Abstract
Rapid detection and enumeration of target microorganisms is considered as a powerful tool for monitoring bioremediation process that typically involves cleaning up polluted environments with functional microbes. A novel colorimetric assay is presented based on immunomagnetic capture and bacterial intrinsic peroxidase activity for rapidly detecting Shewanella oneidensis, an important model organism for environmental bioremediation because of its remarkably diverse respiratory abilities. Analyte bacteria captured on the immunomagnetic beads provided a bacterial out-membrane peroxidase-amplified colorimetric readout of the immunorecognition event by oxidizing 3, 3′, 5, 5′-tetramethylbenzidine (TMB) in the present of hydrogen peroxide. The high-efficiency of immunomagnetic capture and signal amplification of peroxidase activity offers an excellent detection performance with a wide dynamic range between 5.0 × 103 and 5.0 × 106 CFU/mL toward target cells. Furthermore, this method was demonstrated to be feasible in detecting S. oneidensis cells spiked in environmental samples. The proposed colorimetric assay shows promising environmental applications for rapid detection of target microorganisms.
Similar content being viewed by others
Introduction
Shewanella oneidensis is a facultative anaerobic bacterium that widely inhabits both aquatic and terrestrial environments1,2. S. oneidensis is known for versatile electron accepting capacities which allow it to undertake respiration, under anaerobic conditions, by reducing a variety of terminal electron acceptors such as Fe(III), Mn(IV), sulfur and nitrate3,4. The ability to effectively reduce oxidized metals and nitrate has established S. oneidensis as an important model organism for implications with regard to environmental pollutants. For example, S. oneidensis can reduce uranium from the dissolved liquid state (U(VI)) to insoluble oxides (U(IV)), which will facilitate the clean-up of metal pollutants in natural environment5,6. In addition, S. oneidensis is also able to produce large quantities of sulfide from thiosulfate and then immobilize toxic metal ions through forming insoluble metal sulfides7. The environmentally versatile abilities of S. oneidensis rely heavily on its flexible extracellular electron transport system that is composed of c-type cytochromes, most of which are predicted to be located on the outer membrane8. There are extensive studies on this strain but focused primarily on the mechanism of extracellular electron transfer4,9,10. Little research has been conducted on detection method for the functional microbe in microbial remediation. To better monitor bioremediation processes, it is critical to develop rapid and low cost methods to detect target microorganisms that are directly related to the transformation of contaminants.
Immunomagnetic capture (IMC) is a rapid, efficient and simple method that has been widely used in biomedicine, food safety and environmental monitoring11,12. This method uses specific antibodies-coated magnetic beads (MBs) to separate and enrich target analyte from complex environmental samples. In this way, IMC can eliminate the effect of sample matrix on the following protocols while retaining microbial viability. Recently, IMC has been applied in combinations of several modern methods, such as Flow Cytometry13, PCR14 and ELISA15. However, most of these methods are either time-consuming or expensive, requiring complicated instruments.
Colorimetric assays based on horseradish peroxidase/3,3′,5,5′-tetramethyl-benzidine (HRP/TMB) system have recently gained much attention due to their various advantages such as high sensitivity, low toxicity and simple management16,17,18. HRP is a heme-containing protein that can utilize hydrogen peroxide to oxidize a wide variety of compounds. The potent activity of HRP can be attributed to its heme components19,20. Structurally similar to HRP, c-type cytochromes are essential for almost all organisms (particularly for S. oneidensis) and have been demonstrated to be able to catalyze chemical reactions involving oxidation and reduction of substrates. For example, the heme peroxidase activity of c-type cytochromes has been utilized in gel staining to characterize bacterial respiration21,22. Studies on microbial extracellular respiration have demonstrated that 80% of the membrane-bound c-type cytochromes in S. oneidensis cells are located on the outer membrane23. However, coupling of bacterial out membrane peroxidase activity with IMC for colorimetric immunoassay remains unexplored.
The present work describes a new method for fast detection of S. oneidensis. This method combines IMC protocol with microbial intrinsic peroxidase-based colorimetric assay. The coupling of high-efficiency of immunomagnetic separation and signal amplification of bacteria-originating extracellular peroxidase offers a wide dynamic detection range and a low detection limit for S. oneidensis. Furthermore, this strategy was also demonstrated feasible in a real sample by selectively detecting S. oneidensis spiked in a river water sample. The proposed method provides a novel platform for detecting bacteria in the field of bioremediation.
