Abstract
The presence of bacteria from the Dickeya spp. and Pectobacterium spp. in farmlands leads to global crop losses of over $420 million annually. Since 1982, the scientists have started to suspect that the development of disease symptoms in crops might be inhibited by bacteria present in the soil. Here, we characterized in terms of physicochemical properties and the composition of bacterial soil microbiota two fields differing, on the basis of long-term studies, in the occurrence of Dickeya spp.- and Pectobacterium spp.-triggered infections. Majority, i.e. 17 of the investigated physicochemical features of the soils collected from two fields of either low or high potato blackleg and soft rot diseases incidences turned out to be similar, in contrast to the observed 4 deviations in relation to Mg, Mn, organic C and organic substance contents. By performing microbial cultures and molecular diagnostics-based identification, 20 Pectobacterium spp. strains were acquired from the field showing high blackleg and soft rot incidences. In addition, 16S rRNA gene amplicon sequencing followed by bioinformatic analysis revealed differences at various taxonomic levels in the soil bacterial microbiota of the studied fields. We observed that bacteria from the genera Bacillus, Rumeliibacillus, Acidobacterium and Gaiella turned out to be more abundant in the soil samples originating from the field of low comparing to high frequency of pectinolytic bacterial infections. In the herein presented case study, it is shown for the first time that the composition of bacterial soil microbiota varies between two fields differing in the incidences of soft rot and blackleg infections.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Introduction
Pectinolytic bacteria from the Dickeya spp. and Pectobacterium spp., referred as Soft Rot Pectobacteriaceae (SRP), currently comprise 13 Dickeya and 21 Pectobacterium species1,2,3. These phytopathogens cause the symptoms of soft rot on potato or other vegetables and ornamentals, as well as blackleg on potato, cabbage, corn and other economically important plants2,4. The blackleg symptoms can be recognized by blackening of the base of the shoot, which often leads to wilting of the whole plant. Soft rot, on the other hand, is manifested by maceration of the internal tissue of vegetative organs. The global spread of Dickeya spp. and Pectobacterium spp. results not only in significant yield reductions, but also increasing economic losses4. In particular, the presence of SRP in farmlands has been associated with crop damage reaching up to $420 million annually5, with the highest economic impact towards the potato production sector6. In Europe alone, SRP phytopathogens triggered approximately €46 million damage in potato plantations7. In more detail, 32%, 43% and 25% of these losses have been associated with seed, table and processing potatoes, respectively7. In view of high impact on food security, widespread occurrence and expansion of the geographical range of SRP, it needs to be stressed that there are currently no effective plant protection measures against pectinolytic phytopathogens6. Despite the fact that different control approaches relying either on physical, chemical or biological methods have been tested so far4, the potency of these methods is too low to effectively prevent the spread of these microorganisms in the environment. To date, preventive methods, for example applying diagnostics to test seed potatoes for the presence of Dickeya and Pectobacterium spp., cleaning and disinfection of machinery and equipment, limiting irrigation from possibly contaminated waterways, removing diseased plants, minimizing mechanical damage at harvest, and optimizing the storage conditions, have been regarded as the most efficacious plant protection methods6.
The spread of Dickeya spp. and Pectobacterium spp. over long distances was previously associated not only with shipment of latently infected seed potatoes, but also with transferring of contaminated soil8. Indeed, the members of the family Pectobacteriaceae are able to retain viability in soil from 1 week to 6 months, depending on the encountered environmental conditions8. Interestingly, the ability of SRP to survive in soil tends to be species-specific as most of the so-far studied Pectobacterium spp. are capable of surviving under these conditions for up to several months, while Dickeya spp., e.g. Dickeya dianthicola strains, are generally regarded as seed-borne pathogens9. For instance, Pectobacterium spp. strains were able to survive in soil for approximately two months10. At the same time, Dickeya spp. remained viable in soil for up to seven days at 6 °C and 50% soil moisture11. Several factors, like temperature, moisture and pH of the soil, impacts persistence of SPR in this environmental niche12. Microbiological analyses of the soil collected in Australia one year before planting of potatoes showed that 25% of the samples contained the cells from the current Dickeya spp. or Pectobacterium spp.13. Among these bacteria, Erwinia carotovora subsp. carotovora turned out to be the most abundant species as it accounted for 93% of the strains. On the contrary, Erwinia carotovora subsp. atroseptica was found only in 5.6% of the samples. It is worth to underline that in the 90’s, Erwinia carotovora subsp. carotovora corresponded to a consortium including the following recently differentiated species: Pectobacterium brasiliense, Pectobacterium carotovorum, Pectobacterium parmentieri, Pectobacterium parvum, Pectobacterium peruviense, Pectobacterium polaris, Pectobacterium punjabese and Pectobacterium versatile. Pectobacterium atrosepticum was found only in 5.6% of the samples. On the other hand, Dickeya spp. strains were rarely detected in the collected soil samples as these strains amounted to just about 2% of the total number of the identified strains14,15.
