Abstract
Behect’s disease is a chronic vasculitis characterized by complex multi-organ immune aberrations. However, a comprehensive understanding of the gene-regulatory profile of peripheral autoimmunity and the diverse immune responses across distinct cell types in Behcet’s disease (BD) is still lacking. Here, we present a multi-omic single-cell study of 424,817 cells in BD patients and non-BD individuals. This study maps chromatin accessibility and gene expression in the same biological samples, unraveling vast cellular heterogeneity. We identify widespread cell-type-specific, disease-associated active and pro-inflammatory immunity in both transcript and epigenomic aspects. Notably, integrative multi-omic analysis reveals putative TF regulators that might contribute to chromatin accessibility and gene expression in BD. Moreover, we predicted gene-regulatory networks within nominated TF activators, including AP-1, NF-kB, and ETS transcript factor families, which may regulate cellular interaction and govern inflammation. Our study illustrates the epigenetic and transcriptional landscape in BD peripheral blood and expands understanding of potential epigenomic immunopathology in this disease.
Similar content being viewed by others
Introduction
Behcet’s disease (BD) is a systemic inflammatory disorder of unknown etiology affecting blood vessels1. It commonly manifests as inflammation of the intra-ocular structure, and recurrent oral/genital ulceration. BD causes morbidity and mortality, particularly in Asians2. Current treatments for BD are aggressive systemic and topical glucocorticoids, with or without immunosuppressive agents. However, these can lead to undesirable side effects, such as hyperglycemia, osteoporosis, and obesity, related to prolonged drug usage3. Thus, there is a need to develop new targeted therapies for BD. Painful skin lesions, recurrent ulceration, and blindness result from the combination of genetic susceptibility, environmental triggers, and dysregulated immune responses involving T helper 17 (Th17) cells, monocytes, skin CD8+ T cells and pro-inflammatory cytokines4,5,6,7,8. However, to date, the knowledge of genetic contributors and pathogenic cells to BD is still limited.
Over recent decades, progress in single-cell sequencing technologies has enabled profiling of the genetic transcriptomics of peripheral blood mononuclear cells (PBMCs) and skin tissues from BD patients4,8. Although previous studies have examined the single-cell gene expression of BD, heterogeneity in the single-cell epigenomics of PBMCs has not been profiled. Integrating the single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) and single-cell RNA-seq (scRNA-seq) enables the identification of the potential disease-associated regulatory program9,10,11,12,13,14,15,16.
In this study, we aimed to map the cellular landscape of PBMCs in BD patients, with the goal of dissecting disease heterogeneity among patients and identify the underlying cellular and molecular events. To accomplish this, we simultaneously generated both transcriptomic and epigenomic data in BD patients to identify the gene regulatory network. Our analyses uncovered widespread gene expression and chromatin accessibility changes in both BD patients and unaffected controls, including hyperactivation signatures in T cells and monocytes. Notably, we also nominated potential TF activators of chromatin accessibility and gene expression in BD. Moreover, our multi-omics analysis was effective at predicting disease regulatory networks, highlighting the predicted involvement of AP-1, NF-kB, and ETS transcript factor families in BD pathophysiology. Overall, our study provides insights into the understanding of the peripheral immune pathogenesis of BD.
Results
High-resolution single-cell epigenomic and transcriptional peripheral immune cell-type mapping of Behcet disease patients
We performed droplet-based scRNA-seq and scATAC-seq (10X Genomics) to map the immune landscape of PBMCs from 22 BD patients in scATAC-seq dataset, 23 BD patients in scRNA-seq dataset and 8 non-BD individuals in both dataset (Fig. 1a, Supplementary Data 1). After stringent quality control filtration, a total of 152,704 cells of the scATAC-seq dataset and 272,113 cells of the scRNA-seq dataset were retained for downstream analysis, with an average of 8810 unique nuclear fragments and an average of 14.5 in TSS enrichment for scATAC-seq-profiled cells, and an average of 2042.9 UMIs for scRNA-seq-profiled cells (Fig. 1b, Supplementary Fig. 1a–h). We did not detect any potential batch effects in our datasets (Supplementary Fig. 1d–g). Therefore, no batch correction method was applied in our further analysis. The quality control thresholds of the scATAC-seq and scRNA-seq are described in the Methods. The scATAC-seq dataset, aligned using dimension reduction and graph-based clustering, yielded discrete cell clusters, primarily representing T (CD4/CD8) cells, monocytes, dendritic cells (DCs), T cells, natural killer (NK) cells, B lymphocytes, and Hematopoietic stem and progenitor cell (HSPC) (Fig. 1c, Supplementary Data 2). With scATAC-seq, we first manually annotated based on chromatin accessibility at the promoter regions of key lineage markers for six major immune cell lineages of the PBMCs by comparing differentially accessible chromatin regions (DARs): T cells (84,202 cells; 1–4, clusters 7–9); monocytes (29,624 cells; clusters 16–19); DCs (1,861 cells; clusters 11, 15); NKs (22,738 cells; clusters 5, 6); BCs (12,610 cells; clusters 10, 12, 13) and HSPC (1669 cells; 14 clusters)17 (Fig. 1b–d). Open chromatin at known major immune cell lineages specific genes validated our analysis. T cells had high accessibility at cis-elements neighboring CD8A17 and IL7R8,17 (Fig. 1d, Supplementary Fig. 2a, c). NK cells had higher accessibility at GNLY18. Monocytes showed higher accessibility within S100A817 (Fig. 1d, Supplementary Fig. 2a, c). We found that HSPC had higher accessibility at GATA217 (Fig. 1d, Supplementary Fig. 2a, c). B cells had high accessibility at MS4A119 (Fig. 1d, Supplementary Fig. 2a, c). DCs showed higher accessibility within HLA-DQA120 (Fig. 1d, Supplementary Fig. 2a, c). We also used chromVAR21 to compute transcription factor (TF) motif deviation in single cells by estimating the enrichment of TF binding motifs in open chromatin regions and examined the enrichment of TF motifs in immune cell types concerning diagnosis and identified NFKB1 TF motifs with increased enrichment with BD patients in monocytes (cluster 18, Fig. 1e, Supplementary Fig. 2b, d). For example, LEF1 was active in naive T cell lineage and myeloid cells shared the activity of SPI1 factor motif but demonstrated unique activity of the GATA2 factor in HSPC17,22 (Fig. 1e, Supplementary Fig. 2b, d).
a Schematic highlighting design of single-cell multi-omics profiling of PBMCs from BD patients (scATAC: n = 22; scRNA: n = 23) and non-Behect’s disease patients (n = 8) in this study. Cells were then split and profiled using scATAC-seq and scRNA-seq for each condition. b UMAP projections of complete scATAC-seq and scRNA-seq datasets between non-BD and BD patients with cells colored by unsupervised cell clusters. c Total number of six main immune cell types profiled passing quality control filtering for scATAC and scRNA-seq. d Aggregate accessibility profiles for scATAC-seq six main cell type at canonical cell-type marker genes. e UMAP projection of scATAC-seq peripheral blood profiles colored by chromVAR TF motif bias-corrected deviations for the indicated factors. f Row-normalized single-cell gene expression heatmap of six main immune cell-type marker genes. All data are aligned and annotated to hg38 reference genome.
Likewise, we detected similar cell types and annotated them based on known marker genes using scRNA-seq: T cells (152,842 cells; clusters 1–3, 6, 7, 10, 11, 25); monocytes (53573 cells; clusters 5, 9, 12, 19, 21, 26); DCs (3000 cells; clusters 13, 14, 20); NKs (40875 cells; clusters 15, 17, 22); BCs (20971 cells; clusters 4, 8, 16, 18) and HSPC (852 cells; clusters 23, 24) (Fig. 1b, c, f). As expected, many differentially expressed genes (DEGs) in the six major immune cell types agreed with previous literature and our scATAC-seq dataset, such as IL7R for T cells, GNLY for NKs, S100A8 for monocytes, HLA-DQA1 for DC, MS4A1 for BC and GATA2 for HSPC17,23 (Fig. 1f). Altogether, we mapped the high-resolution epigenomic and transcriptional peripheral immune landscape in BD at a single-cell resolution.
