of Chlamydia pneumoniae and Helicobacter pylori DNA in human atherosclerotic plaques by PCR. J. Clin. Microbiol. 38 (12), 4408–4411.
Perschinka H., Wellenzohn B., Parson W., Zee R. van der, Willeit J., Kiechl S., Wick G. 2007. Identification of atherosclerosis-associated conformational heat shock protein 60 epitopes by phage display and structural alignment. Atherosclerosis. 194 (1), 79–87.
Perschinka H., Mayr M., Millonig G., Mayerl C., Zee R. van der, Morrison S.G., Morrison R.P., Xu Q., Wick G. 2003. Cross-reactive B-cell epitopes of microbial and human heat shock protein 60/65 in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 23 (6), 1060–1065.
Wick G., Jakic B., Buszko M., Wick M.C., Grundtman C. 2014. The role of heat shock proteins in atherosclerosis. Nat. Rev. Cardiol. 11 (9), 516–529.
Afek A., George J., Gilburd B., Rauova L., Goldberg I., Kopolovic J., Harats D., Shoenfeld Y. 2000. Immunization of low-density lipoprotein receptor deficient (LDL-RD) mice with heat shock protein 65 (HSP-65) promotes early atherosclerosis. J. Autoimmun. 14 (2), 115–121.
Xu Q., Dietrich H., Steiner H.J., Gown A.M., Schoel B., Mikuz G., Kaufmann S.H., Wick G. 1992. Induction of arteriosclerosis in normocholesterolemic rabbits by immunization with heat shock protein 65. Arterioscler. Thromb. J. Vasc. Biol. 12 (7), 789–799.
Mangione M.R., Vilasi S., Marino C., Librizzi F., Canale C., Spigolon D., Bucchieri F., Fucarino A., Passantino R., Cappello F., Bulone D., San Biagio P.L. 2016. Hsp60, amateur chaperone in amyloid-beta fibrillogenesis. Biochim. Biophys. Acta. 1860 (11, Pt. A), 2474–2483. doi 10.1016/j.bbagen.2016.07.019
Ojha B., Fukui N., Hongo K., Mizobata T., Kawata Y. 2016. Suppression of amyloid fibrils using the GroEL apical domain. Sci. Rep. 6, 31 041.
Padmadas N., Panda P.K., Durairaj S. 2016. Binding patterns associated Aß-HSP60 p458 conjugate to HLA-DR-DRB allele of human in Alzheimer’s disease: An in silico approach. Interdiscip. Sci. Comput. Life Sci. 10 (1), 93–104. doi 10.1007/s12539-016-0170-y
Sievers F., Higgins D.G. 2014. Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 1079, 105–116.
Manning J.R., Jefferson E.R., Barton G.J. 2008. The contrasting properties of conservation and correlated phylogeny in protein functional residue prediction. BMC Bioinformatics. 9, 51.
Kumar S., Stecher G., Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874.
Jones D.T., Taylor W.R., Thornton J.M. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8 (3), 275–282.
Galzitskaya O.V., Garbuzynskiy S.O., Lobanov M.Y. 2006. FoldUnfold: Web server for the prediction of disordered regions in protein chain. Bioinformatics. 22 (23), 2948–2949.
Galzitskaya O.V., Garbuzinskiy S.O., Lobanov M.Yu. 2006. Prediction of natively unfolded regions in protein chain. Mol. Biol. (Moscow). 40 (2), 298‒304.
Lobanov M.Y., Galzitskaya O.V. 2011. The Ising model for prediction of disordered residues from protein sequence alone. Phys. Biol. 8 (3), 035004.
Lobanov M.Y., Sokolovskiy I.V., Galzitskaya O.V. 2013. Is unstruct: Prediction of the residue status to be ordered or disordered in the protein chain by a method based on the Ising model. J. Biomol. Struct. Dyn. 31 (10), 1034–1043.
Galzitskaya O.V., Garbuzynskiy S.O., Lobanov M.Y. 2006. Prediction of amyloidogenic and disordered regions in protein chains. PLoS Comput. Biol. 2 (12), e177.
Garbuzynskiy S.O., Lobanov M.Y., Galzitskaya O.V. 2010. FoldAmyloid: A method of prediction of amyloidogenic regions from protein sequence. Bioinformatics. 26 (3), 326–332.
Klochkov V.I. 2013. Metrologiya, standartizatsiya i sertifikatsiya (Metrology, Standardization, and Certification). Moscow: VLADOS.
