Abstract
HSPA1A, which encodes cognate heat shock protein 70, plays important roles in various cellular metabolic pathways. To investigate its effects on osteogenic differentiation of bone marrow mesenchymal stem cells (BMSCs), its expression level was compared between undifferentiated and differentiated BMSCs. Rat HSPA1A overexpression in BMSCs increased osteoblast-specific gene expression, alkaline phosphatase activity and mineral deposition in vitro. Moreover, it upregulated β-catenin and downregulated DKK1 and SOST. The enhanced osteogenesis due to HSPA1A overexpression was partly rescued by a Wnt/β-catenin inhibitor. Additionally, using a rat tibial fracture model, a sheet of HSPA1A-overexpressing BMSCs improved bone fracture healing, as determined by imaging and histological analysis. Taken together, these findings suggest that HSPA1A overexpression enhances osteogenic differentiation of BMSCs, partly through Wnt/β-catenin.
Similar content being viewed by others
Introduction
Mesenchymal stem cells (MSCs) retain their self-renewal capability and have the potential to differentiate into a variety of cell types1; thus, they have emerged as the most promising candidate for tissue repair. It is essential to obtain a deeper understanding of the osteogenic differentiation of MSCs for clinical applications in skeletal regenerative medicine, as well as of their physical and pathological mechanisms in bone metabolism2. Thus, identifying the genetic factors involved in osteogenic differentiation of MSCs has become a very important area of research.
Heat shock protein (HSP) 70 family is evolutionarily highly conserved molecular chaperone stabilizes existing proteins against aggregation and mediates the folding of newly translated proteins3,4. Although it is transiently expressed in cells following heat shock or other stresses, it is also constitutively expressed in some tissues, correlating with senescence and chemoresistance5. The constitutive member is often referred to as cognate heat shock protein (HSC70), which is encoded by Heat shock protein family A member 1A (HSPA1A)3,6. It could be a post-transcriptional regulator of gene expression that binds and stabilizes select mRNAs containing AU-rich elements7. Increasing evidence has revealed that the appropriate heat stress promotes osteogenic differentiation of MSCs or osteoprogenitor cells8,9,10, which induces the expression of HSPs, including HSC70. In addition, HSC70 is believed to play an important role in bone formation, since it is highly expressed in new bone-generating areas11,12. These findings indicate that intracellular expression of HSPA1A might play a positive role in osteogenic differentiation of MSCs.
Wnt/β-catenin signaling is a crucial regulator of MSCs13 and plays an essential role in osteogenic differentiation14,15. Disruption of β-catenin leads to extensive bone marrow adiposity and low bone mass16. Wnt ligands bind to Frizzled and LRP5/6 receptors and induce stabilization of cytoplasmic β-catenin by inhibiting GSK3β15. β-catenin accumulates in the cytoplasm and travels to the nucleus, where it engages the N-terminus of DNA-binding proteins of the Tcf/Lef family17, thereby affecting target gene transcription. The binding of dickkopf-1 (DKK1) and sclerostin (SOST) to LRP5/6 or Frizzled coreceptor was shown to block Wnt/β-catenin signaling18,19. Interestingly, David et al. revealed crosstalk between HSC70 and Wnt/β-catenin signaling; that is, HSPA1A is upregulated in response to Wnt activation in stem cells20.
In this study, we found the endogenous expression of HSPA1A increased during the process of osteogenic differentiation. Accordingly, we hypothesize that upregulated expression of HSPA1A could promote osteogenic differentiation of MSCs via Wnt/β-catenin signaling. by assessing the expression levels of specific genes and calcium deposition. We revealed that HSPA1A overexpression enhanced osteogenic differentiation of rat bone marrow MSCs (rBMSCs) partly via the Wnt/β-catenin signaling pathway in vitro. Moreover, using a rat tibial fracture model, we found that a sheet of rBMSCs overexpressing HSPA1A improved bone fracture healing in vivo.
Materials and Methods
Cells and reagents
rBMSCs were purchased from Cyagen Biosciences (Guangzhou, China). These cells can differentiate into osteoblasts, adipoblasts and chondrocytes under specific inductive conditions. Adherent cells were trypsinized and passaged after reaching 80% confluence. Cells from passages 3–9 were used in subsequent experiments.
Recombinant DKK1 was purchased from PeproTech (Rocky Hill, NJ, USA). In accordance with a previous study, the applied concentration of DKK1 was 0.5 μg/mL21.
Lentiviral packaging and cell infection
Lentivirus overexpressing HSPA1A (lenti-HSPA1A, pLV[Exp]-Puro-CMV >rat-HSPA1A [NM_031971.2]: IRES: EGFP) particles and lentiviral GFP particles (lenti-control, pLV[Exp]-Puro-CMV >EGFP) were prepared by Cyagen Biosciences. The overexpressing HSPA1A lentivirus particles details are specified in Supplemental material 1. The lentiviral GFP particles were used as control group in this study.