Results
The detection mechanism of colorimetric immunomagnetic assay
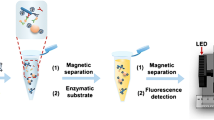
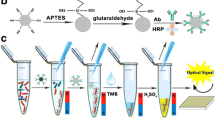
Fig. 1 depicts the principle of the colorimetric immunomagnetic assay for detecting S. oneidensis cells. S. oneidensis cells were first captured by anti-S. oneidensis antibody-conjugated MBs (antibody/MBs) through antigen-antibody reaction, yielding bacteria/MBs complexes. Then, the bacteria/MBs complexes were attracted by a magnet, separated from the sample matrix and finally transferred to a microplate for further colorimetric assay.


In a conventional HRP/TMB system (Equation (1)), HRP (a heme-containing protein) catalyzes the oxidation and reduction reaction between TMB and hydrogen peroxide, yielding a characteristic strong absorption at 650 nm. In this study, c-type cytochromes such as OmcA and MtrC, located at the outer membrane of S. oneidensis cells, are similar in structure to HRP and play a similar role (Equation (2)) in the interaction between TMB and hydrogen peroxide. Consequently, a blue color developed in the S. oneidensis/MBs/TMB/H2O2 system, which provides a colorimetric detection method for S. oneidensis cells. The optical density of color-developing reaction product was measured with a microplate reader. Based on the obtained data, a regression model was developed to describe the relationship between S. oneidensis cell number and optical density.
Characterization of the antibody/MBs-S. oneidensis interaction
As shown in Fig. 2, the isolated S. oneidensis cells were observed with an optical microscope (inset). Attachment of S. oneidensis to antibody/MBs was further confirmed with a scanning electron microscope (SEM). These results suggest that S. oneidensis cells bind to antibody/MBs due to antigen-antibody recognition. To evaluate the capture efficiency of antibody/MBs, S. oneidensis cells at a concentration of 5.0 × 106 CFU/mL were captured with antibody/MBs and then plated on plain LB agar. A capture efficiency of 75.2 ± 4.6% for S. oneidensis cells was found.
In addition, the antibody/MBs-captured S. oneidensis cells were investigated to be capable of a conventional color reaction where peroxidase activity was responsible for catalyzing the oxidization of TMB. As shown in Fig. 3, similar to S. oneidensis alone (Fig. 3a) and cytochrome c control (Fig. 3b), S. oneidensis/MBs complex (Fig. 3c) displayed a color developing ability when immersed in the substrate solution containing H2O2 and TMB. The color product was further characterized by UV–vis spectroscopy. As shown in Fig. 3, for Curve a, b and c, a strong absorption peak appeared at 650 nm, which is the characteristic absorption peak of the product from peroxidase-catalyzed H2O2/TMB reaction. Yet, there was neither blue color developed nor characteristic optical absorption for the MBs control (Fig.3d, Curve d) or the H2O2/TMB solution control (Fig. 3e, Curve e). Although previous work has showed that there are peroxidase-like activities in magnetic particles24, the MBs used in this study presented almost no peroxidase activity. This may be attributed to the special structure of polystyrene MBs which are produced by coating the silica core first with a layer of magnetic particles and then an additional surface modification layer. These results demonstrated the feasibility of the proposed colorimetric immunomagnetic assay for rapid detection of S. oneidensis cells.
Peroxidase-like activities of S. oneidensis.
UV-Vis of color development in (a) S. oneidensis, (b) Cytochrome c, (c) S. oneidensis/MBs, (d) MBs and (e) Pure water. The color developing was performed in substrate solution containing H2O2 and TMB for 30 min. Inset: photographs of the corresponding solutions.