Multi-omic characterization of T cell heterogeneity in BD blood
The high sensitivity of scATAC-seq and scRNA-seq allowed us to further map these major immune cell types into subtypes22,23. As in previous studies, we compared the DARs in the scATAC-seq dataset and DEGs in the scRNA-seq dataset to map T cell subsets17,22. We also used gene activity scores (GAS) for cell type identification in the scATAC-seq dataset due to the sparsity of single-cell cis-element information24. Specifically, we identified T cell subsets into 15 subgroups according to the expression of cell type marker GAS and genes (Fig. 2a, Supplementary Fig. 3a, Supplementary Fig. 4a, b), including CD4 cytotoxic T cells (CD4 CTLs; CD4+GZMK+), CD4 naïve T cells (CD4 Naïve; CD4+SELL++TCF7++), CD4 central memory T cells (CD4 TCMs; CD4+SELL+TCF7+), CD4 T follicular helper (CD4 TFH; CD4+ICOS+CXCR5+), CD4 T helper 1 cells (CD4 Th1; CD4+CXCR3+), CD4 T helper 2 cells (CD4 Th2; CD4+CCR4+), CD4 T helper 17 cells (CD4 Th17; CD4+CCR6+ RORC+), CD4 regulatory T cells (CD4 Treg; CD4+FOXP3+), CD8 mucosal-associated invariant T cells (CD8 MAIT; CD8+CCR6+ RORC+), CD8 naïve T cells (CD8 Naïve; CD8+SELL++TCF7++), CD8 central memory T cells (CD8 TCMs; CD8+SELL+TCF7+), CD8 effector memory T cells (CD8 TEMs; CD8+IFNG+GZMK+), CD8 exhausted T cells (CD8 TEXs; CD8+PDCD1+), CD8 regulatory T cells (CD8 Treg; CD8+KIR3DL2 +KIR2DL2 +) and double negative T cells (DNT, CD3+CD4+CD8−) (Fig. 2b, Supplementary Fig. 3a, Supplementary Fig. 4a, S4b)17,22,23,25,26. CD8 Tregs are rare cell types in PBMCs. Recently, it has been found that they undergo clonally expansion in some autoimmune disorders27. To further identify the regulatory TF in CD8 Treg, we compared the differentially accessible chromatin regions (DARs) and analyzed their enrichment of key TFs. Compared to CD8 naïve T cells, TF Eomesodermin (EOMES) were the top TFs enriched in CD8 Tregs26 (Supplementary Fig. 3b).
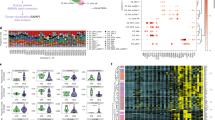
a UMAP projections of T cell subsets of scATAC-seq dataset. b Heatmap visualization of log-normalized gene activity scores of subpopulation-specific genes in T cell subsets. Selected genes are indicated. c Differences in the proportions of CD4Tfh, CD4Th2, CD8 Navie, CD8 Treg and DNT cells among non-BD (n = 8) and BD groups (n = 22). The p values were calculated using two-sided Wilcoxon rank-sum test. The horizontal lines denote median. d Comparison of aggregate TF footprints for JUNB and FOSL2 in CD4Treg cells from non-BD and BD. The p value of the TF footprint was compared by two-sided Wilcoxon rank-sum test. e UpSet plot showing the integrated comparative analysis of upregulated DEGs in T cells between non-BD and BD groups. Upregulated DEGs: upregulated in BD, downregulated in non-BD. The count shows the number of DEGs. f Representative GO terms and KEGG pathways enriched in the nearest genes of upregulated DARs of T cell subsets in the BD/non-BD comparison group. The p values were calculated by hypergeometric test. All data are aligned and annotated to hg38 reference genome.
There has been a relative dearth of deep profiling of T cell subsets from BD patients. To address this, we first examined how BD impacted the composition of peripheral T cells in the scATAC-seq dataset and scRNA-seq dataset (Fig. 2c, Supplementary Fig. 5a, b). We saw similar trends in T cell subsets composition between the multi-omics dataset, including decreased CD4 TFH, and CD4 TH2, as well as increased in CD8 Naïve, CD8 Treg, and DNT in patients with BD (Fig. 2c). DNT cells have been reported to be increased in BD patients, which is consistent with our finding28. TFs tightly control cell fate in immune cells and have been implicated in the pathogenesis of autoimmune diseases, such as BATF in arthritis and PU.1 in systemic lupus erythematosus29,30,31,32,33. We performed TF footprint analysis to further clarify cell-type-specific trans-regulatory elements in BD (Fig. 2d, Supplementary Data 3). We examined the regulatory role of the Treg TF AP-1 family, such as JUNB and FOSL2. JUNB and Fos-related antigen 2 (FOSL2) motif variabilities in our scATAC-seq were increased and upregulated in BD (Fig. 2d). This result suggests that the AP-1 family TFs might have higher accessibility in BD, providing insight into how the AP-1 family contributes to BD pathophysiology34,35. We therefore identified the DEGs and DARs between BD and non-BDs subjects in the scRNA-seq and scATAC-seq dataset (Fig. 2e, Supplementary Fig. 5c, d, Supplementary Fig. 6a, Supplementary Data 4). We also used muscat36 to validate our DEGs result (Supplementary Fig. 7a, Supplementary Data 5). We next applied CHIPseeker37 to find the nearest genes of the DARs and used the DARs to overlap with DEGs (Supplementary Fig. 6b). We observed that 14 genes were both upregulated in T cell subsets, including DUSP238, JUNB39, IRF140, and DDIT439, suggesting T cells might be in proinflammatory state in BD patients. CD541, CD6942, NFKBIA43 were up-regulated in all the CD4 T cell subsets, suggesting CD4 T cell subsets were both highly activated in BD42 (Supplementary Fig. 5c). In contrast, CD744, IL2RG45, IFITM146, IFITM246 were up-regulated in all the CD8 T cell subsets (Supplementary Fig. 5d).
A Gene Ontology (GO) analysis of the DEGs showed that the cellular response to cytokine stimulus was enriched in most T cell lineages, suggesting the immune-activated states in BD patients (Fig. 2f, Supplementary Data 6). In T cells, the GO analysis showed that CD4 CTLs, CD8 Tex, and CD8 Treg were all enriched in the TCR signaling, and costimulation by the CD28 family pathway (Fig. 2f). The IL-4 signaling pathway was among the top enriched pathways in T helper cells, suggesting B cell-induced IL-4 mediated hyper-interplay with T cells in BD47. Th1 cells and Th17 cells were reported to take part in the BD pathogenesis5,48,49 (Fig. 2f). We observed that Th1 cells were involved in the NF-kB signaling, TNF signaling pathway. Th17 cells were involved in the response to the interferon-gamma pathway with interferon signaling-related genes upregulated (ISG15, IFITM1, IFITM2) (Supplementary Fig. 7b). We also shed light on the significant number of rare T cell types, including MAIT and DNT cells. DNT cells were involved in the TCR signaling, T cell activation pathway, and HIF1 TF pathway (Fig. 2f). MAIT cells from BD patients showed increased enrichment for pathways associated with TNF signaling, NF-kB signaling, VEGFA-VEGFR2 signaling, and cellular response to cytokine stimulus (Fig. 2f). MAIT cells have been reported to contribute to the pathogenesis of other forms of vasculitis8,50. Overall, these analyses provide transcriptional and epigenomic evidence that highly activated peripheral T cells may be associated with BD, consistent with previous single-cell reports8.
Multi-omic characterization of NK and B cell heterogeneity in BD blood
Despite studies that show NK cells are involved in the dysregulated immune response in BD, the pathogenesis of NK cell subsets in BD still needs to be explored further. As demonstrated previously, peripheral NK cells were identified into three subsets based on the GAS and mark genes: early NKs (NK1; NCAM1highFCGR3AlowB3GAT1low), intermediate NKs (NK2, NCAM1highFCGR3AlowB3GAT1low) and late NKs (NK3, NCAM1highFCGR3AlowB3GAT1low)17 (Fig. 3a, b, Supplementary Fig. 8a, b). While we did not notice a significant change in the percentage of total NK cells (Supplementary Fig. 8c, d), we noted significant transcriptional and epigenomic reconfiguration in all the NK subsets driven by up-regulated of several canonical NK cell activation genes (Supplementary Fig. 8e–g, Supplementary data 7), including CD6951 as well as interferon-stimulated genes (ISGs) IFITM2, IRF1, and ISG20 (Fig. 3c). NK1 also expressed higher cytotoxic effector molecule-encoding genes GZMB, GZMM, and GZMH (Fig. 3c).
a UMAP projections of NK cell subsets of scATAC-seq dataset. b UMAP projection colored by gene activity scores for the annotated lineage-defining genes of scATAC-seq dataset. The minimum and maximum gene activity scores are shown in each panel. c UpSet plot showing the integrated comparative analysis of upregulated DEGs in NK cells between non-BD and BD groups. Upregulated DEGs: upregulated in BD, downregulated in non-BD. The count showing the number of DEGs. d Representative GO terms and KEGG pathways enriched in the upregulated DEGs of NK cell subsets in the BD/non-BD comparison group. The p values were calculated by hypergeometric test. e UMAP projections of B cell subsets of scATAC-seq dataset. f UMAP projection colored by gene activity scores for the annotated lineage-defining genes of scATAC-seq dataset. The minimum and maximum gene activity scores are shown in each panel. g UpSet plot showing the integrated comparative analysis of upregulated DEGs in NK cells between non-BD and BD groups. h Representative GO terms and KEGG pathways enriched in the upregulated DEGs of B cell subsets in the BD/non-BD comparison group. The p values were calculated by hypergeometric test. All data are aligned and annotated to hg38 reference genome.
