Abstract
Mesenchymal stromal/stem cells (MSC) play a crucial role in promoting neovascularization, which is essential for wound healing. They are commonly utilized as an autologous source of progenitor cells in various stem cell-based therapies. However, incomplete MSC differentiation towards a vascular endothelial cell phenotype questions their involvement in an alternative process to angiogenesis, namely vasculogenic mimicry (VM), and the signal transducing events that regulate their in vitro priming into capillary-like structures. Here, human MSC were primed on top of Cultrex matrix to recapitulate an in vitro phenotype of VM. Total RNA was extracted, and differential gene expression assessed through RNA-Seq analysis and RT-qPCR. Transient gene silencing was achieved using specific siRNA. AG490, Tofacitinib, and PP2 pharmacological effects on VM structures were analyzed using the Wimasis software. In vitro VM occurred within 4 h and was prevented by the JAK/STAT3 inhibitors AG490 and Tofacitinib, as well as by the Src inhibitor PP2. RNA-Seq highlighted STAT3 as a signaling hub contributing to VM when transcripts from capillary-like structures were compared to those from cell monolayers. Concomitant increases in IL6, IL1b, CSF1, CSF2, STAT3, FOXC2, RPSA, FN1, and SNAI1 transcript levels suggest the acquisition of a combined angiogenic, inflammatory and epithelial-to-mesenchymal transition phenotype in VM cultures. Increases in STAT3, FOXC2, RPSA, Fibronectin, and Snail protein expression were confirmed during VM. STAT3 and RPSA gene silencing abrogated in vitro VM. In conclusion, in vitro priming of MSC into VM structures requires Src/JAK/STAT3 signaling. This molecular evidence indicates that a clinically viable MSC-mediated pseudo-vasculature process could temporarily support grafts through VM, allowing time for the host vasculature to infiltrate and remodel the injured tissues.
Similar content being viewed by others
Introduction
Mesenchymal stromal/stem cells (MSC) are a subset of adult multipotent precursors, known for their therapeutic properties in regenerative medicine1. Such properties are mainly sustained through paracrine response to a variety of biologically active molecules including a wide range of soluble protein factors composed of growth factors, cytokines, and vesicular components that transfer proteins and genetic material to modulate the host microenvironment2,3. After their mobilization and tissue engraftment, MSC-derived secretome then appears to mediate the different steps of the angiogenic process, inducing endothelial cell functions in vitro and promoting angiogenesis in vivo4. As a result, MSC have been widely explored as a promising cell-based therapy in diseases caused by insufficient angiogenesis5,6.
Recent emerging pathophysiological evidence, on the other hand, suggest that MSC may exert a significant role in the occurrence and development of solid tumors through tumor immunosuppressive processes7. Whether MSC differentiate into vascular endothelial cells or adopt any angiogenic or proinflammatory phenotypes within the hypoxic solid tumor microenvironment remains poorly understood. The lack of platelet endothelial cell adhesion molecule-1 (PECAM-1) expression in MSC further questions the potential capacity of MSC to differentiate towards an endothelial cell phenotype8. Interestingly, clear differences between MSC and endothelial cells in their ability to form capillary-like networks and express endothelial markers have been reported9. In contrast to endothelial cells, which more strongly express vascular markers such as VE-cadherin and vWF, MSC are rather more capable of migrating through matrices and establishing networks, a feature more typical of stromal than endothelial populations10 prompting for different mechanisms to occur in MSC-mediated neovascularization processes. Alternative to angiogenesis, vasculogenic mimicry (VM) processes within tumor xenografts has been reported through cancer cells’ ability to, in part, attract MSC within primary solid tumors11, where their pro-angiogenic secretome is believed to contribute to tumour neovascularization12,13,14.
VM has been described in PECAM-1-negative, but VE-cadherin-positive cells, and to be highly dependent on matrix metalloproteinase (MMP)-2 production15,16,17. VM is a unique process originally reported to occur within dense tumors to overcome hypoxic stress by creating tumor-lined pseudo-vasculature, and was first evidenced by the demonstration that non-endothelialized microvessels were capable of transporting blood without clotting18. With regards to tissue engineering strategies, VM represents a neovascularization process where a single stromal-type population may be driven by matrix cues and hypoxic stress towards capillary-like structures19,20. Although these non-endothelialized networks are leakier and less efficient than normal vasculature, they appear functional enough to facilitate blood transport21.
With regards to therapeutic neovascularization, endothelial-like networks may provide a clinically feasible pseudo-vasculature for temporarily sustaining grafts until the host vasculature is able to infiltrate and remodel tissue. The fact that fibroblasts can match endothelial-like MSC behavior suggests that this mechanism of tubulogenesis may be shared amongst other cells of stromal lineage22,23. However, stromal tubulogenesis in non-diseased adult biology lacks documentation. If VM-mediated capillary-like structure formation is, in fact, a latent but inducible stromal function, MSC priming has therefore the potential to open up new cell sources as therapeutically valuable for neovascularization strategies.
Results
In vitro mesenchymal stromal/stem cells capillary-like structure formation kinetic on Cultrex
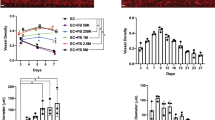
Mesenchymal stromal/stem cells (MSC) in vitro VM capacity to form capillary-like structures in time was first assessed, as representative phase contrast pictures were taken every hour for up to 4 h upon seeding on top of Cultrex (Fig. 1A, upper panels). Pictures were digitized for Wimasis analysis (Fig. 1A, lower panels), and mean loop area (Fig. 1B, left) as well as mean loop perimeter (Fig. 1B, right) respectively quantified from a representative experiment. Altogether, increases in mean loop area and perimeter, and maturation of the structures were found to occur with time confirming the capacity of MSC to generate in vitro VM. The paracrine regulation impact of the growth factors or extracellular matrix (ECM) proteins composing Cultrex was next assessed. Soluble factors were depleted from Cultrex, cells plated on top of either complete or growth factors-depleted Cultrex, and VM structures digitized (Fig. 1C). We found that branching points, loops, and tubes parameters were all reduced when cells adhered to growth factors-depleted Cultrex suggesting that, in addition and beyond ECM protein interaction, VM structures further require to respond to growth factors cues (Fig. 1D) and may require further in-depth investigation in line with the nature of the cell surface actors involved in the recognition of ECM proteins. The nature of the potential signaling events involved was next investigated.