Optimization of immunoassay conditions
To obtain the best detection sensitivity, parameters including antibody/MBs concentration, H2O2 concentration, immune-reaction time and color developing time were optimized using 5.0 × 106 CFU/mL S. oneidensis cells as analyte. Fig. 4a shows the influence of antibody/MBs concentration on detection signal. The OD value first increased and then dropped with increasing antibody/MBs amount, reaching its maximum when 40 μL of the antibody/MBs was used. Hence, 40 μL of antibody/MBs was selected for further studies. The proposed colorimetric assay relies on the bacteria intrinsic peroxidase activity whose sensitivity is affected by the concentration of H2O2. Herein, the effect of H2O2 concentration on the detection signal was evaluated with H2O2 concentration ranging from 0.1 mM to 5 mM. Similarly, the OD value, associated with the amount of target bacteria, first increased and then dropped with increasing H2O2 concentration, reaching its maximum when H2O2 concentration was 2 mM (Fig. 4b). As a result, 2 mM H2O2 was used for subsequent trials. The immunoreaction time can impact the detection sensitivity. As shown in Fig. 4c, the OD value increased gradually with increasing reaction time from 10 min to 50 min and leveled off after that, indicating that the antigen-antibody reaction reached its equilibrium after 50 min. Thus, in following experiments, reactions were allowed to continue for 50 min. For colorimetric immunoassay, the color developing time can strongly influence detection sensitivity. Fig. 4d shows that the peak value increased with color developing time and the maximum absorbance occurred at 30 min. After that, color began to fade due to the precipitation of TMB out of the solution. Consequently, 30 min was selected to be the optimal color developing time.
Specificity and reproducibility
To investigate the specificity of the proposed colorimetric immunomagnetic assay, the proposed detecting procedure was performed on both S. oneidensis and control organisms. Fig. 5 shows the absorbance for samples containing 5.0 × 106 CFU/mL S. oneidensis, Shewanellaputrefaciens (S. putrefaciens), Shewanella decolorationis (S. decolorationis), Pseudomonas agglomerans (P. agglomerans), Aeromonas hydrophila (A. hydrophila), Staphylococcus aureus (S. aureus), E. coli or B. subtilis. There were minor changes in absorbance of the control samples containing P. agglomerans, A. hydrophila, S. aureus, E. coli and B. subtilis as compared with that of the blank control (pure water). For samples containing phylogenetically close Shewanella species such as S. putrefaciens and S. decolorationis, there were only weak detection signals, which can be attributed to the partial cross-reaction between antibody/MBs and bacteria that shared the conservative epitopes with Shewanella species. However, for samples containing S. oneidensis, the response signal increased drastically.
Specificity of the colorimetric immunomagnetic assay.
Blank control (no bacterial cells), E. coli (5.0 × 106 CFU/mL), B. subtilis (5.0 × 106 CFU/mL), S. aureus (5.0 × 106 CFU/mL), P. agglomerans (5.0 × 106 CFU/mL), A. hydrophila (5.0 × 106 CFU/mL), S. decolorationis (5.0 × 106 CFU/mL), S. putrefaciens (5.0 × 106 CFU/mL), S. oneidensis (5.0 × 106 CFU/mL). Immunoreaction time, 50 min; antibody/MBs, 40 μL; color developing time, 30 min. Error bars represent S.D., n = 3.
In order to evaluate the effects of sample matrix components on the proposed assay method, we further challenged the system with river samples spiked with different microorganisms. Analyte solutions were prepared by adding S. oneidensis (5.0 × 106 CFU/mL), E. coli (5.0 × 106 CFU/mL), B. subtilis (5.0 × 106 CFU/mL), or a mixture of the three bacteria (each at 5.0 × 106 CFU/mL) into river water samples collected from a river near our institute. As indicated in Fig. 6, higher absorbance was observed for samples with S. oneidensis than for those without S. oneidensis. The present of other sample matrix components seems to exert no significant interfering effects on the response signal to target S. oneidensis. The results clearly suggested the high selectivity of the proposed immunoassay to S. oneidensis.
In addition, the assay method was tested for its reproducibility. Five repetitive measurements were conducted on S. oneidensis of 5.0 × 106 CFU/mL. The method was demonstrated to be highly reproducible with a relative standard deviation of 5.41%.
Analytical performance
Under the optimized working conditions, the sensitivity and detection range of the colorimetric immunomagnetic assay were determined with varying amounts of S. oneidensis cells. As shown in Fig. 7, the absorbance increased linearly as the amount of S. oneidensis cells increased from 5.0 × 103 to 5.0 × 106 CFU/mL. The relationship can be described as Y = 1.133−7X + 0.027 with a correlation coefficient of 0.9988 (R2), where Y is the absorbance and X is the amount of S. oneidensis cells (CFU/mL). The detection limit was estimated to be 5.0 × 103 CFU/mL according to the signal-to-noise ratio of three. The results suggested that the proposed assay method was sensitive for detecting S. oneidensis cell with a wide detection range.