For infections, rBMSCs were incubated with lentiviral particles and polybrene (5 μg/mL) in growth medium. After 5–6 h, the infection medium was discarded. After 3 days, the cells were screened using puromycin (4 μg/mL; Sigma, Shanghai, China) and then passaged for use in subsequent experiments. The expression of HSPA1A was quantified by quantitative real-time polymerase chain reaction (qPCR) and Western blot analyses.
Cell Counting Kit-8 (CCK-8)
To assess the effect of HSPA1A overexpression on the proliferation of rBMSCs, the cells were seeded into a 96-well plate (5000/well) and allowed to adhere for 24 h. After 24 h, the medium was removed and the cells were treated with 10% CCK-8 (Dojindo, Kumamoto, Japan) in 100 μL low-sugar Dulbecco’s modified Eagle’s medium (L-DMEM) without fetal bovine serum (FBS) for 2 h at 37 °C. Absorbance at 450 nm, which is directly proportional to cell proliferation, was measured using a microplate reader (ELX808; BioTek, Winooski, VT, USA).
Osteogenic differentiation protocol
MSCs were cultured in growth medium [L-DMEM; 10% FBS (1495527; Gibco, Waltham, MA, USA) and 100 IU/mL penicillin/streptomycin] in 6- or 12-well cell culture plates (Corning, Shanghai, China), at a density of 3 × 104/cm2 and incubated for 48 h at 37 °C under 5% CO2. The cells were subsequently cultured in osteogenic induction medium (L-DMEM with 10% FBS, 100 IU/mL penicillin/streptomycin, 100 nM dexamethasone, 0.2 mM ascorbic acid and 10 mM β-glycerophosphate). The cells were maintained by the addition of fresh osteogenic induction medium every 2–3 days.
Measurement of alkaline phosphatase (ALP) activity
For the measurement of ALP activity, cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) and the lysate (10 μL) was incubated with 90 μL fresh solution containing p-nitrophenyl phosphate substrate at 37 °C for 30–60 min. The reaction was stopped by the addition of 0.5 N NaOH (100 μL) and the absorbance was measured at 405 nm using a microplate reader (ELX808; BioTek). The total protein concentration was measured using a BCA protein assay kit (KeyGen BioTECH, Nanjing, China). The relative ALP activity is expressed as the percentage change in optical density (OD) per unit time per milligram protein: (OD/15 min/mg protein) ×100.
Alizarin red staining (ARS)
After the induction of osteogenic differentiation, mineral deposition was assessed by ARS (Cyagen Biosciences). Cells were fixed in 4% paraformaldehyde (Sangon Biotech, Shanghai, China) for 15 min at room temperature and then washed with distilled water. A 1% solution of alizarin red was added and incubated for 30 min at room temperature, followed by rinsing with distilled water. The solution was collected and 200 μL were plated on 96-well plates, which were read at 560 nm using a microplate reader (ELX808; BioTek). The readings were normalized to the total protein concentration.
Immunofluorescence
Cells were cultured in induction medium in a 12-well plate and RUNX2, COL1A1 and β-catenin were detected using a fluorescence microscope (EU5888; Leica, Wetzlar, Germany). Briefly, cells were fixed in 4% paraformaldehyde for 15 min at room temperature, permeabilized and blocked for 30 min in 0.05% Triton X-100 and 2% bovine serum albumin. Fixed cells were washed and incubated overnight with anti-RUNX2 (1:1600; Cell Signaling Technology, Shanghai, China), COL1A1 (1:100; Santa Cruz Biotechnology, Shanghai, China), or β-catenin (1:100; Cell Signaling Technology). Cells were incubated with a fluorescence-conjugated secondary antibody (Beyotime) for 120 min and nuclei were stained with 4′,6-diamidino-2-phenylindole (KeyGen Biotech, Nanjing, China) for 2 min. Samples were observed under a fluorescence microscope (Leica).
RNA isolation and qPCR
Total cellular RNA was isolated using RNAiso reagent (Takara, Dalian, China) and quantified by measuring the absorbance at 260 nm (NanoDrop 2000; Thermo Fisher Scientific, Waltham, MA, USA). Total RNA (≤1000 ng) was reverse-transcribed into cDNA in a reaction volume of 20 μL using the Double-Strand cDNA Synthesis Kit (Takara, Dalian, China). One microliter of cDNA was used as the template for the qPCR reaction. All gene transcripts were quantified by qPCR using the Power SYBR® Green PCR Master Mix (Takara) on the ABI StepOnePlus System (Applied Biosystems, Warrington, UK). The mRNAs of the target genes and the housekeeping gene (18S) were quantified in separate tubes. All primers were synthesized by Sangon Biotech (Shanghai, China). The primer sequences used are shown in Table 1. The cycle conditions were as follows: 95 °C for 30 s and then 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The relative target gene expression levels were calculated using the 2−△△Ct method.