In vitro mesenchymal stem/stromal cells capillary-like structure formation on Cultrex. Mesenchymal stromal/stem cells (MSC) monolayers were trypsinized and seeded on top of Cultrex to generate capillary-like structures as described in the Methods section. (A) Representative phase contrast pictures were taken (upper panels), and WIMASIS analysis of the structures represented in blue (lower panels). (B) Capillary-like structures were quantified using WIMASIS and representative quantification provided for mean loop area (left panel) and mean loop perimeter (right panel) presented. (C) MSC were seeded on top of complete Cultrex or on Cultrex that was depleted from soluble factors as described in the Methods section, and representative phase contrast pictures taken (upper panels) and WIMASIS analysis of the structures represented in blue (lower panels). (D) VM parameters were extracted from the Wimasis analysis of C), and data expressed as percent of the value obtained on complete Cultrex. All experiments were performed in triplicates from different MSC passages.
Transcriptomic analysis reveals the JAK/STAT signaling pathway as a hub in MSC 3D capillary-like structures
RNA-Seq analysis was next used to decipher the molecular signature of human MSC cultured as capillary-like structures and compared to that of cell monolayers to identify signaling pathways which could trigger transcriptional regulation induced by VM. Total RNA was extracted from samples for each condition, and gene expression modulation was assessed through RNA-Seq. Network graph shows enriched pathways and their respective differentially expressed genes (DEGs) in cells cultured on Cultrex versus cells cultured as cell monolayers (Fig. 2A). Upregulated and downregulated genes are color-coded in green and red, respectively. KEGG pathways enrichment analysis of DEGs with an absolute fold change (FC) > 2 and adjusted p-value < 0.001 in each experimental condition is shown (Fig. 2B). Among the several pathways identified, the JAK/STAT signaling pathway was the most induced, followed respectively by the Wnt, TGF-β, and prolactin pathways.
Transcriptomic analysis reveals JAK/STAT signaling pathway as a hub in MSC capillary-like structure formation. Human mesenchymal stromal/stem cells (MSC) were seeded as either cell monolayers or as capillary-like structures on top of Cultrex for 4 h. Total RNA was extracted from triplicate samples for each condition, and gene expression modulation was assessed through RNA-Seq analysis as described in the Methods section. (A) Network graph showing enriched pathways and their respective differentially expressed genes (DEGs) in cells cultured on Cultrex versus cells cultured as monolayers. Upregulated and downregulated genes are color-coded in green and red, respectively. (B) KEGG pathways enrichment analysis of DEGs with an absolute fold change (FC) > 2 and adjusted p-value < 0.001 in each experimental condition68,69,70. Data was analyzed on triplicate samples from a representative experiment.
Protein-protein interaction network predicts STAT3 interrelationship with downstream biomarkers involved in epithelial-to-mesenchymal transition during MSC capillary-like structure formation
A protein-protein interaction (PPI) network of STAT3 was constructed by using the STRING database (https://string-db.org/) and was used to predict and analyze potential STAT3 interactors. Indirect target proteins of STAT3 were retrieved from STRING and predicted STAT3 interrelationship with biomarkers involved in EMT and inflammation during MSC capillary-like structure formation. These included IL6, IL1B, CSF-1 to 3, FOXC2, FN1, and SNAI1 (Fig. 3A). To confirm those interrelationships, MSC were seeded as either monolayers (Plastic, white bars) or as capillary-like structures formed upon 4 h on top of Cultrex (Cultrex, black bars). Total RNA was extracted from triplicate samples for each condition, and gene expression modulation was assessed by RT-qPCR as described in the Methods section. In addition to STAT3, increases in EMT markers encoding Fibronectin (FN1) and Snail (SNAI1) were observed along with predicted FOXC2, an important nuclear factor recently reported to promote VM in ovarian cancer24 (Fig. 3B). Immunoblotting was further performed using protein lysates isolated from 2D and 3D cultures, and confirmed the increases in Fibronectin, Snail, STAT3, and FOXC2 at the protein level (Fig. 3C).
Transcriptomic analysis reveals JAK/STAT signaling pathway as a hub in MSC capillary-like structure formation. (A) Indirect target proteins of STAT3 were retrieved from STRING as described in the Methods section. (B) Human mesenchymal stromal/stem cells (MSC) were seeded as either monolayers (Plastic, white bars) or as capillary-like structures on top of Cultrex (Cultrex, black bars) for 4 h. Total RNA was extracted from triplicate samples for each condition, and gene expression modulation was assessed by RT-qPCR as described in the Methods section. (C) Lysates were isolated from monolayers and from VM cultures, and used to perform immunoblotting analysis of the indicated biomarkers at the protein level. Cropped blots are presented for the sake of clarity and conciseness of data presentation. Uncropped full-length blots are presented in the Supplementary data section, as Supplementary Figure S3C. Protein data are representative from two distinct experiments. RT-qPCR data originate from triplicate sample analysis.
Pharmacological targeting and transient STAT3 gene silencing abrogate MSC capillary-like structure formation, and inhibit the acquisition of an EMT inflammatory molecular signature
STAT3-mediated transcriptional regulation upon VM was next further assessed in relation to EMT and pro-inflammatory molecular signatures. Pharmacological approaches were used to target STAT3, and the impact on MSC capillary-like structures assessed in the absence (Vehicle) or presence of JAK2/STAT3 inhibitor AG490, JAK1 and JAK3 inhibitor Tofacitinib, or Src inhibitor PP2. Representative phase contrast pictures show that VM structures were altered by these inhibitors (Fig. 4A). Next, translocation of STAT3 within the nucleus was assessed in order to document any STAT3-mediated transcriptional activity. Nuclear fractionation was performed as described in the Methods section, and enrichment of the nuclear biomarker Fibrillarin observed with virtually no contamination from cytoplasmic GAPDH (Fig. 4B). Interestingly, significant increases of nuclear FOXC2 and SNAI1 were observed within 3D structures, along with increased STAT3 and its phosphorylated form (Fig. 4C). These increases were abrogated upon AG490 treatment suggesting that STAT3 phosphorylation was a prerequisite for these events to occur.
