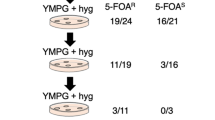
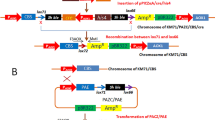
Abstract
Using CRISPR/Cas9 system, the recipient strains K. phaffii VKPM Y-5013 (His– phenotype) and K. phaffii VKPM Y-5014 (Leu– phenotype) were derived from the K. phaffii VKPM Y-4287 strain, which has a high expression potential. Based on the developed recipient strains, markerless producers of heterologous proteins could be obtained. Efficiency of the gene inactivation with different variants of sgRNA ranged from 65 to 98% and from 15 to 72% for the HIS4 and LEU2 genes, respectively. The recipient strains retained growth characteristics of the parent strain and exhibited high expression potential, as estimated by the production of heterologous phytase from Citrobacter gillenii. Average productivity of the transformants based on the K. phaffii VKPM Y-5013 and K. phaffii VKPM Y-5014 strains was 2.1 and 2.0 times higher than productivity of the transformants of the commercial K. phaffii GS115 strain. Method for sequential integration of genetic material into genome of the K. phaffii VKPM Y-5013 strain was proposed. A highly effective multicopy markerless strain producing C. gillenii phytase was obtained.




Similar content being viewed by others
Abbreviations
- FB:
-
fermentation broth
- PAM:
-
protospacer adjacent motif
- sgRNA:
-
single guide RNA
References
Ahmad, M., Hirz, M., Pichler, H., and Schwab, H. (2014) Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production, Appl. Microbiol. Biotechnol., 98, 5301-5317, https://doi.org/10.1007/s00253-014-5732-5.
Luo, H. Y., Yao, B., Yuan, T. Z., Wang, Y. R., Shi, X. Y., Wu, N. F., and Fan, Y. L. (2004) Overexpression of Escherchia coli phytase with high specific activity, Chin. J. Biotechnol., 20, 78-84.
Huang, H., Luo, H., Yang, P., Meng, K., Wang, Y., Yuan, T., Bai, Y., and Yao, B. (2006) A novel phytase with preferable characteristics from Yersinia intermedia, Biochem. Biophys. Res. Commun., 350, 884-889, https://doi.org/10.1016/j.bbrc.2006.09.118.
Xiong, A. S., Yao, Q. H., Peng, R. H., Zhang, Z., Xu, F., Liu, J. G., Han, P. L., and Chen, J. M. (2006) High level expression of a synthetic gene encoding Peniophora lycii phytase in methylotrophic yeast Pichia pastoris, App. Microbiol. Biotechnol., 72, 1039-1047, https://doi.org/10.1007/s00253-006-0384-8.
Pal Roy, M., Mazumdar, D., Dutta, S., Saha, S. P., and Ghosh, S. (2016) Cloning and expression of phytase appA gene from Shigella sp. CD2 in Pichia pastoris and comparison of properties with recombinant enzyme expressed in E. coli, PLoS One, 11, e0145745, https://doi.org/10.1371/journal.pone.0145745.
Zhao, W., Xiong, A., Fu, X., Gao, F., Tian, Y., and Peng, R. (2010) High level expression of an acid-stable phytase from Citrobacter freundii in Pichia pastoris, Appl. Biochem. Biotechnol., 162, 2157-2165, https://doi.org/10.1007/s12010-010-8990-4.
Rozanov, A. S., Pershina, E. G., Bogacheva, N. V., Shlyakhtun, V., Sychev, A. A., and Peltek, S. E. (2020) Diversity and occurrence of methylotrophic yeasts used in genetic engineering, Vavilovskii Zhurn. Genet. Selektsii, 24, 149-157, https://doi.org/10.18699/VJ20.602.
Pronk, J. T. (2002) Auxotrophic yeast strains in fundamental and applied research, App. Environ. Microbiol., 68, 2095-2100, https://doi.org/10.1128/AEM.68.5.2095-2100.2002.
Wang, Y., Yau, Y. Y., Perkins-Balding, D., and Thomson, J. G. (2011) Recombinase technology: applications and possibilities, Plant Cell Rep., 30, 267-285, https://doi.org/10.1007/s00299-010-0938-1.