Western blot analysis
Cells were lysed in RIPA lysis buffer supplemented with a proteasome inhibitor (Beyotime). Total proteins were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to a polyvinylidene fluoride membrane (Millipore, Shanghai, China). After blocking in 5% non-fat milk for 2 h, the membranes were incubated overnight at 4 °C with antibodies specific to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:1500, Cell Signaling Technology), HSPA1A (1:1000; Cell Signaling Technology), Osteocalcin (OCN) (1:1000; abcam, Shanghai, China), RUNX2 (1:1600), COL1A1 (1:1000), or β-catenin (1:1000). Horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:1500; Cell Signaling Technology) was applied as a secondary antibody for 2 h at room temperature. The immunoreactive bands were detected using an enhanced chemiluminescent detection reagent (Millipore, Shanghai, China). Signal intensity was measured using a Bio-Rad XRS chemiluminescence detection system (Bio-Rad, Hercules, CA, USA).
Cell sheet preparation
A cell sheet was fabricated as we reported previously22. Briefly, confluent cells in flasks at 1 × 105/cm2 were cultured in L-DMEM with the addition of vitamin C (20 μg/mL)23 for 2 weeks until a sheet of rBMSCs formed and could be detached intact from the substratum using a cell scraper (Supplemental material 2).
In vivo evaluation in animals
All Sprague Dawley (SD) rats were supplied by the Academy of Medical Sciences of Zhejiang province. All animal experiments were performed in accordance with the Animal Care and Use Committee guidelines of Zhejiang province. All experimental procedures were approved by the Institutional Animal Care and Use Committee at Zhejiang university.
All surgical procedures were performed by two experienced traumatic orthopedic surgeons (Zhijun Pan and Deting Xue). Fractures were generated in 8-week-old male SD rats (weighing approximately 200 g). The rats were anesthetized with 0.3% pentobarbital sodium (Sigma) intraperitoneally at 30 mg/kg body weight. The fracture model was established as reported previously24,25,26. After anesthesia, an incision was made below the knee. A transverse osteotomy from front to back was generated at the tibia (above the fibular junction) using an oscillating mini saw. An intramedullary needle (1.2-mm-diameter stainless steel syringe needle) was inserted inside the medullary canal of the tibia for fixation. The gap was 1.0 mm (Supplemental material 3). The 15 fractures in 15 rats were randomized into three groups. In the blank group (n = 5), nothing was grafted into the fracture site; in the lenti-control group (n = 5), a sheet of lenti-control rBMSCs was wrapped around the fracture site; and in the lenti-HSPA1A group (n = 5), a sheet of rBMSCs overexpressing HSPA1A was wrapped around the fracture site.
Radiographic analysis and micro-computed tomography (CT) evaluation
Animals were euthanized 8 weeks after surgery and samples were collected. Radiographs were taken using a dual-track molybdenum/rhodium+ Mo target mammography machine (22 KV, 250 mAS; GE, Fairfield, CT, USA) 8 weeks after surgery to evaluate callus formation and bridging bone formation at the fracture sites. All samples were scanned for bone formation using a μCT 100 imaging system (Scanco Medical, Brüttisellen, Switzerland) with the following scan parameters: X-ray energy setting of 70 kVp, 1024 reconstruction matrix, slice thickness of 0.0148 mm and integration time of 300 ms. The bone volume fraction (BV/TV) including the area of bone formation was calculated by three-dimensional standard microstructural analysis27.
Histological evaluation
The samples were fixed using 4% paraformaldehyde for 72 h at room temperature and then decalcified using 10% ethylenediaminetetraacetic acid (Sigma), with a solution change once a week for more than 8 weeks, before embedding in paraffin. Serial sections of 3 mm thickness were cut and mounted on polylysine-coated slides. Hematoxylin and eosin, Safranin O, Masson and immunofluorescence staining of COL1A1 were performed separately on consecutive tissue sections in accordance with previous studies24,28 and images were obtained using a microscope.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 software (IBM, Armonk, NY, USA). All experiments were performed at least in triplicate and the data are presented as means ± standard deviation. Statistical significance was determined using a two-tailed Student’s t-test when comparing two groups and one-way ANOVA followed by Bonferroni’s post hoc test when comparing more than two groups. A P value of 0.05 or less was considered to represent a statistically significant difference.
Results
Endogenous HSPA1A expression
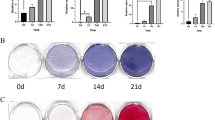
To determine the expression levels of HSPA1A associated with osteogenic differentiation of MSCs, we examined HSPA1A endogenous expression in rBMSCs at days 0, 7 and 14 during the process of osteogenic differentiation. Compared with undifferentiated rBMSCs, the mRNA expression of HSPA1A increased significantly at days 7 and 14 during osteogenic differentiation (Fig. 1A). In addition, the protenin expression of HSPA1A also increased significantly at day 14 during osteogenic differentiation (Fig. 2B,C).
Endogenous HSPA1A expression and the construction of HSPA1A-overexpressing rBMSCs and lenti-control rBMSCs.