STAT3 pharmacological targeting alters MSC capillary-like structure formation and nuclear expression of FOXC2 and SNAIL. (A) Human mesenchymal stromal/stem cells (MSC) were seeded on top of Cultrex and capillary-like structures generated for 4 h in the absence (Vehicle) or presence of 10 µM JAK/STAT3 inhibitors AG490 and Tofacitinib, or Src inhibitor PP2 respectively. Representative phase contrast pictures were taken. (B) Nuclear fractionation protocol was performed to validate by Western blotting nuclear Fibrillarin enrichment, and enriched cytosolic GAPDH. (C) Nuclear fractionation was performed from MSC monolayers or from capillary-like structures treated or not with AG490. Total proteins were extracted and representative blots for FOXC2, Fibrillarin, SNAIL, STAT3, and phosphoSTAT3 are presented. Protein data are representative from three distinct experiments. Cropped blots are presented for the sake of clarity and conciseness of data presentation. Uncropped full-length blots are presented in the Supplementary data section, as Supplementary Figure S4B and S4C.
In addition to the above pharmacological approach, transient gene silencing of STAT3 (siSTAT3) was performed as described in the Methods section and cell lysates assessed for STAT3 and GAPDH protein expression. Immunoblotting confirmed the efficient STAT3 protein knock-down (Fig. 5A), and this resulted in altered capillary-like structures in comparison to control cells which were transfected with a scrambled siRNA sequence (siScrambled; Fig. 5B). Among the cellular processes required for VM structure formation, intrinsic cell migration was found reduced in conditions where STAT3 was repressed (Fig. 5C, closed circles). This observation supports the signal transducing role of STAT3 in cell migration for VM to occur as reported in other cancer cell models25,26,27. Finally, transfected MSC were harvested from either cell monolayers (Plastic, white bars) or from capillary-like structures (Cultrex, black bars). Total RNA was extracted from triplicate samples for each condition, and gene expression modulation assessed by RT-qPCR as described in the Methods section. Interestingly, results demonstrate increased expression of a pro-inflammatory molecular signature as reflected by high IL6, IL1b, FOXC2, CSF1, and CSF2 gene expression in siScrambled-transfected cells (Fig. 5D, siScrambled, black bars). In contrast, upon STAT3 transient silencing, the above observed increases were significantly prevented in response to VM cultures cues (Fig. 5D, siSTAT3, black bars).
STAT3 gene silencing alters MSC capillary-like structure formation, cell chemotaxis, and acquisition of an inflammatory phenotype. (A) Gene silencing of STAT3 (siSTAT3) was performed in human mesenchymal stromal/stem cells (MSC) as described in the Methods section. Control cells were transfected with a scrambled siRNA sequence (siScrambled). Cell lysates were assessed for GAPDH and STAT3 protein expression. Cropped blots are presented for the sake of clarity and conciseness of data presentation. Uncropped full-length blots are presented in the Supplementary data section, as Supplementary Figure S5A. (B) Transfected MSC were seeded on top of Cultrex and 3D capillary-like structures generated for 4 h. Representative phase contrast pictures were taken (upper panels) and Wimasis analysis (lower panels) performed, or (C) real-time cell migration assessed using the xCELLigence as described in the Methods section. (D) Transfected MSC were seeded as either monolayers (Plastic, white bars) or as capillary-like structures on top of Cultrex (Cultrex, black bars) for 4 h. Total RNA was extracted from triplicate samples for each condition, and gene expression modulation was assessed by RT-qPCR as described in the Methods section. Protein and VM data are representative from two distinct experiments. RT-qPCR data were analyzed from triplicate samples.
A role for the 37-kDa laminin receptor precursor/67-kDa laminin receptor (RPSA) in MSC capillary-like structure formation
Considering the induced Snail expression upon VM and the signaling crosstalk recently reported to link Snail to RPSA28, we further tested whether RPSA was regulated upon VM. MSC were cultured as capillary-like structures, total RNA and cell lysates isolated to find that RPSA was induced at both the protein (Fig. 6A) and gene levels (Fig. 6B). We next assessed the transcriptional control STAT3 or FOXC2 potentially exerted on SNAI1 and RPSA gene expression. We found that only STAT3 silencing prevented SNAI1 and RPSA to be induced upon VM, whereas FOXC2 silencing did not affect either genes expression (Fig. 6B). Altogether, this suggests that a role for RPSA in VM may be transcriptionally regulated downstream from Snail. Interestingly, transient repressing of RPSA lead to decreased protein expression (Fig. 6C) and to reduced capillary-like structures formation (Fig. 6D, upper panels). This was further supported upon pharmacological inhibition of RPSA function with NSC4792429 (Fig. 6D, lower panels).
A role for the 37-kDa laminin receptor precursor/67-kDa laminin receptor (RPSA) in MSC 3D capillary-like structure formation. Human mesenchymal stromal/stem cells (MSC) were seeded as either monolayers or as capillary-like structures on top of Cultrex for 4 h. Cell lysates and total RNA were extracted from each condition. (A) Immunoblotting was performed to detect RPSA protein levels. (B) RT-qPCR was performed to assess gene expression in cells cultured as capillary-like structures (Black bars) vs. monolayers (Open bars) upon transient gene silencing of either STAT3 (siSTAT3) or FOXC2 (siFOXC2) as described in the Methods section. (C) Cell lysates were assessed to confirm transient gene silencing of RPSA (siRPSA) at the protein level, and representative capillary-like structures phase-contrast pictures taken in (D) transfected cells (upper panels), or cells treated with a pharmacological RPSA inhibitor (lower panels). Cropped blots are presented for the sake of clarity and conciseness of data presentation. Protein and VM data are representative from two distinct experiments. RT-qPCR data were analyzed from triplicate samples. Uncropped full-length blots are presented in the Supplementary data section, as Supplementary Figure S6A and S6C.
Discussion
Given MSC are easily expandable ex vivo, enhancement strategies, including genetic manipulation, pre-activation, and modification of culture methods, have been investigated to generate highly functional MSC with improved therapeutic efficacy30. Here, we highlight a Src/JAK/STAT3 signaling hub as crucial for VM and for MSC priming upon capillary-like structure formation. Pre-activating MSC upon VM formation mimics their natural tissue engraftment microenvironment in vivo and provides enhanced cell-cell and cell-ECM interactions. This could significantly improve their future biological behaviour including proliferation, immune regulation, and committed differentiation31.