Real sample analysis
In order to evaluate the analytical reliability and potential applications, the proposed colorimetric immunomagnetic assay was used to detect S. oneidensis level in real water samples. A series of samples were prepared by spiking river water samples collected from a local river with different amounts of S. oneidensis cells. Sequentially, the prepared samples were analyzed using the proposed colorimetric assay and the results are summarized in Supplementary Table S1 online. The recovery obtained is in the range between 83.32% and 102.40% with the relative standard deviation between 1.63% and 11.98%. The results indicate that the proposed assay method is feasible for S. oneidensis detection in environmental samples.
Discussion
S. oneidensis inhabits various environments, such as lake25, ocean26 water and marine sediments27. To estimate its numbers, microbiological ecology methods are currently employed based on DNA extracting and PCR testing. Unfortunately, a problem with DNA extraction is that the subsequent PCR testing is easily inhibited by the impurities that are present in the extracted DNA templates28. In spite of high specificity to target cells in environmental samples29, reverse transcription fluorescent in situ hybridization, which utilizes sequence-specific nucleic acid labeled with fluorophores as probes, involves a very time-consuming and labor-intensive sample treatment procedure, including fixation, hybridization and washing. Fortunately, the recently developed IMC technique is not only simple and time-saving, but efficient in removing inhibitory substances and heterogeneous bacteria in samples30,31. In addition, IMC is non-destructive for the target cells. This is a very important characteristic that allows subsequent detection of such bacteria as S. oneidensis with bacterial intrinsic enzyme activity.
This IMC followed by bacterial intrinsic peroxidase activity-catalyzed redox-reduction reaction can be useful to detect S. oneidensis cells in the environment. Our results showed a detection limit of 5.0 × 103 CFU/mL toward S. oneidensis. Compared to recently developed methods such as surface-enhanced Raman spectroscopy and microarray hybridization (see Supplementary Table S2 online), our method showed competitive or better performance. Compared with a previously reported electrochemical immunoassay32, the advantages of this method includes speed and minimum sample manipulation. In addition, a previous study revealed that even low-biomass water samples from a bioremediation site contained cells at a density of 3 × 106 cells/mL33, which indicates the method proposed here is good enough to detect and monitor bioremediation processes. Therefore, the present work provides an alternative approach for S. oneidensis detection.
Our method relies on IMC for concentrating and isolating target organism from sample matrix. Since selective separation is achieved by using specific antibodies-attached magnetic spheres, the efficacy of this method can be affected by the properties of antibodies and magnetic spheres. In the study, commercially available polystyrene magnetic microspheres were employed to prepare immunomagnetic beads. These superparamagnetic magnetic particles with homogeneous sizes ensured the reproducibility of the detection. Polyclonal antibodies raised against S. oneidensis cell, in the study, are directed against multiple epitopes rather than a single epitope, which results in reliable immunological recognition to the target cells. With these magnetic microspheres and polyclonal antibodies, the prepared antibodies/MBs would enable a reliable and reproducible S. oneidensis detection. Virtually, the antibodies/MBs do not react with the bacteria34 usually found in river waters and the environment such as P. agglomerans, A. hydrophila, S. aureus, E. coli and B. subtilis and only partially cross-react with phylogenetically close Shewanella species such as S. putrefaciens and S. decolorationis. In addition, the magnet strength also affects IMC efficacy. In fact, a homemade magnetic separator equipped with strong permanent magnets (8000 Gauss) was employed in the experiment and it was efficient enough to completely separate the magnetic particles from the solution within 5 min. Although previous reports have shown that sample matrix would affect IMC performance35, our study demonstrated that sample matrix (river water spiked with E. coli or B. subtilis) exerts only minor interfering effects on detection.
The color developing procedure was designed for colorimetric detection of the bead-captured S. oneidensis cells. This bacterial outer membrane peroxidase activity-based signal amplification eliminates the need for enzyme or fluorophores-conjugated antibody or PCR amplification to identify the captured cells and enables easy detection of the target cells in a micro-well plate either qualitatively with naked eyes or quantitatively by using an OD reader. Although we have only demonstrated S. oneidensis cell detection, this strategy can be applied to other metal-reducing microorganisms, such as Geobacter sulfurreducens, Shewanella decolorationis and Shewanella putrefaciens, by substituting the antibody immobilized on the magnetic bead with antibodies that are specific to the target bacteria. Coupled with an automated immunomagnetic separation system, this detection strategy can be an alternative method for high throughput analysis of metal-reducing bacteria in environmental samples.