Cregg, J. M., Barringer, K. J., Hessler, A. Y., and Madden, K. R. (1985) Pichia pastoris as a host system for transformations, Mol. Cell. Biol., 5, 3376-3385, https://doi.org/10.1128/mcb.5.12.3376-3385.1985.
Theodorakis, C. W. (2018) Mutagenesis, Encyclopedia Ecology, 2475-2484, https://doi.org/10.1016/b978-008045405-4.00408-0.
Näätsaari, L., Mistlberger, B., Ruth, C., Hajek, T., Hartner, F. S., and Glieder, A. (2012) Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology, PLoS One, 7, e39720, https://doi.org/10.1371/journal.pone.0039720.
Weninger, A., Fischer, J. E., Raschmanova, H., Vogl, T., and Glieder, A. (2018) Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers, J. Cell. Biochem., 119, 3183-3198, https://doi.org/10.1002/jcb.26474.
Weninger, A., Hatzl, A. M., Schmid, C., Vogl, T., and Glieder, A. (2016) Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris, J. Biotechnol., 235, 139-149, https://doi.org/10.1016/j.jbiotec.2016.03.027.
Nemudryi, A. A., Valetdinova, K. R., Medvedev, S. P., and Zakian, S. M. (2014) TALEN and CRISPR/Cas genome editing systems: tools of discovery, Acta Naturae, 6, 20-42, https://doi.org/10.32607/20758251-2014-6-3-19-40.
Mohammadhassan, R., Tutunchi, S., Nasehi, N, Goudarziasl, F., and Mahya, L. (2023) The prominent characteristics of the effective sgRNA for a precise CRISPR genome editing, CRISPR Technol. Recent Adv., IntechOpen, https://doi.org/10.5772/intechopen.106711.
Gordeeva, T. L., Borshchevskaya, L. N., Feday, T. V., Tkachenko, A. A., and Sineoky, S. P. (2021) The expression potential of new yeast strains of the Komagataella genus [in Russian], Biotekhnologiya, 37, 5-13, https://doi.org/10.21519/0234-2758-2021-37-4-5-13.
Tkachenko, A. A., Kalinina, A. N., Borshchevskaya, L. N., Sineoky, S. P., and Gordeeva, T. L. (2021) A novel phytase from Citrobacter gillenii: characterization and expression in Pichia pastoris (Komagataella pastoris), FEMS Microbiol. Lett., 368, fnaa217, https://doi.org/10.1093/femsle/fnaa217.
Gao, Y., and Zhao, Y. (2014) Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing, J. Integr. Plant. Biol., 56, 343-349, https://doi.org/10.1111/jipb.12152.
Heigwer, F., Kerr, G., and Boutros, M. (2014) E-CRISP: fast CRISPR target site identification, Nat. Methods, 11, 122-123, https://doi.org/10.1038/nmeth.2812.
Gassler, T., Heistinger, L., Mattanovich, D., Gasser, B., and Prielhofer, R. (2019) CRISPR/Cas9-mediated homology-directed genome editing in Pichia pastoris, in Recombinant Protein Production in Yeast, Methods Mol. Biol., pp. 211-225, https://doi.org/10.1007/978-1-4939-9024-5_9.
Gasser, B., Prielhofer, R., Marx, H., Maurer, M., Nocon, J., Steiger, M., Puxbaum, V., Sauer, M., and Mattanovich, D. (2013) Pichia pastoris: protein production host and model organism for biomedical research, Fut. Microbiol., 8, 191-208, https://doi.org/10.2217/fmb.12.133.
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn., Cold Spring Harbor, Cold Spring Harbor Laboratory Press, N.Y.
Chen, C. C., Wu, P. H., Huang, C. T., and Cheng, K. (2004) A Pichia pastoris fermentation strategy for enhancing the heterologous expression of an Escherichia coli phytase, Enzyme Microb. Technol., 35, 315-320, https://doi.org/10.1016/j.enzmictec.2004.05.007.
Rebrikov, D. V. (2011) Real time PCR, Binom, Moscow.
Waterham, H. R., Digan, M. E., Koutz, P. J., Lair, S. V., and Cregg, J. M. (1997) Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter, Gene, 186, 37-44, https://doi.org/10.1016/S0378-1119(96)00675-0.