(A) The endogenous expression of HSPA1A mRNA was determined by qPCR at days 0, 7 and 14 of osteogenic differentiation. (B,C) The endogenous expression of HSPA1A protein was determined by Western blot analysis at days 0, 7 and 14 of osteogenic differentiation. (D) rBMSCs after lentiviral transfection and puromycin screening were observed under a normal microscope and a fluorescence microscope. (E) The proliferation rate of rBMSCs was not significantly affected by HSPA1A overexpression. (F,G) Protein levels of HSPA1A were determined by Western blot analysis among the lenti-HSPA1A, mock treated group and lenti-control group. The mRNA expression levels were normalized to that of 18S ribosomal RNA. The data are expressed as means ± standard deviation, n = 3, *P < 0.05 vs. the lenti-control group.
The overexpression of HSPA1A promoted osteogenic differentiation of rBMSCs.
(A) The expression of ALP mRNA was determined by qPCR at days 3, 7 and 14 of osteogenic differentiation. (B) The expression of RUNX2 mRNA (C) The expression of OCN mRNA (D) The expression of COL1A1 mRNA (E) Alizarin red staining in the lenti-control and lenti-HSPA1A overexpression groups at day 28 of osteogenic differentiation. Scale bar = 200 μm (F) Alizarin red staining area determined by measuring the absorbance at 560 nm. (G) ALP activity in the control and HSPA1A overexpression groups at day 14 of osteogenic differentiation. The mRNA expression levels were normalized to that of 18S ribosomal RNA. The data are expressed as means ± standard deviation of three independent experiments and one of three independent experiments is shown. *P < 0.05 vs. the lenti-control group.
The establishment of HSPA1A overexpression in rBMSCs
To clarify the role of HSPA1A during osteogenic differentiation, a lentiviral vector system (Supplemental material 1) was used efficiently to overexpress HSPA1A in >80% of third-generation rBMSCs, which was quantified by evaluating the ratio of green fluorescent protein (GFP)-positive cells to the total cell number (Fig. 1D). HSPA1A expression was quantified by Western blot analyses 1 week after infection and screening. Compared with those in the lenti-control group and mock treated group (without virus), HSPA1A were overexpressed 4.12-fold in the lenti-HSPA1A group (Fig. 1F,G).
HSPA1A overexpression did not affect rBMSC proliferation
To determine whether HSPA1A overexpression influences the proliferation of rBMSCs, CCK8 detection was performed. The effects of HSPA1A overexpression on rBMSC proliferation at days 7, 10 and 14 after infection following culture in normal growth medium are shown in Fig. 1E. No significant difference was detected in the cell proliferation rate between HSPA1A-overexpressing and non-overexpressing rBMSCs.
HSPA1A overexpression increased the levels of osteo-specific genes and proteins
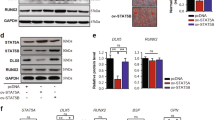
To assess the role of HSPA1A overexpression in osteogenic differentiation, the levels of osteo-specific genes and proteins, including ALP, RUNX2, osteocalcin (OCN) and COL1A1, were detected by qPCR and Western blot analyses. qPCR analysis revealed that ALP, RUNX2, OCN and COL1A1 mRNA levels were significantly higher in rBMSCs overexpressing HSPA1A at days 3, 7 and 14 than in the non-overexpressing rBMSCs (P < 0.05, Fig. 2A–D).
Western blot analysis revealed a higher level of RUNX2 protein expression in rBMSCs overexpressing HSPA1A than in the non-overexpressing cells (Fig. 3D).
The overexpression of HSPA1A upregulated the Wnt/β-catenin signaling pathway during rBMSC osteogenesis.
(A) The expression of β-catenin mRNA was determined by qPCR at days 3, 7 and 14 of osteogenic differentiation. (B) The expression of DKK1 mRNA (C) The expression of SOST mRNA (D,E) The expression of RUNX2 and β-catenin proteins was determined by Western blot analysis after osteogenic differentiation for 7 days. The mRNA expression levels were normalized to that of 18S ribosomal RNA. The protein expression levels were normalized to that of GAPDH. The data are expressed as means ± standard deviation of three independent experiments and one of three independent experiments is shown. *P < 0.05 vs. the lenti-control group.
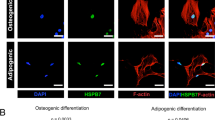
We also used immunofluorescence to confirm the expression of RUNX2 and COL1A1 proteins and showed that these protein expression levels increased at day 7 in HSPA1A-overexpressing cells (Fig. 4A,B).
Immunofluorescence staining showed that the protein levels of COL1A1, RUNX2 and β-catenin were up-regulated by HSPA1A overexpression.
(A) The level of COL1A1 protein (red) at day 7 of osteogenic differentiation. The nuclei are counterstained with DAPI (blue). Scale bar = 200 μm. (B) The level of RUNX2 protein (red) at day 7 of osteogenic differentiation. The nuclei were counterstained with DAPI (blue). Scale bar = 50 μm. (C) The level of β-catenin protein at day 7 of osteogenic differentiation. The nuclei were counterstained with DAPI (blue). Scale bar = 50 μm.