In summary, a novel signal amplification strategy for rapid detection of S. oneidensis was successfully designed. This is the first study that coupled bacterial intrinsic peroxidase activity with IMC. The primary advantages of this method include sensitivity, speed and minimum sample manipulation, which are realized via the high-efficiency of IMC and signal amplification of bacteria extracellular peroxidase. This method offers a technique for determining functional microbes in bioremediation.
Methods
Microbial strains and culture
S. oneidensis (ATCC700550) was grown from a glycerol stock in Luria-Bertani broth as described in a previous report32. The S. oneidensis cell culture was centrifuged at 5000 g for 20 min and washed three times with 0.01 M phosphate buffer solution (PBS, pH 7.4). The collected cell precipitate was resuspended in PBS and stored at 4°C for further use. Before each measurement, cell solution was serially diluted with PBS and determined via agar plate count for cell number. E. coli and B. subtilis were used as control.
Antibody production and antibody/MBs preparation
S. oneidensis cells were processed to yield whole-cell antigens. Briefly, cell solution was centrifuged at 5000 g for 20 min and cells were resuspended to a concentration of approximately 109 cells/mL. The cells were treated with formaldehyde for 30 min at room temperature, washed three times with PBS, resuspended in PBS to a concentration of 108 cells/mL and stored at 4°C. Using the prepared whole-cell antigen, a female New Zealand white rabbit weighing 2 kg was immunized intramuscularly for a total of four times at intervals of ten days for producing polyclonal antibody against S. oneidensis. The obtained antisera were purified with ammonium sulfate precipitation and characterized by enzyme-linked immunosorbent assay according to a previous protocol36. The purified anti-S. oneidensis antibodies were stored at −20°C.
To prepare antibody/MBs, anti-S. oneidensis antibodies were attached to carboxylate- functionalized MBs (MBs-COOH) (Affimag PSC, 1–2 μm in diameter, Tianjin BaseLine ChromTech Research Centre, Tianjin, China) using an N-ethyl-N′-(3-(dimethylamino) propyl) carbodiimide and N-hydroxysuccinimide (EDC/NHS) amidization protocol (Fig. 1a) according to a previous report37. Briefly, MBs-COOH was first washed three times with 2-(N-morpholino) ethanesulfonic acid (MES) buffer (0.05 M, pH 5.5), then mixed with 1 mL of MES buffer (containing 20 mg of EDC and 20 mg of NHS) and activated for 30 min. The MES buffer was later removed by attracting the beads to the wall of the vial with a magnet. The MBs were washed with MES buffer for three times to remove excess EDC and NHS. Subsequently, the resulting functionalized-beads were dispersed in 0.3 mL of PBS and sonicated for 5 min to obtain a homogeneous dispersion. Then 0.3 mL of anti-S. oneidensis antibody at 1.4 mg/mL was added to the dispersion. After mixed thoroughly, the mixture was stirred for 3 h at room temperature. The reaction mixture was placed on a magnet for 5 min and the pellet was collected after removal of supernatant and then washed three times with PBS. The resulting antibody/MBs complex was redispersed in 1.0 mL of PBS containing 1% BSA and stored at 4°C.
Immunomagnetic capture of S. oneidensis
Serial dilutions of bacterial sample were made for IMC. To begin with, 500 μL of diluted sample was added to a 1.5 mL centrifuge tube and then 40 μL of antibody/MBs was added and mixed with the bacterial sample in the 1.5-mL tube. The tube was rotated for 1 h, allowing the target bacteria to attach to the antibody/MBs. Bacteria/beads complexes were then magnetically separated by placing the tube on a magnet. After removal of supernatant, the collected complexes were washed for three times, resuspended in approximately 100 μL of pure water and transferred to a microplate.
Colorimetric quantification of S. oneidensis
Colorimetric assays were performed on a 96-well plate as illustrated in Fig. 1b. In short, 100 μL of bacteria/MBs complex was mixed with equal volume of substrate solution (containing 100 mM citric acid, 200 mM Na2HPO4, 0.32 mM TMB and 2 mM H2O2, pH 4.3). The plate was incubated for 30 min at room temperature for color developing. After color development, the reaction was stopped with 2 M sulfuric acid (50 μL/well) and then the solution in the wells was transferred to a new plate with the aid of a magnetic separator. Optical density (OD) was measured at 450 nm using a microplate reader (iMark, Bio-rad).