To further elucidate the pathogenic pathways, we next examined DEGs of NK cell subsets for GO analysis (Fig. 3d). The top signaling pathways of NK1 in BD included the cytokine signaling pathway, regulation of T cell activation, IL-18 signaling pathway, CXCR4 pathway, costimulation by the CD28 family, and VEGFA-VEGFR2 signaling pathway (Fig. 3d). The pathway enrichment result implicates that NK1 cells in BD might be involved in the pro-angiogenic process. T-cell activation, CD28 co-stimulation, T-cell receptor signaling pathway, IL-6 signaling pathway, transcription regulation by RUNX1, and CD8 TCR downstream pathway were up-regulated in NK2, while regulation of the viral process, regulation of cell-cell adhesion, Th17 cell differentiation, cytokine-mediated signaling pathway, TNF signaling pathway, and FCGR3A-mediated phagocytosis were upregulated in NK3 (Fig. 3d). Both NK cell subsets in BD were enriched in the T cell activate and cytokine-related pathway with increased cytotoxic activity and up-regulated TNF pathway. The differences between non-BD suggest that NK cells in BD were in proinflammatory state52. The above findings suggested that NK-mediated immunity was activated and pro-inflammatory in BD patients.
We next interrogated changes in the B cells of BD patients, because B cells may play an eminent role in the pathogenesis53 (Fig. 3e, Supplementary Fig. 9a). Embedding of B cells alone was manually identified into 4 subsets: naïve B cells (NB; MS4A1+TCL1A+), memory B cells (MB, MS4A1+TCL1A+), double negative B ells (DN2B; CD19+ITGAX+CR2−), and plasma B cells (PB, CD38+)17 (Fig. 3f, Supplementary Fig. 9b). As observed in the transcriptional data, all the B cell subsets in BD highly expressed interferon-stimulated genes IRF140, IFITM246, and antigen processing and presentation-related molecules HLA-DQB154, and the cytokine IL2RG55, as well as AP-1 family genes JUNB56, MAPK signaling, and NF-kB signaling related genes DUSP138, NFKBIA43 (Fig. 3g, Supplementary Fig. 9c–f, Supplementary Data 7). MB expressed higher RELA, BACH1, SPIB, JUN and JUND. DN2B showed higher expression on ISG20, TNFRSF13C and BTG1 (Fig. 3g, Supplementary Data 7). By overlapping the DARs and DEGs, the activation marker CD69 was highly expressed in DN2B57, and the antigen-presenting and activation marker CD83 was highly expressed in MB and NB, suggesting a highly activated state of B cells in BD patients58 (Supplementary Fig. 9e, f).
Next, we performed GO analysis on upregulated genes in BD patients (Fig. 3h). The GO results showed that DN2B in BD patients were enriched in positive regulation of T cell activation, regulation of IL-2 production, activator protein 1 (AP-1) pathway, PD-1 signaling, and costimulation by the CD28 family59, while PB were enriched more in IL-1 signaling FCERI mediated NF-kB activation, Dectin-1 signaling and cellular response to hypoxia (Fig. 3h). Moreover, GO analysis suggested that MB from BD patients were enriched in the AP-1 pathway, IL-18 signaling, MAPK signaling, signaling by interleukins, and response to the interferon-beta pathway (Fig. 3h). As for NB, Antigen processing and presentation, major histocompatibility complex (MHC) protein complex assembly, regulation of T cell activation, response to the virus, and TCR signaling pathway were up-regulated (Fig. 3h). Taken together, these findings indicated common and distinct functions of B cell subsets among non-BDs and BD patients and suggested an enhanced humoral immunity is developed in BD patients.
Multi-omic characterization of myeloid cells heterogeneity in BD blood
To increase cell-level resolution and dissect myeloid cells, we first clustered DCs and identified 3 DC subtypes: type 1 classical dendritic cells (cDC1; HLA+CLEC9A+), type 2 classical dendritic cells (cDC2; HLA+ITGAX+), and plasmacytoid DCs (pDCs; HLA+IRF8+CLEC4C+)17,20 (Fig. 4a, Supplementary Fig. 10a–c). We first examined how BD impacted the composition of peripheral DCs in two datasets (Supplementary Fig. 10D, S10E). We saw cDC2 significantly increased in the scRNA-seq dataset (Supplementary Fig. 10d). Next, we analyzed the difference between BD and non-BD in transcription and epigenomic profiling (Fig. 4b–e, Supplementary Fig. 10f, g, Supplementary Data 5). Notably, cDC2 in BD patients showed significantly higher chromatin accessibility on the IL-1B locus and CD83 locus, suggesting increased cytokine secretion and enhanced antigen presentation in DCs (Fig. 4b, c). To further access the TF that drove differences in cDC2, we conducted a TF footprint analysis compared to BD and non-BD in cDCs (Fig. 4d, Supplementary Data 1). The results showed that NFKB family TF (RELB, NFKB1) showed high accessibility in BD patients.
a UMAP projections of DC cell subsets of scATAC-seq dataset. b Genome browser tracks showing single-cell chromatin accessibility of cDC2 cells in the IL1B loci. c Genome browser tracks show single-cell chromatin accessibility of cDC2 cells in the CD83 loci. d Comparison of aggregate TF footprints for RELB and NFKB1 in cDC2 cells from non-BD and BD. The p value of the TF footprint was compared by two-sided Wilcoxon rank-sum test. e UpSet plot showing the integrated comparative analysis of upregulated DEGs in DC subsets between non-BD and BD groups. Upregulated DEGs: upregulated in BD, downregulated in non-BD. The count showing the number of DEGs. f Representative GO terms and KEGG pathways enriched in the upregulated DEGs of DC cell subsets in the BD/non-BD comparison group. The p values were calculated by hypergeometric test. g UMAP projections of monocyte subsets of scATAC-seq dataset. h UMAP projection colored by gene activity scores for the annotated lineage-defining genes of scATAC-seq dataset. The minimum and maximum gene activity scores are shown in each panel. i Dot plots of gene expression of the marker genes of monocyte subsets in scRNA-seq dataset. The dot size indicates the percentage of the cells in each cluster in which the gene of interest. The standardized gene expression level was indicated by color intensity. j Differences in the proportions of monocyte subsets among non-BD (n = 8) and BD groups (n = 22). The p values were calculated using two-sided Wilcoxon rank-sum test. The horizontal lines denote median. k Representative GO terms and KEGG pathways enriched in the marker genes of the ActCM and ActNCM. The p values were calculated by hypergeometric test. l TF footprints with motifs in the indicated scATAC-seq monocyte subsets. The p value of the TF footprint was compared by one-way ANOVA. All data are aligned and annotated to hg38 reference genome.
Consistent with our epigenomic dataset, the cytokines and chemokines CCL3L1, CCL3, CCL4L2, IL1B, and ISGs IFITM3, IRF1, IFITM1, ISG20, as well as activation marker CD83 and NF-kB signaling related genes (RELB) were up-regulated in cDC2 (Fig. 4e). Interestingly, HIF1A was also up-regulated in cDC2, suggesting epigenetic reprogramming and metabolism changes in cDC260 (Fig. 4e). Furthermore, cDC2 with high expression of NFKBIA, JUNB, and MAP3K8 showed GO enrichment in TNF signaling, regulation of T cell activation, interferon signaling, IL-18 signaling, and AP-1 pathway based on GO analysis (Fig. 4e, f)61,62. The GO analysis showed that the DEGs of cDC1 in BD patients were enriched in the Dectin-1 signaling, gluconeogenesis, interleukin-1 signaling, MAPK1/MAPK3 signaling, and signaling by interleukins pathways. While the DEGs of pDC were enriched in TCR signaling, glycolysis, and gluconeogenesis, Fc epsilon receptor signaling (Fig. 4e, f). Our analysis of DCs showed that DC subsets in BD patients varied greatly compared to those in non-BD, suggesting enhanced DC function and further directed T cells differentiation in BD patients63.
It has been reported that monocytes play vital roles in BD4, however, the epigenomic changes of monocytes in BD have not been well characterized. To address this, we sub-grouped monocytes into 4 subclusters, which consisted of classical monocytes based on the GAS and DEGs between each cluster. The classical monocytes (CM) were identified based on the expression of CD14+FCGR3A-, while the non-classical monocytes (NCM) expressed more FCGR3A (Fig. 4g–i, Supplementary Fig. 11a, b, Supplementary Data 8). Notably, cell subsets proportions were altered in BD patients. BD patients had a large proportion of monocytes that highly expressed cytokines and chemokines IL1B, CXCL8, CCL4L2, and CCL3 with CD14+S100A12+FCGR3A-, which we identified as activated classical monocytes (ActCM)43. We also identified previously reported C1Q+ monocytes with a high expression on ISG IFITM2, and pro-inflammatory genes TNF, IL1B, CXCL8, CCL4L2, CCL3, as well as FCGR3A, RHOC, which we identified as activated non-classical monocytes (ActNCM)4,43,64 (Fig. 4g–i). Comparing the relative cell proportions in BD and non-BD groups, we observed a similar trend to what was previously reported in single-cell profiling4, a significant expansion in ActCM and ActNCM, and a notable decrease in NCM in the scATAC-seq dataset, while the proportion of CM was comparable between BD and non-BD groups (Fig. 4j, Supplementary Fig. 11c, Supplementary Data 2). Although we did not notice significant changes in ActNCM in the scRNA-seq dataset, we still observed a significant increase in ActCM (Supplementary Fig. 11c).