As demonstrated here, STAT3 signalling appears crucial for MSC migration, capillary-like structure formation, and transcriptional regulation of EMT, inflammatory, and angiogenic biomarkers upon VM. Accordingly, MSC mobilization and priming within their perivascular/vascular location is thought to regulate vascular function, as well as new blood vessel formation32,33,34. While no single marker currently defines native MSC in vivo, their phenotype appears however mainly determined through the specific microenvironment location where they engraft and promote tissue regeneration/repair. MSC can regulate different processes including inflammation and angiogenesis, the production of ECM, and the regeneration of functional cells through cell differentiation. Not surprisingly, those MSC therapeutic properties have recently supported their need for cell priming in regenerative medicine6,35, in part through their angiogenic properties36,37,38.
STAT3 repression further appeared to downregulate VM-induced 37/67 kDa laminin receptor (RPSA) expression highlighting a new signalling crosstalk linking capillary-like structure formation to RPSA, a molecule that acts as a key player in tumorigenesis, affecting cell growth, adhesion, migration, invasion and cell death processes. Noteworthy, roles of the cellular prion protein (PrPC) and RPSA interaction in cancer biology have recently emerged39. In addition, hypoxia preconditioning was found to enhance MSC survival and their angiogenic properties, potentially through induced up-regulation of PrPC which, in turn, enhanced MSC proliferation via PrPC-dependent JAK2/STAT3 activation40. In line with such evidence, vascular progenitors derived from murine MSC were avidly recruited within hypoxic solid tumors xenografts11, and hypoxia found to trigger adaptive metabolic mechanisms41,42 and to accelerate capillary-like structure formation43. Interestingly, homing of MSC towards the hypoxic solid tumor microenvironment in vivo was reported to correlate with IL6-mediated STAT3 activation of survival pathways which facilitated tumor progression44. In line with the induced STAT3 expression observed upon VM, prosurvival STAT3 activation was found to prevent MSC apoptosis and to improve infarct repair45. Whether PrPC is regulated in VM remains to be explored, although PrPC has high binding affinity for RPSA39,46. Elucidation of the role played by PrPC/RPSA interaction in regulating VM may represent a very promising avenue to gain insight into tissue regeneration processes and potentially solid tumor development.
STAT3 is recognized as a critical transcription factor in angiogenesis through, in part, protein stabilisation of HIF-1α47. Accordingly, STAT3 deficiency resulted in decreased MSC production of VEGF48. Such STAT3 involvement was also found to play a role in maintaining the self-renewal properties of cancer stem cells49, where VEGF-mediated angiogenesis was further reported to link EMT-induced cancer stemness to tumor initiation50. Here, we demonstrate that expression of EMT biomarkers Snail, Fibronectin, and RPSA was induced at both the gene and protein levels in VM cultures. More importantly, STAT3 repression or pharmacological inhibition prevented their expression and correlated with decreased VM. In support, phosphorylation of STAT3 was found to promote VM by inducing EMT in colorectal cancer25. Finally, selective JAK/STAT3 signalling was reported in the regulation of CSF-2 and CSF-3 in Concanavalin-A-activated MSC51,52 confirming that transcriptional up-regulation of CSF-2 and CSF-3 in activated MSC may contribute to their immunomodulatory and proangiogenic phenotype.
The mechanistic parallels and signal transducing pathways between MSC capillary-like structure formation and cancer cells VM bring a novel perspective towards understanding their use in therapy and tissue regeneration. In cancer, VM is often associated with the classical EMT process where epithelial cells exhibit increased migration without complete loss of cell-cell adhesion and polarity53. Here, we better defined the molecular signature associated with 3D structure formation by MSC, and highlighted the importance that STAT3 signaling crosstalk provides in the regulation of the pro-inflammatory/angiogenic/EMT phenotype associated to VM (Fig. 7). Interestingly, regulation of pathophysiological and tissue regenerative functions of MSC were recently found mediated via the Wnt signaling pathway54. STAT3 plays a role in tissue regeneration by MSC, and it has been suggested that STAT3 activation or inactivation can modulate MSC trophic effects55. In addition, the transcriptomic screen further supports the fact that both the response to, and production of TGF-β, an inducer of the epithelial-to-mesenchymal transition (EMT) process, by MSC may regulate their VM capacity as well as immunomodulatory and angiogenic molecular signature56.
Scheme summarizing the STAT3 signaling hub and regulatory impact on capillary-like structure formation, and on the acquisition of an angiogenic, inflammatory, and EMT phenotype. MSC monolayers can recapitulate in vitro VM when cultured on Cultrex. Capillary-like structures can be inhibited in the presence of AG490 and PP2, respectively JAK/STAT3 and Src inhibitors. MSC priming on Cultrex triggers STAT3 expression which, in turn, can regulate the acquisition of an EMT phenotype (increased SNAIL, FOXC2, and RPSA), as well as an angiogenic/inflammatory molecular signature (increased IL6/IL1b/CSF-1, -2). Collectively, this phenotype may form pseudo-vasculature and sustain early pro-angiogenic processes physiologically.
In conclusion, the present study reveals that ECM cues that trigger in vitro VM appear to involve a crucial role for Src/JAK/STAT3 signaling in priming MSC to generate capillary-like structures. With regards to therapeutic neovascularization, it is tempting to hypothesize that, upon mobilization and engraftment to the injured tissues, circulating MSC primed into capillary-like networks may support a clinically feasible pseudo-vasculature to temporarily sustain grafts through VM, until the host vasculature is able to infiltrate and remodel the injured tissues. Furthermore, an inflammatory/angiogenic/EMT molecular signature associated to MSC VM also supports their role in promoting angiogenesis.