References
Jiang, J. & Kappler, A. Kinetics of microbial and chemical reduction of humic substances: implications for electron shuttling. Environ. Sci. Technol. 42, 3563–3569 (2008).
Hau, H. H. & Gralnick, J. A. Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 61, 237–258 (2007).
Shi, L., Squier, T. C., Zachara, J. M. & Fredrickson, J. K. Respiration of metal (hydr) oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes. Mol. Microbiol. 65, 12–20 (2007).
Beliaev, A. S. et al. Global transcriptome analysis of Shewanella oneidensis MR-1 exposed to different terminal electron acceptors. J. Bacteriol. 187, 7138–7145 (2005).
Heidelberg, J. F. et al. Genome sequence of the dissimilatory metal ion–reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20, 1118–1123 (2002).
Marshall, M. J. et al. c-Type cytochrome-dependent formation of U (IV) nanoparticles by Shewanella oneidensis. PLoS Biol. 4, e268 (2006).
Lee, J. H. et al. Manganese sulfide formation via concomitant microbial manganese oxide and thiosulfate reduction. Environ. Microbiol. 13, 3275–3288 (2011).
Shi, L. et al. Isolation of a high-affinity functional protein complex between OmcA and MtrC: two outer membrane decaheme c-type cytochromes of Shewanella oneidensis MR-1. J. Bacteriol. 188, 4705–4714 (2006).
Xiao, X. et al. Biodecolorization of Naphthol Green B dye by Shewanella oneidensis MR-1 under anaerobic conditions. Bioresour. Technol. 110, 86–90 (2012).
Gralnick, J. A., Vali, H., Lies, D. P. & Newman, D. K. Extracellular respiration of dimethyl sulfoxide by Shewanella oneidensis strain MR-1. Proc. Natl. Acad. Sci. U S A 103, 4669–4674 (2006).
Kobayashi, S., Natori, K., Takeda, N. & Sakae, K. Immunomagnetic capture rt-PCR for detection of norovirus from foods implicated in a foodborne outbreak. Microbiol. Immunol. 48, 201 (2004).
Gatto-Menking, D. L. et al. Sensitive detection of biotoxoids and bacterial spores using an immunomagnetic electrocheminescence sensor. Biosens. Bioelectron. 10, 501–507 (1995).
Sousa, S. et al. Development of a Fluorescent Based Immunosensor for the Serodiagnosis of Canine Leishmaniasis Combining Immunomagnetic Separation and Flow Cytometry. PLoS Negl. Trop. Dis. 7, e2371 (2013).
Jeníková, G., Pazlarová, J. & Demnerová, K. Detection of Salmonella in food samples by the combination of immunomagnetic separation and PCR assay. Int. Microbiol. 3, 225–229 (2010).
Cudjoe, K. S. et al. Detection of Clostridium perfringens type A enterotoxin in faecal and food samples using immunomagnetic separation (IMS)-ELISA. Int. J. Food Microbiol. 12, 313–321 (1991).
Xianyu, Y. et al. Enzymatic Assay for Cu (II) with Horseradish Peroxidase and Its Application in Colorimetric Logic Gate. Anal. Chem. 85, 7029–7032 (2013).
Gao, Z., Xu, M., Hou, L., Chen, G. & Tang, D. Irregular-shaped platinum nanoparticles as peroxidase mimics for highly efficient colorimetric immunoassay. Anal. Chim. Acta 766, 79–86 (2013).
Le Goff, G. C., Blum, L. J. & Marquette, C. A. Enhanced colorimetric detection on porous microarrays using in situ substrate production. Anal. Chem. 83, 3610–3615 (2011).
Veitch, N. C. Horseradish peroxidase: a modern view of a classic enzyme. Phytochemistry 65, 249–259 (2004).
Dawson, J. H. Probing structure-function relations in heme-containing oxygenases and peroxidases. Science 240, 433–439 (1988).
Vargas, C., McEwan, A. & Downie, J. Detection of c-type cytochromes using enhanced chemiluminescence. Anal. Biochem. 209, 323–326 (1993).