HSPA1A overexpression enhanced ALP activity and calcium deposit formation
We evaluated ALP activity, an early marker of osteogenesis, at day 14 during osteogenic differentiation. Compared with the non-overexpression group, higher ALP activity was observed in the HSPA1A overexpression group (P < 0.05, Fig. 2G). Calcium deposits were also examined by ARS and the staining areas were quantified by measuring the absorbance at 560 nm. More calcium deposits appeared in the HSPA1A-overexpressing than in the non-overexpressing rBMSCs at day 28 (Fig. 2E,F).
HSPA1A overexpression activated the Wnt/β-catenin signaling pathway
To confirm the above findings suggesting that Wnt/β-catenin signaling is involved in the observed phenomena, the expression of β-catenin was determined by qPCR, Western blot analysis and immunofluorescence. The expression of DKK1 and SOST was also detected by qPCR. The results of qPCR and Western blot analyses demonstrated higher expression of β-catenin in the rBMSCs overexpressing HSPA1A (Fig. 3A,D,E). Compared with those in the non-overexpression group, DKK1 and SOST levels were significantly reduced in the rBMSCs overexpressing HSPA1A (Fig. 3B,C). Moreover, using immunofluorescence, we found lower levels of β-catenin accumulation, the majority of which was in the cytoplasm in the non-overexpression group (Fig. 4C).
The increased osteogenic differentiation of rBMSCs due to HSPA1A overexpression was partially rescued by the addition of a Wnt/β-catenin signaling inhibitor (DKK1)
To verify the involvement of the Wnt/β-catenin signaling pathway, we evaluated the inhibitory effect of this pathway on osteogenesis in the HSPA1A-overexpressing group. After the addition of DKK1 for 1 h, osteo-specific genes and proteins were examined. As shown in Fig. 5, lower expression levels of the osteo-specific genes ALP, RUNX2, OCN and COL1A1 were identified in the inhibitor-treated cells (lenti-HSPA1A+DKK1) than in the cells with HSPA1A overexpression alone.
The increased osteogenesis caused by HSPA1A overexpression could be rescued partially by the addition of a Wnt/β-catenin signaling inhibitor (DKK1).
(A) The expression of ALP, RUNX2, OCN and COL1A1 mRNA in the lenti-control, lenti-control + DKK1, lenti-HSPA1A and lenti-HSPA1A + DKK1 groups were determined by qPCR. The mRNA expression levels were normalized to that of 18S ribosomal RNA. (B,C) The expression of β-catenin, RUNX2, OCN and COL1A1 in the lenti-control, lenti-control + DKK1, lenti-HSPA1A and lenti-HSPA1A + DKK1 groups were determined by Western blot analysis. The protein expression levels were normalized to that of GAPDH. (D) Alizarin red staining in the lenti-control, lenti-control + DKK1, lenti-HSPA1A and lenti-HSPA1A + DKK1 groups at day 28 of osteogenic differentiation. Scale bar = 200 μm (E) Alizarin red staining area determined by measuring the absorbance at 560 nm. (F) ALP activity in the lenti-control, lenti-control + DKK1, lenti-HSPA1A and lenti-HSPA1A + DKK1 groups at day 14 of osteogenic differentiation. The data are expressed as means ± standard deviation of three independent experiments and one of three independent experiments is shown. *P < 0.05 vs. the lenti-control group. #P < 0.05 vs. the lenti-HSPA1A group.
Moreover, ALP activity detection demonstrated higher ALP activity in MSCs overexpressing HSPA1A than in the lenti-HSPA1A+DKK1 group (Fig. 5F). There was also less matrix mineralization at day 28 of osteogenic differentiation in the lenti-HSPA1A+DKK1 group than in rBMSCs overexpressing HSPA1A (Fig. 5D,E).
A sheet of rBMSCs overexpressing HSPA1A accelerated bone fracture healing in a rat tibial fracture model
Radiographs taken at 8 weeks showed that the cortical gap was clearly present in the blank group. In the lenti-control group, this gap was obscure and more bridging callus formation was evident at the fracture site compared with the blank group. In the rBMSCs overexpressing HSPA1A, the gap had disappeared (Fig. 6A).
Radiographic and micro-computed tomography (CT) analyses of the fracture site at 8 weeks after surgery in each group.
(A) Radiographic analysis of the fracture sites at 8 weeks after surgery in each group. (B,C) Micro-CT images of the fracture sites in each group at 8 weeks after surgery. Scale bar = 1 mm.
The results of micro-CT indicated significantly more bone formation in the rBMSCs overexpressing HSPA1A than in the lenti-control group; moreover, among the three groups, the gap was widest in the blank group (Fig. 6B). Quantitatively, the fractures treated with a sheet of rBMSCs overexpressing HSPA1A displayed a significant increase in the BV/TV compared with those of the other two groups (Fig. 6C).
Histological analysis showed that the gaps were filled with fibrous tissue and a few chondrocytes; moreover, no bridging bone formation at the fracture site was observed in the blank group (Fig. 7A,D). In the lenti-control group, a thick callus consisting of newly formed woven bone tissue was detected at the fracture site and the callus was undergoing remodeling (Fig. 7B,E). In the rBMSCs overexpressing HSPA1A, the fracture sites were sealed and the remodeling of the callus was almost complete, indicating bony healing of the fracture (Fig. 7C,F).