Furthermore, within these subpopulations, we conducted GO analysis (Fig. 4k). Both ActCM and ActNCM were enriched in cytokine signaling in the immune system pathway (Fig. 4k, Supplementary Data 6). GO enrichment analysis of upregulated genes in ActCM highlighted strong signatures for IL-18 signaling, NF-kB signaling, regulation of myeloid cell differentiation, TNF signaling, Toll-like receptor signaling, and positive regulation of cell death pathway. Relative to other sub-population, the GO analysis showed ActNCM were enriched for signaling by interleukins, positive regulation of cytokine production, regulation of T cell activation, regulation of cell-cell adhesion, the VEGFA-VEGFR2 signaling pathway, and regulation of tumor necrosis factor production (Fig. 4k).
We next performed TF deviation analysis on monocyte subsets, we noticed monocyte subsets exhibited deviated variations in different TF family members from homeostasis to the activated effector state (Supplementary Fig. 11d). For example, both ActCM and ActNCM were associated with high levels of activity of TF involved in NF-kB signaling signatures and myeloid differentiation, including REL, RELA, NFKB1, and NFKB264,65 (Supplementary Fig. 11d). Interestingly, ActCM had increased enrichment in TFs that represent activation and maturation stages, such as AP-1 family members, FOS, and JUN56,64,66. In addition, CM was also enriched in the activation of AP-1 family TFs (Fig. 4l). Meanwhile, ActNCM showed high activity of TFs involved in haematopoetic commitment and survival of monocytes, including NR4A1 and NR4A264,67 (Fig. 4l, Supplementary Fig. 11d, e, Supplementary Data 1). Collectively, myeloid subsets maintained chromatin reprogramming and transcription changes that promote a rapid inflammatory response in BD.
Multi-omic integration mapping enables cellular annotation and analysis
We reasoned that integrating data from scATAC-seq and scRNA-seq datasets may enable the determination of gene-regulatory networks (GRNs) by data integration, facilitating the interpretation of the key regulatory processes underlying the pathogenesis of BD. Based on the sub-clustering and manual cellular annotation we described above, we identified 29 immune cell subsets in 20,000 cells (Fig. 5a). We also automated annotated cell types using clustifyr68 package based on Hao et al.23 to validate our manual annotation based on known marker genes (Supplementary Fig. 12a, b). Next, we interrogated the BD and non-BD datasets from epigenomic and transcriptomic data and utilized the current frameworks supporting the integration of scATAC-seq and scRNA-seq data, relying on identifying mutual nearest neighbors cells – cells, which represent shared biological states in a common lower-dimensional space - to then find representative cells from one dataset in the other (Supplementary Fig. 13a–c). The whole process was parallelized and separately aligned using the ArchR24 and Seurat69,70 pipelines by separating cells into smaller groups. This procedure enabled us to accurately integrate the transcriptomic data from the scRNA-seq dataset with the chromatin accessibility data from the scATAC-seq dataset by mapping the GAS and gene expression to generate an integration matrix. As expected, the GAS and gene expression were matched, which allowed us to distinguish the 29 immune cell types (Fig. 5b, Supplementary Fig. 13d).
a Schematic for multiomic integration strategy for processing the scRNA-seq and scATAC-seq dataset. Following integration and label transfer. Dots represent individual cells, and colors indicate immune cell types (labeled on the right). b Dot plots of gene activity scores (left) and gene expression (right) of the marker genes in scATAC-seq and scRNA-seq dataset. The dot size indicates the percentage of the cells in each cluster in which the gene of interest. The standardized gene activity score level (left) and gene expression level (right) were indicated by color intensity. c Enrichment for autoimmune disease-associated SNPs performed by g-ChromVAR. Color indicates enrichment score. The adjusted p values were calculated using Mann–Whitney U test and Benjamini–Hochberg test. d Cis-regulatory architecture at the following GWAS loci and cell types in PBMCs: KLRC4-KLRK1. Only connections originating in the loci with peak-to-gene accessibility above 0.4 are shown. All data are aligned and annotated to hg38 reference genome.
Dissecting the molecular mechanisms behind autoimmune disease complex phenotypes identified by genome-wide association studies (GWAS) requires pinpointing disease-relevant cell types. However, nearly 90% of causal genetic variants lie in noncoding regions31. In addition, much work has shown that the resolution of intersecting GWAS signals with bulk data is impeded by cell-type heterogeneity71,72,73. We wondered if we could use our single-cell data to better dissect the cell-type-specific effects of genetic variations underlying complex human autoimmune disease traits. To address these issues, we adopted the g-chromVAR74 method on our global single-cell chromatin data to identify trait-cell type associations in a peripheral immune cell subsets-dependent manner (Fig. 5c).
We used a publicly available database for autoimmune and non-immune disorders from a previous study31 and calculated the enrichment of disease-related SNPs in 29 peripheral immune cell types using the g-chromVAR31,74. The majority of the autoimmune associations were strongly enriched for a corresponding trait association. For example, BD was significantly enriched in CD4 CTL, CD8TEM, DNT, and CD8TCM cells75,76, while type 1 diabetes was most strongly enriched in Th17 cells (Fig. 5c)77. Within the open chromatin region of our broad cell subsets, g-chromVAR enrichment revealed significant T cell subsets, reinforcing the previous study22,75,76. T cells with cytotoxicity have been reported to contribute to BD pathogenesis in both skin and circulation8. Additional trends of T cells enrichment are also observed here for type 1 diabetes and asthma, although no statistically significant. Most of these non-autoimmune disease GWAS were not apparent in the immune cell peaks, demonstrating GWAS enrichment was consistent with our expectations. Although not the focus of our current study, we observed that our generated PBMC chromatin data could provide cellular-specific enrichment of human autoimmune disease heritability.
We compiled a list of 66 index SNPs from Farh et al.31 representing GWAS hits for BD. We then identified all the SNPs in scATAC peaks and focused on their nearest genes. Furthermore, we calculated peak-to-gene connections for these gene locus using ArchR (Fig. 5d, Supplementary Fig. 13e–g). We noticed the rs20198574378, that confers the risk of BD was in the KLRC4-KLRK1 enhancer region, which was opened in CD8 T cell subsets and NK cells subsets79. This enhancer was highly accessible in NK and CD8 T subsets, but not in T cells, B cells or monocytes, demonstrating NK and CD8 T specificity. In KLRC4-KLRK1 enhancer region showed statistically increasingly strong peak-to-gene linkages in non-BDs compared to BD patients (Fig. 5d, Supplementary Fig. 13g). There are no differences in the expression of KLRC4-KLRK1 in NK and T cell subsets in the scRNA-seq dataset (Supplementary Data 4, 7, 9). However, we did not notice other SNP loci showed stronger predictive linkages between BD and non-BDs. The low peak accessibility and gene expression in the BD state illustrates chromatin dynamic regulation in the KLRC4-KLRK1 locus. Since Killer cell lectin-like receptor subfamily (KLRC) regulates NK and CD8T function79,80,81, it is possible that this SNP contribute to the pathogenesis of BD by dysregulating KLRC4-KLRK1 in NK and CD8 T function. Therefore, our multi-omic integration analysis could provide predictive disease mechanisms that involve alterations in BD chromatin and gene regulatory regions.
Multi-omic integration analysis identifies candidate TF regulators of DORC activity
The modulation of gene expression by changes in chromatin accessibility is crucial to understating the pathogenesis of autoimmune disease. To address this, we have created well-integrated multi-omic data and used FigR82 to deduce key transcriptional regulatory networks that are required for BD pathogenesis. We first used our scATAC-seq and scRNA-seq data for ChromVAR enrichment of TF motifs among predetermined cis-regulatory elements, as well as the correlation of TF deviation score with the overall chromatin accessibility level for gene activity scores of TF genes, to infer likely positive TF regulators24 (Fig. 6a). We noted that in our dataset, ETS family TFs such as SPIB and SPI1 (also known as PU.1) ranked as the top two TFs related to IFN stimulation and MHC class II gene expression83,84 (Fig. 6a). In addition, AP-1 family TFs JUNB and FOS, and NF-kB family TFs NKFB1 and RELB, also showed high activity in our multi-omic dataset.
a Volcano plot of positive TF regulators using gene expression of the TF and inferred gene activity score. b Top hits based on the number of significant gene-peak correlation across all cell types. Genes with >7 linkages are defined as domains of regulatory chromatin (DORCs). c Scatterplot showing all DOC-to-TF associations, colored by the signed regulation score. d. Candidate TF regulators of HLA-DQB1. Highlighted points are TFs with abs(regulation score) >=1 (−log10 scale), with all other TFs shown in gray. e TF-DORC network visualization for candidate TF positive regulators-implicated DORCs (green nodes) and their associated TFs (red nodes). Edges are scaled and colored by the signed regulation score. Highlighted points in red are TFs with abs(regulation score) >=1 (−log10 scale). All data are aligned and annotated to hg38 reference genome.