Wong, N., Liu, W., and Wang, X. (2015) WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system, Genome Biol., 16, 218, https://doi.org/10.1186/s13059-015-0784-0.
Cho, S. W., Kim, S., Kim, Y., Kweon, J., Kim, H. S., Bae, S., and Kim, J. S. (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases, Genome Res., 24, 132-141, https://doi.org/10.1101/gr.162339.113.
Yang, Y., Liu, G., Chen, X., Liu, M., Zhan, C., Liu, X., and Bai, Z. (2020) High efficiency CRISPR/Cas9 genome editing system with an eliminable episomal sgRNA plasmid in Pichia pastoris, Enzyme Microb. Technol., 138, 109556, https://doi.org/10.1016/j.enzmictec.2020.109556.
Doench, J. G., Hartenian, E., Graham, D. B., Tothova, Z., Hegde, M., Smith, I., Sullender, M., Ebert, B. L., Xavier, R. J., and Root, D. E. (2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation, Nat. Biotechnol., 32, 1262-1267, https://doi.org/10.1038/nbt.3026.
Liu, Q., Shi, X., Song, L., Liu, H., Zhou, X., Wang, Q., Zhang, Y., and Cai, M. (2019) CRISPR–Cas9-mediated genomic multiloci integration in Pichia pastoris, Microb. Cell Factories, 18, 144, https://doi.org/10.1186/s12934-019-1194-x.
Tkachenko, A. A., Gordeeva, T. L., Sineoky, S. P., Borshchevskaya, L. N., Feday, T. D., National Research Center “Kurchatov Institute” (2022) Komagataella phaffii yeast strain with inactive HIS4 gene – recipient for the construction of marker-free strains producing heterologous proteins, The Federal Service for Intellectual Property, Moscow, Patent no. 2787584.
Tkachenko, A. A., Gordeeva, T. L., Sineoky, S. P., Borshchevskaya, L. N., National Research Center “Kurchatov Institute” (2022) Yeast strain Komagataella phaffii with an inactivated LEU2 gene as a recipient for constructing strains producing heterologous proteins, The Federal Service for Intellectual Property, Moscow, Patent no. 2788528.
Hsu, P. D., Lander, E. S., and Zhang, F. (2014) Development and applications of CRISPR-Cas9 for genome engineering, Cell, 157, 1262-1278, https://doi.org/10.1016/j.cell.2014.05.010.
Zhang, X. H., Tee, L. Y., Wang, X. G., Huang, Q. S., and Yang, S. H. (2015) Off-target effects in CRISPR/Cas9-mediated genome engineering, Mol. Ther. Nucleic. Acids, 4, E264, https://doi.org/10.1038/mtna.2015.37.
Zhu, T., Guo, M., Tang, Z., Zhang, M., Zhuang, Y., Chu, J., and Zhang, S. (2009) Efficient generation of multi-copy strains for optimizing secretory expression of porcine insulin precursor in yeast Pichia pastoris, J. Appl. Microbiol., 107, 954-963, https://doi.org/10.1111/j.1365-2672.2009.04279.x.
Funding
This work was made with financial support of the Ministry of Science and Higher Education of the Russian Federation grant for Kurchatov Center of Genome Research (Agreement no. 075-15-2019-1659), and was performed using the resources of the Bioresource Center – All-Russian Collection of Industrial Microorganisms.
Author information
Authors and Affiliations
Contributions
T.L.G., S.P.S. conceived and supervised the study; A.A.T. carried out experiments; T.L.G., A.A.T., L.N.B. discussed the results of experiments; A.A.T., T.L.G. wrote the manuscript; T.L.G., L.N.B., A.A.T. edited the manuscript.
Corresponding author
Ethics declarations
The authors declare no conflict of interest in financial or any other sphere. This article does not contain descriptions of studies performed by the authors with participation of humans or using animals as objects.
Rights and permissions
About this article
Cite this article
Tkachenko, A.A., Borshchevskaya, L.N., Sineoky, S.P. et al. CRISPR/Cas9-Mediated Genome Editing of the Komagataella phaffii to Obtain a Phytase-Producer Markerless Strain. Biochemistry Moscow 88, 1338–1346 (2023). https://doi.org/10.1134/S0006297923090134
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0006297923090134