Methods
Reagents
Micro bicinchoninic acid (BCA) protein assay reagents were from Pierce (Micro BCA™ Protein Assay Kit; Thermo Fisher Scientific, Waltham, MA, USA). The polyclonal antibodies against STAT3 (12640 S), Snail (3879 S), FOXC2 (12974 S), and Fibronectin (30903 S), as well as the monoclonal antibody against GAPDH (D4C6R) were all from Cell Signaling Technology (Danvers, MA, USA). The antibodies against 37-kDa laminin receptor precursor/67-kDa laminin receptor (RPSA, Ab133645) and β-Actin were from Abcam (Cambridge, UK). HRP-conjugated donkey anti-rabbit and anti-mouse immunoglobulin (Ig) G secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). The JAK family tyrosine kinase inhibitor Tofacitinib (CP-690550) and NSC47924 were from Cedarlane (Burlington, ON), while AG490 was from Calbiochem (La Jolla, CA). All other reagents were from Sigma-Aldrich Corp (St-Louis, MO, USA).
Cell culture and capillary-like structure formation assay
Human bone marrow-derived mesenchymal stromal/stem cells (MSC) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cell culture media was from Life Technologies Corp (Carlsbad, CA, USA). Cells were plated in high glucose αMEM supplemented with 10% FBS and 50 units/mL penicillin/streptomycin and cultured in a humidified incubator at 37 °C with 5% CO2. MSC were kept subconfluent and expanded in number over 10 passages by a 1:2 split on a weekly basis. VM was assessed in vitro using Cultrex (3432-010-01, R&D Systems) to monitor capillary-like structures formation43. In brief, each well of a 96-well plate was pre-coated with 50 µl of Cultrex. MSC suspension in culture media (104 cells/100 µl) was then seeded on top of polymerized Cultrex. Tested compounds were added to the cell culture media and incubated at 37 °C in a CO2 incubator. Pictures were taken over time using a digital camera coupled to a phase-contrast inverted microscope. Mean loop area: For each loop, the area (number of pixels) enclosed by it is considered as its area. The mean loop area is the arithmetic mean of all loop areas. Mean loop perimeter: For each loop, the pixels that belong to its edge are considered its border or perimeter. The mean loop perimeter is the arithmetic mean of all loop perimeters. The number of loops and area covered upon tube branching formed by the cells were quantified using either the Wimasis analysis software (https://www.wimasis.com; Cordoba, Spain) or the ImageJ software (https://imagej.net)57.
Total RNA isolation, cDNA synthesis, and real-time quantitative PCR
Total RNA was extracted from cell monolayers using 1 mL of TriZol reagent for a maximum of 3 × 106 cells as recommended by the manufacturer (Life Technologies, Gaithersburg, MD). For cDNA synthesis, 2 µg of total RNA was reverse-transcribed using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA). The cDNA was stored at -20oC prior to PCR. Gene expression was quantified by real-time quantitative PCR using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA). DNA amplification was carried out using the CFX Connect Real-Time PCR System (Bio-Rad) and product detection was performed by measuring the binding of the fluorescent dye SYBR Green I to double-stranded DNA. The following primer sets were from QIAGEN: FOXC2 (Hs_FOXC2_1_SG, QT00220871), IL1b (Hs_IL1B_1_SG, QT00021385), IL6 (Hs_IL6_1_SG, QT00083720), STAT3 (Hs_STAT3_1_SG, QT00068754) SNAI1 (Hs_SNAI1_SG, QT00010010), Fibronectin (Hs_FN1_1_SG, QT00038024), CSF-1 (Hs_CSF1_1_SG, QT00035224), CSF-2 (Hs_CSF2_1_SG, QT00000896), RPSA (Hs_RPSA_1_SG, QT00044310), GAPDH (Hs_GAPDH_1_SG, QT00079247) and Peptidylprolyl Isomerase A (PPIA) (Hs_PPIA_4_SG, QT01866137). The relative quantities of target gene mRNA were normalized against internal housekeeping genes PPIA and GAPDH. The RNA was measured by following a ∆CT method employing an amplification plot (fluorescence signal vs. cycle number). The difference (∆CT) between the mean values in the triplicate samples of the target gene and the housekeeping genes was calculated with the CFX manager Software version 2.1 (Bio-Rad) and the relative quantified value (RQV) was expressed as 2−∆CT. Single amplicons and appropriate melting curves were indicative of specific qPCR conditions and efficacy (not shown).
Transfection method and RNA interference
For gene silencing experiments, MSC were transiently transfected with siRNA sequences using Lipofectamine-2000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA). Gene silencing was performed using 20 nM siRNA against RPSA (Hs_RPSA_1 siRNA, SI03045371), STAT3 (Hs_STAT3_7 siRNA, S102662338), or scrambled sequences (AllStar Negative Control siRNA, 1027281). The above small interfering RNA and mismatch siRNA were all synthesized by QIAGEN and annealed to form duplexes. Gene silencing efficacy was assessed by RT-qPCR as described above.
Total RNA library preparation
Total RNA library preparation was conducted as previously described by us58. In brief, 500 ng of total RNA was extracted from MSC cultures for library preparation. RNA quality was assessed with the Bioanalyzer RNA 6000 Nano assay on the 2100 Bioanalyzer system (Agilent technologies, Mississauga, ON), ensuring all samples had an RNA integrity number (RIN) above eight. The KAPA mRNA-Seq HyperPrep kit (KAPA, Cat no. KK8581) was used for library preparation. Ligation was performed with Illumina dual-index UMI, and 10 PCR cycles to amplify cDNA libraries. Libraries were quantified by QuBit and BioAnalyzer DNA1000. All libraries were diluted to 10 nM and normalized by qPCR using the KAPA library quantification kit (KAPA; Cat no. KK4973). Finally, libraries were pooled to equimolar concentrations, and three biological replicates were generated.
RNA sequencing
High RNA quality was confirmed as previously described58. The samples were then sequenced at the Genomics Core Facility of the Institute for Research in Immunology and Cancer (IRIC, Montreal, QC).
Reads alignment and differential expression analysis
Reads were aligned and sorted by coordinates to the human genome build 38 (GRCh38.p13) with version 37 of Gencode gene annotations, using the STAR aligner (STAR_2.7.1a)59,60. Quantification of genes was performed during alignment by STAR. Differentially expressed genes among groups were identified using the R packages DESeq2 (v 1.30.1)61. After analysis, only genes with adjusted p-values (adjp) < 0.05, and log2 fold change (FC) ≥ 1.0 were considered significant. Hierarchical clustering of differentially expressed genes was used to represent the results (R package ggplot2) and heatmaps generated62. For all statistical analyses, differences were considered statistically significant if the adjp value calculated by Student’s t-test with Bonferroni correction were < 0.05.