To connect distal cis-regulatory elements to genes and infer a GRN, we first utilized our scATAC-seq and scRNA-seq data. We next defined domains of regulatory chromatin (DORCs). We used a computational approach (n = 150,000 cells per assay), FigR framework, to determine DORC within a fixed window (100 kb) around the transcription start site of each gene (Fig. 6b–d). In this way, we identified a total of 23,627 unique cis-regulatory associations genome-wide, showing significant chromatin accessibility peaks with gene expression (permutation p <=0.05). We defined 202 regions with an exceptionally large (>7) number of significant peak-gene associations as DORCs, identified as those exceeding an inflection point (Fig. 6b, Supplementary Data 10). It has previously been reported that DORCs are highly cell-type specific85. We noted that the top genes were SEMA7A, PIK3R2, HLA-DQB1, and IL10, which included many well-known mediators of immunological response associated with innate and adaptive immune response pathways (Supplementary Fig. 14a). For example, the top GO enrichment pathways were regulation of T cell activation, B cell receptor signaling pathway, costimulation by the CD28 family, cytokine signaling, and TCR signaling pathway. Notably, we also observed that the VEGF signaling pathway and positive regulation of leukocyte adhesion to vascular endothelial cells were also on top of the GO analysis, suggesting the possible pathogenesis of vasculitis in BD patients (Supplementary Fig. 14a). Although the top-ranked gene was SEMA7A, we did not observe a significant difference between BD and non-BD (Supplementary Fig. 14B). However, we noted that HLA-DQB1 was highly expressed in the BD group compared to non-BD (Supplementary Fig. 14B). Next, we calculated the correlation of a given DORC gene to TF expression and further queried putative TF regulators for a given DORC (Fig. 6c, d). We identified known activators of HLA-DQB1, including the CCAAT/enhancer-binding protein (CEBP) family TF members: CEBPA, NF-kB family TF members: NFKB2, and RELB (Fig. 6d)86. Importantly, RELB has been reported to be a key activator in DC maturation87.
Based on the above analysis, we inferred that NF-kB family TFs, AP-1 family TFs, and ETS family TFs might work as key pro-inflammatory factors in BD. Leveraging DORCs and the TFs that regulate them, we constructed a GRN that underlies the peripheral blood immunity of BD, relating each TF to each DORC (Fig. 6e, Supplementary Data 11). Consistent with previous reports, AP-1 family TFs are central components of regulatory factors for IL-10 expression88. Using a computational method, we demonstrated the ability to identify DORCs and GRN to determine disease-specific chromatin accessibility profiles relevant to autoimmunity.
Discussion
BD is a complex immunogenic and systemic disease for which the pathogenic cell type and pathway have not yet been identified, leading to a lack of targeted treatment for this disease1,2. Studies of autoimmunity in BD pathogenesis have been particularly limited by mouse models that do not have equivalent clinical phenotypes89. Leveraging scATAC-seq and scRNA-seq to accurately map chromatin accessibility and gene transcription and predict underlying TF regulators and GRN within BD blood samples, provides a unique and physiological approach to elucidate the autoimmunity landscape in this complex disease.
Here, we analyzed the chromatin and transcript landscape underlying human peripheral blood BD heterogeneity by delineating the repertoire of accessible cis-elements and genes in a multi-omic manner. Through our single-cell atlas of peripheral blood and multi-model analysis approach, our work revealed: (1) single-cell epigenomic and transcriptomic profiles of peripheral blood in BD patients; (2) widespread activation of peripheral autoimmunity profiles in BD patients; (3) putative TF activators that drive the changes chromatin accessibility in BD patients; and (4) potential GRN of BD-associated regulatory interactions within putative TF regulators. Importantly, we described the single-cell regulatory peripheral immunity atlas of BD patients, which provides insights into the chromatin level of blood autoimmunity landscape of BD.
Previous single-cell studies on BD included scRNA-seq and scTCR-seq but did not involve scATAC-seq4,8. Both single-cell studies that described the peripheral immunity of the BD have reported enhanced interferon signaling, which also was noted in our data. C1q+ monocytes have been reported to expand in the blood of BD patients by activated IFN-signaling4. CD4+ Treg cells have also been reported to increase in the affected skin tissue of BD patients8. Although we did not notice a significant expansion of CD4+Treg cells in our data, we noticed CD8+Treg cells and Act NCM have significantly higher proportions compared to non-BD. Moreover, we did not notice the frequency of cytotoxicity CD8+ T cell expanded. However, our computational analysis predicted that the GWAS enrichment of open chromatin analysis linked the probable causal BD variants to specific blood cytotoxic CD8+ T cell subsets. It has been previously reported that circulating CD8+ T cell might share a clonal origin with skin-filtrating CD8 T cells and acquire tissue-residential features leading skin lesions8. CD8+ Treg cells expressing inhibitory killer cell immunoglobulin-like receptors (KIRs) have been described as increased in the blood and inflamed tissues of patients with a variety of autoimmune disorders and reported to elevate in COVID-19 patients27. The elevated levels of CD8+ Tregs were also related to COVID-19 vasculitis27. Although the expansion of CD8+Treg in BD may act as a negative feedback mechanism to ameliorate inflammation in peripheral blood, this indicates that CD8+Tregs represent an important element in peripheral tolerance and BD pathophysiology. In total, we provided a more comprehensive blood immune cell landscape of BD.
Our analysis enabled identification of BD-associated SNPs that lie in the regulatory regions of cytotoxicity subsets. Furthermore, we observed that the predictive peak-to-gene linkages near the rs201985743 loci within the KLRC4-KLRK1 region showed significantly stronger linkages in non-BDs compared to BD. Killer cell lectin-like receptor subfamily member 4 (KLRC4, belonging to the NKG2 receptor family known to play an important role in regulating NK and T cell functions81,90, has previously been linked to BD91. The interactions between peaks and genes may indicate physical interaction of the regulatory region affecting its target genes in NK and CD8+ T92,93,94. The differential interaction between the KLRC4-KLRK1 locus in NK cells and CD8+ T cells between non-BD and BD groups may suggest gene expression effects of the causal variant93. However, the molecular cause of NK and CD8 T dysregulation is still unknown. This approach can allow us to predict gene and cellular targets in BD and nominate the most disease-relevant cell types and meriting functional validation.
Importantly, we also generalized multi-model datasets assaying chromatin accessibility and gene expression to infer possible TF activators and their potential GRN that might drive disease-associated phenotypes. This method has been utilized in anticipating in cells from diverse stimuli, cellular differentiation, and even in disease states, providing additional insights into the regulatory mechanisms underlying gene priming. PU.1, AP-1, and NF-kB TFs were predicted and nominated as putative TFs that induced peripheral inflammation of BD, These TFs have been reported to be essential transcription factors regulating multiple inflammatory pathways and contributing to autoimmunity31.
While the causative molecular mechanisms of BD remain elusive, our work offers insights into elucidating the nature of gene regulation in BD, especially regarding the comprehensive single-cell multi-omic landscape of epigenetic and transcriptional patterns. The dataset presented here is a valuable resource for understanding the regulatory relationship in human autoimmunity, and our analysis makes discoveries in chromatin accessibility and gene expression using single-cell multi-omic data. Finally, we represent peripheral immune responses in BD and enhance our understanding of epigenomic pathological immune responses in BD.
Methods
Human subjects
The study was approved by the Ethics Committee of Zhongshan Ophthalmic Center (Guangzhou, China, 2019KYPJ114), and followed the relevant ethical regulations for human research participants according to the Declaration of Helsinki. Written informed consent was obtained from all participating individuals, who were recruited from Zhongshan Ophthalmic Center. Exclusion criteria for the study included comorbid conditions such as cancer, immunocompromising disorders, hypertension, diabetes, and steroid use. The non-BD group consisted of eight individuals, 4 men and 4 women, with an average age of 46.8 years (Supplementary Data 1). The BD patient cohort (Supplementary Data 1) comprised 13 men and 10 women, with an average age of 33.3 years, diagnosed based on the revised diagnostic criteria established by the 2013 International Criteria for BD95.
Cell isolation
To isolate peripheral blood mononuclear cells (PBMCs), peripheral venous blood samples were taken from non-BD individuals or BD patients and treated with a Ficoll-Hypaque density solution and heparin. The mixture was then centrifuged for 30 min. The single-cell suspensions were stained with Trypan blue to assess viability and quantity. Only those samples with cell viability over 90% were selected for subsequent experiments. For each sample that contained over 1 × 107 viable cells, a portion of PBMCs was extracted for scRNA-seq analysis while reserving another fraction for scATAC-seq assays.
scATAC-seq processing
The single-cell nuclei were isolated, washed and counted following the manufacturer’s protocols. To obtain the desired final concentration based on the number of cells, an appropriate volume of Diluted Nuclei Buffer (10x Genomics; PN-2000153) was utilized to resuspend nuclei. The nuclei concentration was then determined using a Countess II FL Automated Cell Counter. Isolated nuclei were promptly used to create 10x single-cell ATAC libraries at Berry Genomics Co., Ltd. (Beijing, China). Each library was uniquely barcoded and quantified by RT-qPCR before being loaded onto an Illumina Novaseq 6000 with a loading concentration of 3.5 pmol/L in pair-end mode. Sequencing was performed until 90% saturation or an average of 30,000 unique reads per cell were acquired. The protocols for sample processing, library preparation, and instrument and sequencing settings on the 10x Chromium were adhered to as described in https://support.10xgenomics.com/single-cell-atac. Raw sequencing data were demultiplexed to fastq format using ‘cellranger-atac-mkfastq’ (10x Genomics, v.1.0.0). Subsequently, the scATAC-seq data reads were aligned to the GRCh38 (hg38) reference genome and quantified using the ‘cellranger-atac count’ function (10x Genomics, v.1.0.0).