Immunofluorescence analysis showed that significantly more COL1A1 appeared at the fracture site in the rBMSCs overexpressing HSPA1A than in the other two groups. The lowest expression of COL1A1 was detected in the blank group. Sporadic fluorescence of GFP was found in the lenti-control group and in the rBMSCs overexpressing HSPA1A (Fig. 8).
Discussion
To our knowledge, this is the first study to investigate the effects of HSPA1A on MSC osteogenic differentiation. We found that endogenous expression of HSPA1A was upregulated during osteogenesis in rBMSCs. Moreover, according to the results of previous studies, HSPA1A is highly expressed in new bone-healing areas11,12. Thereupon, we want to use an overexpression of HSPA1A strategy to accelerate osteogenesis of MSCs. We found that HSPA1A overexpression promoted osteogenic differentiation of rBMSCs partly via the Wnt/β-catenin signaling pathway in vitro. Moreover, a sheet of rBMSCs overexpressing HSPA1A accelerated bone fracture healing in rat fracture models. These findings indicate that HSPA1A overexpression enhances osteogenic differentiation of rBMSCs, at least partly through activation of the Wnt/β-catenin signaling pathway.
Mounting evidence has revealed that increased expression of HSPA1A is closely associated with osteogenesis. Tiffee et al. found that HSC70 was localized in the osteoblasts lining new bone in the primary spongiosa, which might have biosynthetic functions within osteoblasts12. Continuous exposure to mild heat stress (39–41 °C) significantly enhanced the ability of BMSCs to form mineralizing nodules, which were associated with HSPA1A constitutive expression10. Aisha et al. revealed that chaperone proteins, including HSC70, stabilized mRNA transcripts and enhanced ALP and OCN gene transcription29. Likewise, our results demonstrated that the expression of HSPA1A plays a positive role in osteogenic differentiation. We found that HSPA1A was upregulated during osteogenesis in rBMSCs. RUNX2 is a master transcription factor involved in osteogenic differentiation30. In our study, its expression was significantly increased following overexpression of HSPA1A. The levels of an early marker of osteogenic differentiation (ALP) and late markers of osteogenic differentiation (OCN, COL1A1) were also increased due to HSPA1A overexpression. In addition, this overexpression significantly enhanced mineral deposition. Meanwhile, HSPA1A overexpression had no adverse effects on the metabolic activity or cell proliferation of rBMSCs. These results indicate that HSPA1A overexpression promoted the osteogenic differentiation of rBMSCs in vitro.
A previous study reported crosstalk between HSC70 and Wnt/β-catenin signaling in stem cells; that is, HSPA1A upregulation was closely related to activation of the Wnt/β-catenin signaling pathway20. Wnt/β-catenin signaling is an essential pathway in the osteogenic differentiation of MSCs14,15. Wnt signaling results in cellular accumulation of Wnt/β-catenin, followed by nuclear translocation of β-catenin and activation of target genes31. In this study, we detected higher expression of β-catenin following overexpression of HSPA1A during osteogenesis. Moreover, higher β-catenin accumulation was observed, the majority of which was in the nucleus upon HSPA1A overexpression, suggesting that HSPA1A overexpression activates β-catenin-mediated transcription. Meanwhile, lower levels of DKK1 and SOST were observed, which inhibited Wnt/β-catenin by binding to the third β-propeller domain of LRP5/6 via interference of Wnt/LRP/Fz trimolecular complex formation18. Furthermore, the increased osteogenesis of MSCs by HSPA1A overexpression was rescued partly by an inhibitor of Wnt/β-catenin (DKK1). These findings indicate that overexpression of HSPA1A regulates the Wnt/β-catenin signaling pathway during osteogenesis of MSCs.
It has been demonstrated that MSC sheets can enhance bone regeneration26,32,33. Cultured cells can be harvested as intact sheets, thereby decreasing cell damage and maintaining the majority of the extracellular matrix, which can attach to host tissues and fracture sites, covering the surface with minimal cell loss34. In our study, the use of a rBMSC sheet promoted bone formation in a rat tibial fracture model. Better bone healing was also observed when rBMSCs overexpressing HSPA1A were present in the sheet. Additionally, these HSPA1A-overexpressing rBMSCs did not cause localized tumors or malignancies. However, there were only a few cells exhibiting GFP fluorescence in the rBMSC sheet groups, which suggested that only a fraction of the implanted rBMSCs directly differentiated into osteocytes and produced bone matrix. The majority of new bone formation arose from exogenous cells, which is consistent with previous studies28,35. Transplanted MSCs increase bone formation through the production of factors that promote osteogenic differentiation of host cells35. A paracrine effect might have been involved in bone healing in vivo, when the sheet of rBMSCs overexpressing HSPA1A was implanted for fracture repair. Guang et al. revealed that HSC70 plays a significant role in endothelial cell development and angiogenesis36. Aparna et al. also reported that HSC70 could bind and stabilize endogenous AU-rich element-containing mRNAs encoding vascular endothelial growth factor7. These studies indicated that HSC70 might be closely related to vasculogenesis and neovascularization of endothelial cells, which could play an important role in bone healing.