Goodhew, C., Brown, K. & Pettigrew, G. Haem staining in gels, a useful tool in the study of bacterial c-type cytochromes. Biochimica et Biophysica Acta (BBA)-Bioenergetics 852, 288–294 (1986).
Myers, C. R. & Myers, J. M. Localization of cytochromes to the outer membrane of anaerobically grown Shewanella putrefaciens MR-1. J. Bacteriol. 174, 3429–3438 (1992).
Wei, H. & Wang, E. Fe3O4 magnetic nanoparticles as peroxidase mimetics and their applications in H2O2 and glucose detection. Anal. Chem. 80, 2250–2254 (2008).
Myers, C. & Nealson, K. H. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240, 1319–1321 (1988).
Fredrickson, J. K. et al. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6, 592–603 (2008).
Abboud, R. et al. Low-temperature growth of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 71, 811–816 (2005).
Watson, R. & Blackwell, B. Purification and characterization of a common soil component which inhibits the polymerase chain reaction. Can. J. Microbiol. 46, 633–642 (2000).
Nielsen, J. L., Juretschko, S., Wagner, M. & Nielsen, P. H. Abundance and phylogenetic affiliation of iron reducers in activated sludge as assessed by fluorescence in situ hybridization and microautoradiography. Appl. Environ. Microbiol. 68, 4629–4636 (2002).
Christensen, B., Torsvik, T. & Lien, T. Immunomagnetically captured thermophilic sulfate-reducing bacteria from North Sea oil field waters. Appl. Environ. Microbiol. 58, 1244–1248 (1992).
Mullins, P. H., Gürtler, H. & Wellington, E. M. Selective recovery of Streptosporangium fragile from soil by indirect immunomagnetic capture. Microbiology 141, 2149–2156 (1995).
Wen, J., Zhou, S. & Yuan, Y. Graphene oxide as nanogold carrier for ultrasensitive electrochemical immunoassay of Shewanella oneidensis with silver enhancement strategy. Biosens. Bioelectron. 52, 44–49 (2014).
Chakraborty, R. et al. Use of immunomagnetic separation for the detection of Desulfovibrio vulgaris from environmental samples. J. Microbiol. Methods 86, 204–209 (2011).
Niewolak, S. & Opieka, A. Potentially pathogenic microorganisms in water and bottom sediments in the Czarna Hańcza River. Pol. J. Environ. Stud. 9, 183–194 (2000).
Monceyron, C. & Grinde, B. Detection of hepatitis A virus in clinical and environmental samples by immunomagnetic separation and PCR. J. Virol. Methods 46, 157–166 (1994).
Wen, J. et al. Preparation and characterization of egg yolk immunoglobulin Y specific to influenza B virus. Antivir. Res. 93, 154–159 (2012).
Guven, B., Basaran-Akgul, N., Temur, E., Tamer, U. & Boyacı, İ. H. SERS-based sandwich immunoassay using antibody coated magnetic nanoparticles for Escherichia coli enumeration. Analyst 136, 740–748 (2011).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (41222006) and the Science and Technology Planning Project of Guangdong Province, China (2012B091100242).
Author information
Authors and Affiliations
Contributions
J.W. and S.Z. designed the experiments. J.W. prepared the anti-S. oneidensis antibody. J.W. and J.C. carried out the fabrication of immunomagnetic bead and assay of S. oneidensis. J.C. assisted in the material characterizations. J.W. analyzed the data and wrote the paper, S.Z. revised the manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images in this article are included in the article's Creative Commons license, unless indicated otherwise in the image credit; if the image is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the image. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Wen, J., Zhou, S. & Chen, J. Colorimetric detection of Shewanella oneidensis based on immunomagnetic capture and bacterial intrinsic peroxidase activity. Sci Rep 4, 5191 (2014). https://doi.org/10.1038/srep05191
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05191
- Springer Nature Limited
This article is cited by
-
Mixed Consortium of Salt-Tolerant Phosphate Solubilizing Bacteria Improves Maize (Zea mays) Plant Growth and Soil Health Under Saline Conditions
Molecular Biotechnology (2024)
-
Success of microbial genes based transgenic crops: Bt and beyond Bt
Molecular Biology Reports (2021)
-
Simultaneous and quantitative monitoring of co-cultured Pseudomonas aeruginosa and Staphylococcus aureus with antibiotics on a diffusometric platform
Scientific Reports (2017)