scATAC-seq quality control
To generate Arrow files, ArchR v0.9.524 was utilized to analyze the accessible read fragments of each sample, following the default settings. To ensure high signal and sequencing quality, cells with less than 2500 unique fragments and TSS enrichments below 9 were filtered out. Doublets were inferred and eliminated using ArchR with default parameter, while cells that mapped to blacklist regions based on the ENCODE project reference were also excluded.
scATAC-seq dimensionality reduction and clustering
To reduce dimensionality, we employed a layered approach to reduce dimensionality using techniques such as latent semantic indexing (LSI) and singular value decomposition (SVD). The single-cell accessibility profiles were clustered utilizing Seurat’s shared nearest neighbor (SNN) 21 graph clustering with a default resolution of 0.8. We then reclustered using ‘FindClusters’ at a resolution of 0.8 to improve the identification of small clusters. Finally, we utilized uniform manifold approximation and projection (UMAP) to visualize all data in two-dimensional space. We did not detect potential batch effects in our dataset. Therefore, no batch correction method was applied in our further analysis.
scATAC–seq gene activity scores
To calculate gene activity scores, we utilized ArchR17 v.0.9.5 with default parameters, correlating accessibility at the gene body, promoter, and distal regulatory elements with gene expression. The MAGIC96 imputed weight method was then applied to the resulting gene activity scores to reduce noise due to the sparsity of scATAC-seq data.
scATAC–seq pseudobulk replicate generation and peak calling
To enable differential comparisons of clusters, cell types, and clinical states, we created non-overlapping pseudobulk replicates from groups of cells using the ‘addGroupCoverages’ function, by varying the arguments. These pseudobulk replicates were then employed to create the peak matrix using ‘addReproduciblePeakSet’ function. To identify peaks, we utilized MACS297 tool for peak calling. Finally, we utilized the pseudobulk peak set for downstream analysis.
scATAC–seq genomic regions annotation
In the differential analysis, we utilized the ChIPseeker37 package’s “annotatePeak” function with default arguments to annotate the nearest genes in the peak region.
scATAC motif enrichment and motif deviation analysis
Motif enrichment and motif deviation analyses were conducted on the pseudobulk peak set. We employed the Catalog of Inferred Sequence Binding Preferences (CIS-BP) motif (from ChromVAR)24,98, JASPAR2020 motif99 and HOMER100 to perform peak annotation. Furthermore, we utilized the ArchR implementation to calculate the chromVAR deviation scores for these motifs.
scATAC–seq TF Foot-print analysis
To conduct motif footprint analysis, we measured Tn5 insertions in genome-wide motifs, normalized by subtracting the Tn5 bias from the footprinting signal. For each peak set, we employed CIS-BP motifs98 (from chromVAR motifs human_pwms_v1) to calculate motif positions. Normalization of these footprints involved using mean values ± 200–250 from the motif center, after which we plotted the mean and standard deviation for each footprint pseudo-replicate. Comparison of the TF footprint between groups was conducted using the Wilcoxon rank sum test or one-way ANOVA followed by Tukey’s multiple comparison test. A p-value less than 0.05 was considered statistically significant.
scATAC-seq peak to gene linkage analysis
We utilized the ‘addPeak2GeneLinks’ function in ArchR to predict peak-to-gene links, setting the ‘corCutOff’ parameter to 0.4 and ‘reducedDims’ to the dimensionality reduction. The resulting ‘GRanges’ object was utilized for visualization.
Autoimmune SNPs analysis
Pre-computed fine-mapped autoimmune-disease-associated SNPs were downloaded from Farh et al.31. We used the g-chromVAR74 algorithm to identify enrichment of disease variants in each cell type. In brief, the summary statistics we downloaded were converted to hg38 coordinates using the UCSC liftover tool (v377) and formatted for g-chromVAR74. The methodology of g-chromVAR was previously described in detail74. Briefly, g-chromVAR weights chromatin accessibility features by fine-mapped GWAS variants posterior probabilities and calculates the enrichment for each cell type feature intensity. We first binarized the scATAC-seq dataset matrix with one column per cell type. We then followed the recommended guidelines for GWAS enrichment using ‘computeWeightedDeviations’ with default parameters. We then applied the Mann–Whitney U test and the Benjamini-Hochberg procedure for multiple-testing correction to compute enrichment p values101.
scRNA-seq processing
All the samples were processed with the Chromium Single Cell Library, Gel Bead, and Multiplex Kit, and Chip Kit (10x Genomics) to barcode and convert the libraries on the 10x Genomics chromium platform. The Single-cell RNA libraries were prepared using the Chromium Single Cell 5 v2 Reagent (10x Genomics, 1000263) kit, following the manufacturer’s instructions. For sequencing, the scRNA-seq libraries were sequenced on Illumina NovaSeq6000 in pair-end mode, and their quality was checked using FastQC software. Raw data was processed and aligned o the GRCh39 reference by the cellranger software with default parameters (https://support.10xgenomics.com, version 3.1.0) for each sample.
To demultiplex and barcode the sequences obtained from the 10x Genomics single-cell RNA-seq platform, we employed the ‘cellranger-count’ function in the cellranger Software Suite (10x Genomics). To aggregate all the samples, we used ‘cellranger-count’ function.
scRNA-seq quality control
For quality control, we filtered the low-quality cells with greater than 11% of mitochondrial genes and fewer than 200 or more than 3000 detected genes using Seurat V3. We further filtered the cell populations identified as red blood cells and platelets that expressed HBB, HBA1, PPBP, and PF4 genes17.
scRNA-seq dimensionality reduction and clustering
Downstream analysis of scRNA-seq dataset was performed using Seurat v369 as previously described17.To account for technical noise, we choose the top 5000 most variable genes calculated by ‘FindVariableFeatures’ function were used for normalization and scaled. We performed principal component analysis (PCA) on the highly variable genes. The first 30 principal components were further analyzed. We then performed cell clustering based on KNN graphs using the ‘FindNeighnors’ and the ‘FindClusters’ with resolution set as 0.8 in Seurat. We did not use any batch correct method. We further performed the UMAP analysis, a dimensionality-reducing visualization tool, was used to embed the dataset into two dimensions. We did not detect potential batch effects in our dataset. Therefore, no batch correction method was applied in our further analysis.
scRNA–seq differential analysis
For scRNA-seq differential expression analysis, we used the “FindAllMarkers” function from the Seurat package with default parameters. Wilcox rank-sum test was used. The DEGs with logFC > = 0.25 were shown. A p value of less than 0.05 was considered statistically significant. To validate our results of differential analysis between BD and nonBDs, we also used the Muscat R package36. We followed the Muscat tutorial, and used the ‘aggregateData’ function to aggregate our scRNA count assay by cell clusters and samples. The R package limma102 was to make a contrast matrix for BD and nonBDs. Once we have assembled the data, we used the ‘pbDS’ function with parameters set as default. In line with the Seurat method, DEGs with logFC > = 0.25 were shown. A p value of less than 0.05 was considered statistically significant. The R package GeneOverlap103 was utilized to identify (1) the number of overlapping DEGs from muscat method and Seurat method, and (2) the statistical significance of this overlap based on list size and total number from both methods.
scRNA–seq cellular annotation
To validate our manual annotation, we first downloaded the PBMC multimodal single-cell dataset23 as a reference and used the R package clustifyR68 to perform cellular annotation. We chose the ‘celltype. l2’ from the reference as our label and calculated the average expression of the assay of the reference as input reference data. Then we followed the tutorial of clustifyR website with the parameter set as default.
Multiomics data processing
To integrate scRNA-seq and scATAC-seq dataset, we followed the pipeline outlined on ArchR24, Seurat69 and Signac70 websites. First, ArchR24 was used to split the complete dataset into smaller subsets of cells, enabling separate alignments and reducing computational RAM. We then used Seurat’s canonical correlation analysis (CCA) to integrate the epigenetic and transcriptomic data. No additional batch correction methods were implemented. For this integration, the log-normalized and scaled scATAC-seq gene score matrix was aligned with the scRNA-seq gene expression matrix. By directly aligning cells from scATAC-seq with cells from scRNA-seq, the union of the 2,000 most variable genes in each modality as input for Seurat’s “FindTransferAnchors” function and “TransferData” function, using default parameters. To find the nearest neighbor cell in the other modality for each profiled scRNA-seq and scATAC-seq cell, nearest-neighbor search was conducted in the joint CCA L2 space. These modality-spanning nearest-neighbor cell matches from all gestational timepoints were then combined to obtain dataset-wide cell matching.
Gene-regulatory network workflow
We used FigR to infer transcriptional regulators of target genes and construct GRN. We first used the single-cell peaks matrix from scATAC-seq and the count matrix from scRNA-seq (NscATAC-seq = 150,000 cells, NscRNA-seq = 150,000 cells, cells were sampled from the dataset) and used the “runGenePeakcorr” with hg38 genome to determine cis-regulatory associations. Next, we filtered the correlations that p > 0.05 and defined DORC genes as those with more than seven significant peak-gene associations. We used the “runFigRGRN” function with scRNA-seq matrix and DORC data to generate GRN and selected TFs using ggplot2 to visualize the data.