The present study has some limitations. First, although we indicated that HSPA1A overexpression mediates the Wnt/β-catenin signaling pathway to promote osteogenic differentiation of rBMSCs, it is probably involved in the activation of other signaling pathways as well. Second, the mechanisms of bone healing acceleration using a sheet of rBMSCs overexpressing HSPA1A have not been clarified completely and thus further studies are needed.
Conclusion
Based on our data, we found that HSPA1A overexpression enhanced osteogenic differentiation of rBMSCs, partly through activation of the Wnt/β-catenin signaling pathway. The use of HSPA1A-overexpressing MSCs to enhance fracture repair has potential as a new MSC-based therapy.
Additional Information
How to cite this article: Zhang, W. et al. Overexpression of HSPA1A enhances the osteogenic differentiation of bone marrow mesenchymal stem cells via activation of the Wnt/β-catenin signaling pathway. Sci. Rep. 6, 27622; doi: 10.1038/srep27622 (2016).
References
Bianco, P., Riminucci, M., Gronthos, S. & Robey, P. G. Bone marrow stromal stem cells: nature, biology and potential applications. Stem Cells 19, 180–192 (2001).
Hakelien, A. M. et al. The regulatory landscape of osteogenic differentiation. Stem Cells 32, 2780–2793 (2014).
McCallister, C., Kdeiss, B. & Nikolaidis, N. HspA1A, a 70-kDa heat shock protein, differentially interacts with anionic lipids. Biochem Biophys Res Commun 467, 835–40 (2015).
Kirkegaard, T. et al. Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Cah Rev The 463, 549–553 (2010).
Gabai, V. L., Yaglom, J. A., Waldman, T. & Sherman, M. Y. Heat shock protein Hsp72 controls oncogene-induced senescence pathways in cancer cells. Mol Cell Biol 29, 559–569 (2009).
Sasi, B. K., Sonawane, P. J., Gupta, V., Sahu, B. S. & Mahapatra, N. R. Coordinated transcriptional regulation of Hspa1a gene by multiple transcription factors: crucial roles for HSF-1, NF-Y, NF-kappaB and CREB. J Mol Biol. 426, 116–135 (2014).
Kishor, A. et al. Hsp70 is a novel posttranscriptional regulator of gene expression that binds and stabilizes selected mRNAs containing AU-rich elements. Mol Cell Biol 33, 71–84 (2013).
Norgaard, R., Kassem, M. & Rattan, S. I. Heat shock-induced enhancement of osteoblastic differentiation of hTERT-immortalized mesenchymal stem cells. Ann N Y Acad Sci. 1067, 443–447 (2006).
Li, M. et al. Mild heat stress enhances angiogenesis in a co-culture system consisting of primary human osteoblasts and outgrowth endothelial cells. Tissue Eng Part C Methods 20, 328–339 (2014).
Shui, C. & Scutt, A. Mild heat shock induces proliferation, alkaline phosphatase activity and mineralization in human bone marrow stromal cells and Mg-63 cells in vitro. J Bone Miner Res 16, 731–741 (2001).
Yoshida, K., Uoshima, K., Oda, K. & Maeda, T. Influence of heat stress to matrix on bone formation. Clin Oral Implants Res 20, 782–790 (2009).
Tiffee, J. C., Griffin, J. P. & Cooper, L. F. Immunolocalization of stress proteins and extracellular matrix proteins in the rat tibia. Tissue Cell 32, 141–147 (2000).
Reya, T. & Clevers, H. Wnt signalling in stem cells and cancer. Cah Rev The 434, 843–850 (2005).
Ahmadzadeh, A., Norozi, F., Shahrabi, S., Shahjahani, M. & Saki, N. Wnt/beta-catenin signaling in bone marrow niche. Cell Tissue Res 363, 321–335 (2016).
MacDonald, B. T. & He, X. Frizzled and LRP5/6 receptors for Wnt/beta-catenin signaling. Cold Spring Harb Perspect Biol 4 (2012).
Song, L. et al. Loss of wnt/beta-catenin signaling causes cell fate shift of preosteoblasts from osteoblasts to adipocytes. J Bone Miner Res 27, 2344–2358 (2012).
Eastman, Q. & Grosschedl, R. Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr Opin Cell Biol 11, 233–240 (1999).
Balemans, W. et al. The binding between sclerostin and LRP5 is altered by DKK1 and by high-bone mass LRP5 mutations. Calcif Tissue Int 82, 445–453 (2008).
Mao, B. Y. et al. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Cah Rev The 411, 321–325 (2001).
Duffy, D. J., Millane, R. C. & Frank, U. A heat shock protein and Wnt signaling crosstalk during axial patterning and stem cell proliferation. Dev Biol 362, 271–281 (2012).
Yun, H. M. et al. 2,4,5-Trimethoxyldalbergiquinol promotes osteoblastic differentiation and mineralization via the BMP and Wnt/beta-catenin pathway. Cell Death Dis 6, e1819 (2015).