Brocchieri L., Karlin S. 2000. Conservation among HSP60 sequences in relation to structure, function, and evolution. Protein Sci. 9 (3), 476–486.
Drozdetskiy A., Cole C., Procter J., Barton G.J. 2015. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 43 (W1), 389–394. doi 10.1093/nar/gkv332
Skvortsov V.S., Alekseytchuk N.N., Khudyakov D.V., Reyes I.V.R. 2015. pIPredict: A computer tool for prediction of isoelectric points of peptides and proteins. Biochemistry (Moscow), Suppl. Ser. B: Biomed. Chem. 9 (3), 296–303.
Fukasawa Y., Tsuji J., Fu S.-C., Tomii K., Horton P., Imai K. 2015. MitoFates: Improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol. Cell. Proteomics MCP. 14 (4), 1113–1126.
Fenton W.A., Kashi Y., Furtak K., Horwich A.L. 1994. Residues in chaperonin GroEL required for polypeptide binding and release. Nature. 371 (6498), 614–619.
Singh B., Patel H.V., Ridley R.G., Freeman K.B., Gupta R.S. 1990. Mitochondrial import of the human chaperonin (HSP60) protein. Biochem. Biophys. Res. Commun. 169 (2), 391–396.
Voos W., Röttgers K. 2002). Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim. Biophys. Acta—Mol. Cell Res. 1592 (1), 51–62.
Dyson H.J., Wright P.E., Scheraga H.A. 2006. The role of hydrophobic interactions in initiation and propagation of protein folding. Proc. Natl. Acad. Sci. U. S. A. 103 (35), 13 057–13 061.
Ryabova N.A., Marchenkov V.V., Marchenkova S.Y., Kotova N.V., Semisotnov G.V. 2013. Molecular chaperone GroEL/ES: Unfolding and refolding processes. Biochemistry (Moscow). 78 (13), 1405–1414.
Boisvert D.C., Wang J., Otwinowski Z., Horwich A.L., Sigler P.B. 1996. The 2.4 Å crystal structure of the bacterial chaperonin GroEL complexed with ATP gamma S. Nat. Struct. Biol. 3 (2), 170–177.
Seale J.W., Horowitz P.M. 1995. The C-terminal sequence of the chaperonin GroES Is required for oligomerization. J. Biol. Chem. 270 (51), 30 268–30 270.
Goh Y.C., Yap C.T., Huang B.H., Cronshaw A.D., Leung B.P., Lai P.B.S., Hart S.P., Dransfield I., Ross J.A. 2011. Heat-shock protein 60 translocates to the surface of apoptotic cells and differentiated megakaryocytes and stimulates phagocytosis. Cell. Mol. Life Sci. 68 (9), 1581–1592.
Hayoun D., Kapp T., Edri-Brami M., Ventura T., Cohen M., Avidan A., Lichtenstein R.G. 2012. HSP60 is transported through the secretory pathway of 3‑MCA-induced fibrosarcoma tumour cells and undergoes N-glycosylation. FEBS J. 279 (12), 2083–2095.
Blom N., Sicheritz-Pontén T., Gupta R., Gammeltoft S., Brunak S. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 4 (6), 1633–1649.
Chiti F., Dobson C.M. 2009. Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 5 (1), 15–22.
Rambaran R.N., Serpell L.C. 2008. Amyloid fibrils. Prion. 2 (3), 112–117.
Chen J., Yagi H., Sormanni P., Vendruscolo M., Makabe K., Nakamura T., Goto Y., Kuwajima K. 2012. Fibrillogenic propensity of the GroEL apical domain: A Janus-faced minichaperone. FEBS Lett. 586 (8), 1120–1127.
ACKNOWLEDGMENTS
The authors are grateful to all reviewers for their comments on this paper. The work was supported by the Russian Science Foundation, project no. 18-14-00321.
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated by E. Puchkov
1Abbreviations: Hsp, heat shock protein; MMP, matrix metalloproteinase; Icp55, intermediate cleaving peptidase 55, cleaving the intermediates of posttranslational protein modification; TOM20, translocase of the mitochondrial outer membrane; PKA, protein kinase A; PKG, protein kinase G.
Rights and permissions
About this article
Cite this article
Tikhomirova, T.S., Galzitskaya, O.V. Functionally Significant Amino Acid Motifs of Heat Shock Proteins: Structural and Bioinformatics Analyses of Hsp60/Hsp10 in Five Classes of Chordata. Mol Biol 52, 761–778 (2018). https://doi.org/10.1134/S0026893318050138
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893318050138