Statistics and reproducibility
Statistical analysis of the frequencies of immune cell subpopulations between groups was performed using two-sided pairwise Wilcoxon test with Bonferroni’s post-hoc correction with GraphPad Prism 8.0. Two-sided p values of less than 0.05, were considered statistically significant. All the statistical details for the statistical tests can be found in the figure legends as well as in the Method Details section. In estimating the GO biological process and pathway, p values were derived by a hypergeometric test with the default parameters in the Metascape webtool104. Each figure legend includes the details of the size of biological replicates and the assays. Values of p <= 0.05 were considered statistically significant. p values are denoted as *p < =0.05, **p < =0.01, ***p < =0.001, ****p < =0.0001 in the figures. The statistical tests employed are referred to in the respective figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The scRNA-seq, scATAC-seq and bulk RNA-seq data analyzed in the article are available from the corresponding author upon request under the Project Accession No. PRJCA004696 and the GSA Accession No. HRA004778 in https://ngdc.cncb.ac.cn/gsa-human/. Source data underlying Figs. 1c, 2c, 4j data are provided in Supplementary Data 2; Source data underlying Fig. 2e data is in Supplementary Data 4; Source data underlying Figs. 3c, g, h, 4e, Supplementary Figs. 8f, 9c, 10g, data are in Supplementary Data 7; Source data underlying Figs. 3d, 4f, k data are in Supplementary Data 6; Fig. 4i data is in Supplementary Data 8; Source data underlying Figs. 2d, 4d, l are in Supplementary Data 3; Source data underlying Supplementary Fig. 7a is in Supplementary Data 5; Source data underlying Fig. 6b is in Supplementary Data 10; Source data underlying Fig. 6e is in Supplementary Data 11.
Code availability
All custom code used in this work is desposited in Zenodo https://doi.org/10.5281/zenodo.8348340105.
References
Yazici, H., Seyahi, E., Hatemi, G. & Yazici, Y. Behcet syndrome: a contemporary view. Nat. Rev. Rheumatol. 14, 107–119 (2018).
Yazici, Y. et al. Behcet syndrome. Nat. Rev. Dis. Prim. 7, 67 (2021).
Evereklioglu, C. Current concepts in the etiology and treatment of Behcet disease. Surv. Ophthalmol. 50, 297–350 (2005).
Zheng, W. et al. Single-cell analyses highlight the proinflammatory contribution of C1q-high monocytes to Behcet’s disease. Proc. Natl Acad. Sci. USA 119, e2204289119 (2022).
Nanke, Y., Yago, T. & Kotake, S. The role of Th17 cells in the pathogenesis of Behcet’s disease. J. Clin. Med. 6 (2017).
Okubo, M. et al. Transcriptome analysis of immune cells from Behcet’s syndrome patients: the importance of IL-17-producing cells and antigen-presenting cells in the pathogenesis of Behcet’s syndrome. Arthritis Res. Ther. 24, 186 (2022).
Tong, B., Liu, X., Xiao, J. & Su, G. Immunopathogenesis of Behcet’s Disease. Front. Immunol. 10, 665 (2019).
Chang, L. et al. Single cell transcriptome and T cell repertoire mapping of the mechanistic signatures and T cell trajectories contributing to vascular and dermal manifestations of Behcet disease. bioRxiv https://doi.org/10.1101/2022.03.22.485251 (2022).
Granja, J. M. et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 37, 1458–1465 (2019).
Morabito, S. et al. Single-nucleus chromatin accessibility and transcriptomic characterization of Alzheimer’s disease. Nat. Genet. 53, 1143–1155 (2021).
Ranzoni, A. M. et al. Integrative single-cell RNA-Seq and ATAC-seq analysis of human developmental hematopoiesis. Cell Stem Cell 28, 472–487.e477 (2021).
Trevino, A. E. et al. Chromatin and gene-regulatory dynamics of the developing human cerebral cortex at single-cell resolution. Cell 184, 5053–5069.e5023 (2021).
Chiou, J. et al. Single-cell chromatin accessibility identifies pancreatic islet cell type- and state-specific regulatory programs of diabetes risk. Nat. Genet. 53, 455–466 (2021).
Cusanovich, D. A. et al. A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324.e1318 (2018).
Corces, M. R. et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nat. Genet. 52, 1158–1168 (2020).
Chiou, J. et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594, 398–402 (2021).
Shi, W. et al. Chromatin accessibility analysis reveals regulatory dynamics and therapeutic relevance of Vogt-Koyanagi-Harada disease. Commun. Biol. 5, 506 (2022).
Tewary, P. et al. Granulysin activates antigen-presenting cells through TLR4 and acts as an immune alarmin. Blood 116, 3465–3474 (2010).
Stewart, A. et al. Single-cell transcriptomic analyses define distinct peripheral B cell subsets and discrete development pathways. Front Immunol. 12, 602539 (2021).
Leylek, R. et al. Chromatin Landscape Underpinning Human Dendritic Cell Heterogeneity. Cell Rep. 32, 108180 (2020).
Schep, A. N., Wu, B., Buenrostro, J. D. & Greenleaf, W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017).
Satpathy, A. T. et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
Zhang, L. et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564, 268–272 (2018).
Mishra, S. et al. TGF-beta and Eomes control the homeostasis of CD8+ regulatory T cells. J. Exp. Med. 218 (2021).
Li, J. et al. KIR(+)CD8(+) T cells suppress pathogenic T cells and are active in autoimmune diseases and COVID-19. Science 376, eabi9591 (2022).
Ling, E., Shubinsky, G. & Press, J. Increased proportion of CD3+CD4-CD8- double-negative T cells in peripheral blood of children with Behcet’s disease. Autoimmun. Rev. 6, 237–240 (2007).
Fang, Y. et al. The role of a key transcription factor PU.1 in autoimmune diseases. Front. Immunol. 13, 1001201 (2022).
Barnabei, L., Laplantine, E., Mbongo, W., Rieux-Laucat, F. & Weil, R. NF-kappaB: at the borders of autoimmunity and inflammation. Front Immunol. 12, 716469 (2021).
Farh, K. K. et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015).
Xu, W. D., Pan, H. F., Ye, D. Q. & Xu, Y. Targeting IRF4 in autoimmune diseases. Autoimmun. Rev. 11, 918–924 (2012).
Park, S. H. et al. BATF regulates collagen-induced arthritis by regulating T helper cell differentiation. Arthritis Res. Ther. 20, 161 (2018).
Katagiri, T. et al. JunB plays a crucial role in development of regulatory T cells by promoting IL-2 signaling. Mucosal Immunol. 12, 1104–1117 (2019).
Koizumi, S. I. et al. JunB regulates homeostasis and suppressive functions of effector regulatory T cells. Nat. Commun. 9, 5344 (2018).
Crowell, H. L. et al. muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nat. Commun. 11, 6077 (2020).
Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Lang, R. & Raffi, F. A. M. Dual-specificity phosphatases in immunity and infection: an update. Int. J. Mol. Sci. 20 (2019).
Huang, Z. et al. Effects of sex and aging on the immune cell landscape as assessed by single-cell transcriptomic analysis. Proc. Natl Acad. Sci. USA 118 (2021).
Forero, A. et al. Differential activation of the transcription factor IRF1 underlies the distinct immune responses elicited by type I and type III interferons. Immunity 51, 451–464.e456 (2019).
Schuster, C. et al. CD5 controls gut immunity by shaping the cytokine profile of intestinal T cells. Front. Immunol. 13, 906499 (2022).
Maecker, H. T., McCoy, J. P. & Nussenblatt, R. Standardizing immunophenotyping for the Human Immunology Project. Nat. Rev. Immunol. 12, 191–200 (2012).
Ma, P. et al. Immune cell landscape of patients with diabetic macular edema by single-cell RNA analysis. Front. Pharm. 12, 754933 (2021).
Png, Y. T. et al. Blockade of CD7 expression in T cells for effective chimeric antigen receptor targeting of T-cell malignancies. Blood Adv. 1, 2348–2360 (2017).
Chin, S. S. et al. T cell receptor and IL-2 signaling strength control memory CD8+ T cell functional fitness via chromatin remodeling. Nat. Commun. 13, 2240 (2022).
Gómez-Herranz, M., Taylor, J. & Sloan, R. D. IFITM proteins: Understanding their diverse roles in viral infection, cancer, and immunity. J. Biol. Chem. 299, 102741 (2023).
Fairfax, K. C. et al. IL-4-secreting secondary T follicular helper (Tfh) cells arise from memory T cells, not persisting Tfh cells, through a B cell-dependent mechanism. J. Immunol. 194, 2999–3010 (2015).
Ben Ahmed, M., Houman, H., Miled, M., Dellagi, K. & Louzir, H. Involvement of chemokines and Th1 cytokines in the pathogenesis of mucocutaneous lesions of Behcet’s disease. Arthritis Rheum. 50, 2291–2295 (2004).