Gene set enrichment analysis
The gene set enrichment analysis (GSEA) was done as described previously58 and performed with the GSEA software version 4.2.3 (https://www.gsea-msigdb.org)63 with the complete set of normalized input values, using the Hallmark, canonical pathway gene sets (chemical and genetic perturbations, BioCarta, Reactome, and Kegg), and Gene Ontology (GO) gene sets (Biological process). Molecular Signatures Database (MSig-DB; https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp), version 7.5.1 was applied to genes modulated in 3D differentiated MSC compared to 2D cells (values (adjp) < 0.05, and log2 FC ≥ 1.0 were considered as significant) to generate a signature list of the top modulated genes from the following Curated gene sets and Gene Ontology. Software used for the analysis of data during this project included GraphPad Prism and R (version > 3.4). For all statistical analyses, differences were considered statistically significant if the adjp value calculated by Student’s t-test with Bonferroni correction were < 0.05.
Data preparation
The associative relationships of STAT3 were retrieved from STRING v11 database (https://string-db.org)64, with a confidence score setting of 0.4, and the maximum number of interactions to show was no more than 10.
Nuclear extraction
Nuclear extraction was performed as described by us previously65. Briefly, cell monolayers were first lysed with a cytoplasmic buffer and then with a nuclear buffer according to the manufacturer’s instructions (Invent Biotechnologies, SC-003). In the case of the cells cultured on Cultrex, they were first detached from the matrix using a non-enzymatic Cultrex organoid harvesting and dissociation solution (3700-100-01) from R&D Systems (Toronto, ON).
Western blot
Electrophoresis reagents origin, total cell lysis procedure, SDS-polyacrylamide gel electrophoresis (PAGE), electro transfer to low-fluorescence polyvinylidene difluoride membranes, and immunodetection were conducted as described in detail previously66. Immunoreactive material was visualized by enhanced chemiluminescence (ECL).
Real-time cell migration assay
Real-time cell migration assays were conducted as described in detail by us previously using the Real-Time Cell Analyser (RTCA) Dual-Plate (DP) instrument and the xCELLigence system (Roche Diagnostics, QC)66. MSC were transfected with 20 nM siRNAs (siScrambled and siSTAT3) as described above. After transfection, 25,000 cells per well were seeded in a CIM-plate 16 (Roche Diagnostics) and incubated at 37 °C under a humidified atmosphere containing 5% CO2 for 24 h. Cell migration was monitored every 2 h for 20 h. The impedance value was measured by the RTCA DP Instrument and expressed as an arbitrary unit called the Cell Index. Each experiment was performed in quadruplicate wells.
Statistical data analysis
All statistical analyses were conducted using the GraphPad Prism 7 software (https://www.graphpad.com; San Diego, CA). Data and error bars are presented as the mean ± standard error of the mean (SEM) from three or more independent experiments, unless otherwise specified. Hypothesis testing was performed using the Mann-Whitney test (two group comparisons). Probability values of less than 0.05 (*) or 0.01 (**) were considered significant.
Data availability
The RNA sequencing dataset generated during the current study is available on the NCBI GEO platform (GSE267328). Other data are available at the relevant referenced web locations or are available from the corresponding author on reasonable request.
Abbreviations
- BSA:
-
Bovine serum albumin
- DEG:
-
Differentially expressed genes
- ECM:
-
Extracellular matrix
- EMT:
-
Epithelial-to-mesenchymal transition
- FC:
-
Fold change
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- MSC:
-
Mesenchymal stromal/stem cells
- PECAM-1:
-
Platelet endothelial cell adhesion molecule-1
- PPI:
-
Protein-protein interaction
- PrPc :
-
Cellular prion protein
- SDS:
-
Sodium dodecyl sulfate
- VM:
-
Vasculogenic mimicry
References
Ulpiano, C., da Silva, C. L. & Monteiro, G. A. Mesenchymal stromal cells (MSCs): a promising tool for cell-based angiogenic therapy. Curr. Gene Ther.21(5), 382–405 (2021).
Thalakiriyawa, D. S., Jayasooriya, P. R. & Dissanayaka, W. L. Regenerative potential of mesenchymal stem cell-derived extracellular vesicles. Curr. Mol. Med.22(2), 98–119 (2022).
Zriek, F., Di Battista, J. A. & Alaaeddine, N. Mesenchymal stromal cell secretome: Immunomodulation, tissue repair and effects on neurodegenerative conditions. Curr. Stem Cell. Res. Ther.16(6), 656–669 (2021).
Maacha, S. et al. Paracrine mechanisms of mesenchymal stromal cells in angiogenesis. Stem Cells Int.2020, 4356359 (2020).
Bronckaers, A. et al. Mesenchymal stem/stromal cells as a pharmacological and therapeutic approach to accelerate angiogenesis. Pharmacol. Ther.143(2), 181–196 (2014).
Miceli, V. et al. Therapeutic properties of mesenchymal stromal/stem sells: the need of cell priming for cell-free therapies in regenerative medicine. Int. J. Mol. Sci.22(2), 763 (2021).
Tian, Y. et al. The role of hypoxic mesenchymal stem cells in tumor immunity. Int. Immunopharmacol.112, 109172 (2022).
Pusztaszeri, M. P., Seelentag, W. & Bosman, F. T. Immunohistochemical expression of endothelial markers CD31, CD34, Von Willebrand factor, and Fli-1 in normal human tissues. J. Histochem. Cytochem.54(4), 385–395 (2006).
Crisan, M. Transition of mesenchymal stem/stromal cells to endothelial cells. Stem Cell. Res. Ther.4(4), 95 (2013).
Ghajar, C. M. et al. Mesenchymal cells stimulate capillary morphogenesis via distinct proteolytic mechanisms. Exp. Cell. Res.316(5), 813–825 (2010).
Annabi, B., Naud, E., Lee, Y. T., Eliopoulos, N. & Galipeau, J. Vascular progenitors derived from murine bone marrow stromal cells are regulated by fibroblast growth factor and are avidly recruited by vascularizing tumors. J. Cell. Biochem.91(6), 1146–1158 (2004).
Li, G. C. et al. Mesenchymal stem cells promote tumor angiogenesis via the action of transforming growth factor β1. Oncol. Lett.11(2), 1089–1094 (2016).