Qi, Y. et al. Cartilage repair using mesenchymal stem cell (MSC) sheet and MSCs-loaded bilayer PLGA scaffold in a rabbit model. Knee Surg Sports Traumatol Arthrosc 22, 1424–1433 (2014).
Wei, F. et al. Vitamin C treatment promotes mesenchymal stem cell sheet formation and tissue regeneration by elevating telomerase activity. J Cell Physiol 227, 3216–3224 (2012).
Xu, L. et al. Sox11-modified mesenchymal stem cells (MSCs) accelerate bone fracture healing: Sox11 regulates differentiation and migration of MSCs. Faseb J 29, 1143–1152 (2014).
El-Hoss, J. et al. A Combination of rhBMP-2 (Recombinant Human Bone Morphogenetic Protein-2) and MEK (MAP Kinase/ERK Kinase) Inhibitor PD0325901 Increases Bone Formation in a Murine Model of Neurofibromatosis Type I Pseudarthrosis. J Bone Joint Surg Am 96, e117 (2014).
Qi, Y. et al. Mesenchymal stem cell sheet transplantation combined with locally released simvastatin enhances bone formation in a rat tibia osteotomy model. Cytotherapy 15, 44–56 (2013).
Bouxsein, M. L. et al. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res 25, 1468–1486 (2010).
Sun, H., Wu, Y., Fu, D., Liu, Y. & Huang, C. SIRT6 regulates osteogenic differentiation of rat bone marrow mesenchymal stem cells partially via suppressing the nuclear factor-kappaB signaling pathway. Stem Cells 32, 1943–1955 (2014).
Aisha, M. D. et al. Short-term moderate hypothermia stimulates alkaline phosphatase activity and osteocalcin expression in osteoblasts by upregulating Runx2 and osterix in vitro. Exp Cell Res. 326, 46–56 (2014).
Komori, T. et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 (1997).
Rudnicki, M. A. & Williams, B. O. Wnt signaling in bone and muscle. Bone 80, 60–66 (2015).
Qi, Y. et al. Combined mesenchymal stem cell sheets and rhBMP-2-releasing calcium sulfate-rhBMP-2 scaffolds for segmental bone tissue engineering. Cell Transplant 21, 693–705 (2012).
Wang, Z. et al. Osteoblastic mesenchymal stem cell sheet combined with Choukroun platelet-rich fibrin induces bone formation at an ectopic site. J Biomed Mater Res B Appl Biomater 103, 1204–1216 (2015).
Chen, G. et al. Application of the cell sheet technique in tissue engineering. Biomed Rep 3, 749–757 (2015).
Pauley, P. et al. Local transplantation is an effective method for cell delivery in the osteogenesis imperfecta murine model. Int Orthop 38, 1955–1962 (2014).
Hu, G. et al. A novel endothelial-specific heat shock protein HspA12B is required in both zebrafish development and endothelial functions in vitro. J Cell Sci 119, 4117–4126 (2006).
Acknowledgements
We thank Qubo Ni for his expert technical assistance. This work was supported by a grant from the National Natural Science Foundation of China (No.81271973 and No.81201397), the Zhejiang Provincial Natural Science Foundation of China (No. LY15H060001; No. LY15H060002 No. LY16H060003; and No. LY13H060002) and Zhejiang medical and health science and technology plan project (No. 2015KYB182).
Author information
Authors and Affiliations
Contributions
Z.P. contributed design and funding sources to this study. W.Z. and D.X. drafted the manuscript. H.Y., C.L., S.W., Q.Z. and W.Z. preformed the in vivo experiments. W.Z., D.X., E.C., X.G., D.H. and S.W. did all the in vitro parts of the study. Y.T. and J.Y. carried out statistical work. All authors have contributed significantly. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, W., Xue, D., Yin, H. et al. Overexpression of HSPA1A enhances the osteogenic differentiation of bone marrow mesenchymal stem cells via activation of the Wnt/β-catenin signaling pathway. Sci Rep 6, 27622 (2016). https://doi.org/10.1038/srep27622
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep27622
- Springer Nature Limited
This article is cited by
-
A novel technique to harvest bone autografts with mild local hyperthermia and enhanced osteogenic bone quality: a preclinical study in dogs
BMC Oral Health (2023)
-
Comparative transcriptome analysis provides insight into the molecular targets and signaling pathways of deer TGF-1 regulating chondrocytes proliferation and differentiation
Molecular Biology Reports (2023)
-
Knockdown of FOXA1 enhances the osteogenic differentiation of human bone marrow mesenchymal stem cells partly via activation of the ERK1/2 signalling pathway
Stem Cell Research & Therapy (2022)
-
Cryptic genetic variation in a heat shock protein modifies the outcome of a mutation affecting epidermal stem cell development in C. elegans
Nature Communications (2021)
-
Extracellular IL-37 promotes osteogenic differentiation of human bone marrow mesenchymal stem cells via activation of the PI3K/AKT signaling pathway
Cell Death & Disease (2019)