Koarada, S. et al. Increased entry of CD4+ T cells into the Th1 cytokine effector pathway during T-cell division following stimulation in Behcet’s disease. Rheumatology (Oxford) 43, 843–851 (2004).
Braudeau, C. et al. Persistent deficiency of circulating mucosal-associated invariant T (MAIT) cells in ANCA-associated vasculitis. J. Autoimmun. 70, 73–79 (2016).
Borrego, F., Robertson, M. J., Ritz, J., Pena, J. & Solana, R. CD69 is a stimulatory receptor for natural killer cell and its cytotoxic effect is blocked by CD94 inhibitory receptor. Immunology 97, 159–165 (1999).
Wang, R., Jaw, J. J., Stutzman, N. C., Zou, Z. & Sun, P. D. Natural killer cell-produced IFN-gamma and TNF-alpha induce target cell cytolysis through up-regulation of ICAM-1. J. Leukoc. Biol. 91, 299–309 (2012).
Eksioglu-Demiralp, E. et al. Phenotypic characteristics of B cells in Behcet’s disease: increased activity in B cell subsets. J. Rheumatol. 26, 826–832 (1999).
Zajacova, M., Kotrbova-Kozak, A. & Cerna, M. Expression of HLA-DQA1 and HLA-DQB1 genes in B lymphocytes, monocytes and whole blood. Int J. Immunogenet. 45, 128–137 (2018).
Le Floch-Ramondou, A. et al. Blockade of common gamma chain cytokine signaling with REGN7257, an Interleukin 2 Receptor Gamma (IL2RG) monoclonal antibody, protected mice from inflammatory and autoimmune diseases. Blood 140, 473–474 (2022).
Fontana, M. F. et al. JUNB is a key transcriptional modulator of macrophage activation. J. Immunol. 194, 177–186 (2015).
Vazquez, B. N., Laguna, T., Carabana, J., Krangel, M. S. & Lauzurica, P. CD69 gene is differentially regulated in T and B cells by evolutionarily conserved promoter-distal elements. J. Immunol. 183, 6513–6521 (2009).
Krzyzak, L. et al. CD83 modulates B cell activation and germinal center responses. J. Immunol. 196, 3581–3594 (2016).
Klaus, S. J. et al. Costimulation through CD28 enhances T cell-dependent B cell activation via CD40-CD40L interaction. J. Immunol. 152, 5643–5652 (1994).
Netea, M. G. et al. Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 20, 375–388 (2020).
Hemann, E. A. et al. Interferon-lambda modulates dendritic cells to facilitate T cell immunity during infection with influenza A virus. Nat. Immunol. 20, 1035–1045 (2019).
Maney, N. J., Reynolds, G., Krippner-Heidenreich, A. & Hilkens, C. M. U. Dendritic cell maturation and survival are differentially regulated by TNFR1 and TNFR2. J. Immunol. 193, 4914–4923 (2014).
Gringhuis, S. I. et al. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-kappaB activation through Raf-1 and Syk. Nat. Immunol. 10, 203–213 (2009).
You, M. et al. Single-cell epigenomic landscape of peripheral immune cells reveals establishment of trained immunity in individuals convalescing from COVID-19. Nat. Cell Biol. 23, 620–630 (2021).
Ichiyama, T. et al. NF-kappaB activation in peripheral blood monocytes/macrophages and T cells during acute Kawasaki disease. Clin. Immunol. 99, 373–377 (2001).
Behmoaras, J. et al. Jund is a determinant of macrophage activation and is associated with glomerulonephritis susceptibility. Nat. Genet. 40, 553–559 (2008).
Hanna, R. N. et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat. Immunol. 12, 778–785 (2011).
Fu, R. et al. clustifyr: an R package for automated single-cell RNA sequencing cluster classification. F1000Res 9, 223 (2020).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Pickrell, J. K. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 94, 559–573 (2014).
Ulirsch, J. C. et al. Interrogation of human hematopoiesis at single-cell and single-variant resolution. Nat. Genet. 51, 683–693 (2019).
Yang, J. Y., Park, M. J., Park, S. & Lee, E. S. Increased senescent CD8+ T cells in the peripheral blood mononuclear cells of Behcet’s disease patients. Arch. Dermatol. Res. 310, 127–138 (2018).
Yasuoka, H. et al. Preferential activation of circulating CD8+ and gammadelta T cells in patients with active Behcet’s disease and HLA-B51. Clin. Exp. Rheumatol. 26, S59–S63 (2008).
Shao, S. et al. Th17 cells in type 1 diabetes. Cell Immunol. 280, 16–21 (2012).
Kirino, Y. et al. Genome-wide association analysis identifies new susceptibility loci for Behcet’s disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet. 45, 202–207 (2013).
Bauer, S. et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285, 727–729 (1999).
Guma, M. et al. The CD94/NKG2C killer lectin-like receptor constitutes an alternative activation pathway for a subset of CD8+ T cells. Eur. J. Immunol. 35, 2071–2080 (2005).
Kim, D. K. et al. Human NKG2F is expressed and can associate with DAP12. Mol. Immunol. 41, 53–62 (2004).
Kartha, V. K. et al. Functional inference of gene regulation using single-cell multi-omics. Cell Genom. 2 (2022).
Kitamura, N. et al. Role of PU.1 in MHC class II expression through transcriptional regulation of class II transactivator pI in dendritic cells. J. Allergy Clin. Immunol. 129, 814–824.e816 (2012).
Kuwata, T. et al. Gamma interferon triggers interaction between ICSBP (IRF-8) and TEL, recruiting the histone deacetylase HDAC3 to the interferon-responsive element. Mol. Cell Biol. 22, 7439–7448 (2002).
Ma, S. et al. Chromatin potential identified by shared single-profiling RNA chromatin. Cell 183, 1103–1116.e1120 (2020).
Lee, K. W., Lee, Y., Kim, D. S. & Kwon, H. J. Direct role of NF-kappaB activation in Toll-like receptor-triggered HLA-DRA expression. Eur. J. Immunol. 36, 1254–1266 (2006).
Shih, V. F. et al. Control of RelB during dendritic cell activation integrates canonical and noncanonical NF-kappaB pathways. Nat. Immunol. 13, 1162–1170 (2012).
Hu, X. et al. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity 24, 563–574 (2006).
Charles, J., Castellino, F. J. & Ploplis, V. A. Past and present Behcet’s disease animal models. Curr. Drug Targets 21, 1652–1663 (2020).
Kucuksezer, U. C. et al. The role of natural killer cells in autoimmune diseases. Front. Immunol. 12, 622306 (2021).
Karasneh, J., Gul, A., Ollier, W. E., Silman, A. J. & Worthington, J. Whole-genome screening for susceptibility genes in multicase families with Behcet’s disease. Arthritis Rheum. 52, 1836–1842 (2005).
Kumasaka, N., Knights, A. J. & Gaffney, D. J. High-resolution genetic mapping of putative causal interactions between regions of open chromatin. Nat. Genet. 51, 128–137 (2019).
Mumbach, M. R. et al. Enhancer connectome in primary human cells identifies target genes of disease-associated DNA elements. Nat. Genet. 49, 1602–1612 (2017).
Broekema, R. V., Bakker, O. B. & Jonkers, I. H. A practical view of fine-mapping and gene prioritization in the post-genome-wide association era. Open Biol. 10, 190221 (2020).
International Team for the Revision of the International Criteria for Behcet’s, D. The International Criteria for Behcet’s Disease (ICBD): a collaborative study of 27 countries on the sensitivity and specificity of the new criteria. J. Eur. Acad. Dermatol. Venereol. 28, 338–347 (2014).
van Dijk, D. et al. Recovering gene interactions from single-cell data using data diffusion. Cell 174, 716–729.e727 (2018).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014).
Fornes, O. et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Duong, T. E. et al. A single-cell regulatory map of postnatal lung alveologenesis in humans and mice. Cell Genom. 2, 100108 (2022).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Shen, L. J. R. P. GeneOverlap: an R package to test and visualize gene overlaps. R Package 3 (2014).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Shi, W. et al. Single-cell chromatin accessibility and transcriptomic characterization of Behcet’s disease. https://doi.org/10.5281/zenodo.8348340 (2023).
Acknowledgements
This study was funded by the National Natural Science Foundation of China (81721003); Local Innovative and Research Teams Project of Guangdong Pearl River Talents Programme (2017BT01S138); CAMS Innovation Fund for Medical Sciences (2019-I2M-5-005); the State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-sen University. The funding body had no role in study design, collection, management, analysis and interpretation of data, writing of the manuscript, and the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
Y.Z., Y.L. and L.D. designed research; W.S. and J.Y. analyzed data; Z.S., C.P., Q.Z. and Y.L. processed the human samples and library construction; W.S. wrote the paper. X.L. provided comments and reviews during the revision process.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: George Inglis. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shi, W., Ye, J., Shi, Z. et al. Single-cell chromatin accessibility and transcriptomic characterization of Behcet’s disease. Commun Biol 6, 1048 (2023). https://doi.org/10.1038/s42003-023-05420-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-023-05420-x
- Springer Nature Limited