Sun, C. et al. Mesenchymal stem cells promote glioma neovascularization in vivo by fusing with cancer stem cells. BMC Cancer. 19(1), 1240 (2019).
Wang, J. et al. Bone marrow-derived mesenchymal stem cell-secreted IL-8 promotes the angiogenesis and growth of colorectal cancer. Oncotarget. 6(40), 42825–42837 (2015).
Folberg, R. & Maniotis, A. J. Vasculogenic mimicry. APMIS. 112(7–8), 508–525 (2004).
Hendrix, M. J. et al. Expression and functional significance of VE-cadherin in aggressive human melanoma cells: role in vasculogenic mimicry. Proc. Natl. Acad. Sci. U S A. 98(14), 8018–8023 (2001).
Hess, A. R., Seftor, E. A., Seftor, R. E. & Hendrix, M. J. Phosphoinositide 3-kinase regulates membrane type 1-matrix metalloproteinase (MMP) and MMP-2 activity during melanoma cell vasculogenic mimicry. Cancer Res.63(16), 4757–4762 (2003).
Maniotis, A. J. et al. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am. J. Pathol.155(3), 739–752 (1999).
Paulis, Y. W., Soetekouw, P. M., Verheul, H. M., Tjan-Heijnen, V. C. & Griffioen, A. W. Signalling pathways in vasculogenic mimicry. Biochim. Biophys. Acta. 1806(1), 18–28 (2010).
Zhao, N. et al. Hypoxia-induced vasculogenic mimicry formation via VE-cadherin regulation by Bcl-2. Med. Oncol.29(5), 3599–3607 (2012).
Rytlewski, J. A., Alejandra Aldon, M., Lewis, E. W. & Suggs, L. J. Mechanisms of tubulogenesis and endothelial phenotype expression by MSCs. Microvasc Res.99, 26–35 (2015).
Lubarsky, B. & Krasnow, M. A. Tube morphogenesis: making and shaping biological tubes. Cell. 112(1), 19–28 (2003).
Nagle, R. B. & Cress, A. E. Metastasis update: human prostate carcinoma invasion via tubulogenesis. Prostate Cancer. 2011, 249290 (2011).
Recouvreux, M. S. et al. FOXC2 promotes vasculogenic mimicry in ovarian cancer. Cancers (Basel). 14(19), 4851 (2022).
Han, C. et al. Phosphorylation of STAT3 promotes vasculogenic mimicry by inducing epithelial-to-mesenchymal transition in colorectal cancer. Technol. Cancer Res. Treat.16(6), 1209–1219 (2017).
de la Hernández, O. N. et al. Regulation networks driving vasculogenic mimicry in solid tumors. Front. Oncol.9, 1419 (2020).
Zhou, H., Yuan, Y. & Qian, H. Expression of STAT3 and vasculogenic mimicry in gallbladder carcinoma promotes invasion and metastasis. Exp. Ther. Med.22(1), 738 (2021).
Gresseau, L., Roy, M. E., Duhamel, S. & Annabi, B. A signaling crosstalk links SNAIL to the 37/67 kDa Laminin-1 Receptor Ribosomal Protein SA and regulates the acquisition of a cancer stem cell molecular signature in U87 glioblastoma neurospheres. Cancers (Basel). 14(23), 5944 (2022).
Pesapane, A. et al. Discovery of new small molecules inhibiting 67 kDa laminin receptor interaction with laminin and cancer cell invasion. Oncotarget. 6(20), 18116–18133 (2015).
Li, M. et al. Potential pre-activation strategies for improving therapeutic efficacy of mesenchymal stem cells: current status and future prospects. Stem Cell. Res. Ther.13(1), 146 (2022).
Nicolas, J. et al. 3D extracellular matrix mimics: fundamental concepts and role of materials chemistry to influence stem cell fate. Biomacromolecules. 21(6), 1968–1994 (2020).
Au, P., Tam, J., Fukumura, D. & Jain, R. K. Bone marrow-derived mesenchymal stem cells facilitate engineering of long-lasting functional vasculature. Blood. 111(9), 4551–4558 (2008).
Kachgal, S. & Putnam, A. J. Mesenchymal stem cells from adipose and bone marrow promote angiogenesis via distinct cytokine and protease expression mechanisms. Angiogenesis. 14(1), 47–59 (2011).
Zhou, B. et al. A novel function for the haemopoietic supportive murine bone marrow MS-5 mesenchymal stromal cell line in promoting human vasculogenesis and angiogenesis. Br. J. Haematol.157(3), 299–311 (2012).
Merimi, M. et al. The therapeutic potential of mesenchymal stromal cells for regenerative medicine: current knowledge and future understandings. Front. Cell. Dev. Biol.9, 661532 (2021).
Evans, C. E., Iruela-Arispe, M. L. & Zhao, Y. Y. Mechanisms of endothelial regeneration and vascular repair and their application to regenerative medicine. Am. J. Pathol.191(1), 52–65 (2021).
Lee, E. J., Park, H. W., Jeon, H. J., Kim, H. S. & Chang, M. S. Potentiated therapeutic angiogenesis by primed human mesenchymal stem cells in a mouse model of hindlimb ischemia. Regen Med.8(3), 283–293 (2013).
Watt, S. M. et al. The angiogenic properties of mesenchymal stem/stromal cells and their therapeutic potential. Br. Med. Bull.108(1), 25–53 (2013).
Limone, A., Maggisano, V., Sarnataro, D. & Bulotta, S. Emerging roles of the cellular prion protein (PrPC) and 37/67 kDa laminin receptor (RPSA) interaction in cancer biology. Cell. Mol. Life Sci.80(8), 207 (2023).
Han, Y. S. et al. Hypoxia-induced expression of cellular prion protein improves the therapeutic potential of mesenchymal stem cells. Cell. Death Dis.7(10), e2395 (2016).
Lord-Dufour, S. et al. Evidence for transcriptional regulation of the glucose-6-phosphate transporter by HIF-1alpha: Targeting G6PT with mumbaistatin analogs in hypoxic mesenchymal stromal cells. Stem Cells. 27(3), 489–497 (2009).
Proulx-Bonneau, S., Guezguez, A. & Annabi, B. A concerted HIF-1α/MT1-MMP signalling axis regulates the expression of the 3BP2 adaptor protein in hypoxic mesenchymal stromal cells. PLoS One. 6(6), e21511 (2011).
Annabi, B. et al. Hypoxia promotes murine bone-marrow-derived stromal cell migration and tube formation. Stem Cells. 21(3), 337–347 (2003).
Rattigan, Y., Hsu, J. M., Mishra, P. J., Glod, J. & Banerjee, D. Interleukin 6 mediated recruitment of mesenchymal stem cells to the hypoxic tumor milieu. Exp. Cell. Res.316(20), 3417–3424 (2010).
Xuan, Y. T. et al. STAT3 activation prevents mesenchymal stem cell apoptosis and improves infarct repair. Circulation. 126(21S), A12591 (2012).
Van den Broeke, A., Van Poucke, M., Van Zeveren, A. & Peelman, L. J. Ribosomal protein SA and its pseudogenes in ruminants: an extremely conserved gene family. Czech J. Anim. Sci. Czech Acad. Agricultural Sci.58(2), 79–90 (2013).
Dinarello, A. et al. STAT3 and HIF1α cooperatively mediate the transcriptional and physiological responses to hypoxia. Cell. Death Discov. 9(1), 226 (2023).
Wang, M. et al. STAT3 mediates bone marrow mesenchymal stem cell VEGF production. J. Mol. Cell. Cardiol.42(6), 1009–1015 (2007).
Zhao, D. et al. VEGF drives cancer-initiating stem cells through VEGFR-2/Stat3 signaling to upregulate Myc and Sox2. Oncogene. 34(24), 3107–3119 (2015).
Fantozzi, A. et al. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res.74(5), 1566–1575 (2014).
Zgheib, A., Pelletier-Bonnier, É., Levros, L. C. Jr, Annabi, B. & Selective JAK/STAT3 signalling regulates transcription of colony stimulating factor-2 and – 3 in Concanavalin-A-activated mesenchymal stromal cells. Cytokine. 63(2), 187–193 (2013).
Zgheib, A., Lamy, S. & Annabi, B. Epigallocatechin gallate targeting of membrane type 1 matrix metalloproteinase-mediated Src and Janus kinase/signal transducers and activators of transcription 3 signaling inhibits transcription of colony-stimulating factors 2 and 3 in mesenchymal stromal cells. J. Biol. Chem.288(19), 13378–13386 (2013).
Kalluri, R. & Weinberg, R. A. The basics of epithelial-mesenchymal transition. J. Clin. Invest.119(6), 1420–1428 (2009). Erratum in: J Clin Invest. 2010;120(5):1786.
Zhang, Q. et al. Regulation of pathophysiological and tissue regenerative functions of MSCs mediated via the WNT signaling pathway (review). Mol. Med. Rep.24(3), 648 (2021).
Ayala-Cuellar, A. P., Kang, J. H., Jeung, E. B. & Choi, K. C. Roles of mesenchymal stem cells in tissue regeneration and immunomodulation. Biomol. Ther. (Seoul). 27(1), 25–33 (2019).
de Araújo Farias, V., Carrillo-Gálvez, A. B., Martín, F. & Anderson, P. TGF-β and mesenchymal stromal cells in regenerative medicine, autoimmunity and cancer. Cytokine Growth Factor. Rev.43, 25–37 (2018).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9(7), 671–675 (2012).
Gonzalez Suarez, N. et al. EGCG inhibits the inflammation and senescence inducing properties of MDA-MB-231 triple-negative breast cancer (TNBC) cells-derived extracellular vesicles in human adipose-derived mesenchymal stem cells. Cancer Cell. Int.23(1), 240 (2023).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17), i884–i890 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29(1), 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15(12), 550 (2014).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 32(18), 2847–2849 (2016).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 102(43), 15545–15550 (2005).
Szklarczyk, D. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. ;49(D1):D605-D612. Erratum in: Nucleic Acids Res. 2021;49(18):10800. (2021).
Roy, M. E., Veilleux, C., Paquin, A., Gagnon, A. & Annabi, B. Transcriptional regulation of CYR61 and CTGF by LM98: a synthetic YAP-TEAD inhibitor that targets in-vitro vasculogenic mimicry in glioblastoma cells. Anticancer Drugs. 35(8), 709–719 (2024).
Pratt, J., Haidara, K. & Annabi, B. MT1-MMP expression levels and catalytic functions dictate LDL receptor-related protein-1 ligand internalization capacity in U87 glioblastoma cells. Int. J. Mol. Sci.23(22), 14214 (2022).
Torabidastgerdooei, S., Roy, M. E. & Annabi, B. A molecular signature for the G6PC3/SLC37A2/SLC37A4 interactors in glioblastoma disease progression and in the acquisition of a brain cancer stem cell phenotype. Front. Endocrinol. (Lausanne). 14, 1265698 (2023).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of genes and genomes. Nucleic Acids Res.28, 27–30 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci.28, 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res.51, D587–D592 (2023).
Acknowledgements
We thank Dr Yuniel Fernandez-Marrero for his expert support in the bio-informatics analysis.
Funding
This work was funded by the Institutional Research Chair in Cancer Prevention and Treatment held by Dr Borhane Annabi at UQAM, and by a grant from the Natural Sciences and Engineering Research Council of Canada (NSERC, RGPIN-2024-04541) to BA. MER holds a CERMO-FC Fellowship. CV holds a NSERC and Fond de Recherche du Québec – Nature et Technologie (FRQNT) Fellowship.
Author information
Authors and Affiliations
Contributions
Conceptualization, M.E.R., B.A.Data curation, M.E.R., C.V.Formal analysis, M.E.R., B.A.Funding acquisition, B.A.Investigation, M.E.R., C.V.Methodology, M.E.R.Supervision, B.A.Writing – original draft, M.E.R, B.A.Writing – review & editing, M.E.R., C.V., B.A.All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Roy, ME., Veilleux, C. & Annabi, B. In vitro biomaterial priming of human mesenchymal stromal/stem cells : implication of the Src/JAK/STAT3 pathway in vasculogenic mimicry. Sci Rep 14, 21444 (2024). https://doi.org/10.1038/s41598-024-72862-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-72862-6
- Springer Nature Limited