Pectinolytic bacteria originating from soil can penetrate host plant tissue through natural openings, such as lenticels or mechanical wounds, leading to colonization of the intercellular spaces of the roots prior to invading the xylem and causing systemic infection4. It needs to be underlined that majority of the previous research focused on studying the occurrence and persistence of pectinolytic bacteria in the rhizosphere of the crops16 or weeds, while in terms of bulk soil, the presence of these microorganisms was mostly investigated in association with plant debris17,18,19.
Since 1982, it has been suggested that the composition of natural soil microbiota may influence the development of diseases caused by phytopathogens20,21. So far, there have been several reports on bacterial taxa identified in soils that inhibit development of certain plant diseases22,23. For instance, soil containing bacteria from the genera Streptomyces, Pseudomonas and Bacillus was described to suppress Fusarium wilts22. Furthermore, the soil-borne bacterial pathogen Ralstonia solanacearum, which is responsible for bacterial wilt of tomato, was noted to trigger less devastating disease symptoms in soil containing highly diverse bacterial communities enriched with bacteria from the genera Olivibacter, Flavihumibacter and Flavobacterium23. Similarly, a drop in the relative abundance of R. solancearum was associated with an elevation in Arthrobacter, Streptomyces, and Nocardioides detection frequencies. Additionally, the presence of Gracilibacillus, Cellvibrio, Bacillus, and Paenibacillus decreased the number of R. solancearum cells, while the occurrence of Propionibacterium contributed to the raise in this parameter24. Interestingly, Weller et al. hypothesized that beneficial microbiota is transferable between diverse soils25. This concept was proven by performing sterilization and later on inoculation of four agricultural soils with favorable bacteria that led to successive suppression of wheat root disease caused by Fusarium culmorum26. Nowadays, bacterial community profiling methods offer more precise views on the structural diversity of the soil microbiota in terms of detection and identification of non-culturable bacteria27.
The goal of this case study was to evaluate if there are differences in the composition of bacterial community of the soils collected from two fields of highly various incidences of soft rot and blackleg diseases. To achieve this aim, the soils have been characterized in terms of physicochemical properties, the occurrence of SRP was investigated by classical culture methods, while bacterial diversity and abundance of specific taxa were addressed by undertaking 16S rRNA gene amplicon sequencing followed by bioinformatic analyses.
Results and discussion
The incidence of blackleg and soft rot diseases on the studied potato farmlands
The types of the soils from both the studied fields (located in Siemysl and Bonin, Poland) were defined as clay according to the classification of the Polish Soil Science Society: Grading classification of soils and mineral formations. Regarding the potato field located in Siemysl, there were no blackleg symptoms identified throughout the entire growing season of 2021 (Table 1). On this basis, this field was selected for further analyses as of low blackleg and soft rot diseases incidences. In the case of the field of high diseases incidences situated in Bonin, the frequency of blackleg symptoms observed in this area equaled 2% in July and subsequently increased to 20% in the month of the harvest, i.e. August. Concerning the rates of diseases incidences noted during the previous five years, no blackleg nor soft rot symptoms were observed on plants growing at the Siemysl site, in contrast to the Bonin location. In the latter area also the symptoms of other plant diseases were noted in the last 5 years (Table 1).
As cropping and management practices are important factors influencing the occurrence of plant pathogens28, surveys on fertilizers and pesticides applied throughout the growing season on the two fields of interest were performed (Table 1). Both potato fields were amended with the same types of pesticides and mineral fertilizers with the sole difference of application of the aftercrop at the Bonin site. Considering that SRP were proven to be able to utilize organic fertilizers as sources of nutrients29 and to survive better in association with plant residues30, we anticipate that addition of the aftercrop might have had some impact on higher blackleg and soft rot diseases incidences recorded in the field in Bonin than at the Siemysl site.
Physicochemical characteristics of the collected soil samples
Afterwards, physicochemical properties, i.e. pH, salinity, organic matter and the contents of microelements and macroelements, of the soils collected from two fields of diverse blackleg and soft rot diseases incidences were analyzed (Table 2). Previously, the temperature, moisture and pH have been shown to influence the survival of SRP in soil31. In the current study, the soils originating from two fields either of low or high diseases incidences exhibited pH values of 6.1 and 6.4, respectively, which allowed for classification of these soils as “slightly acidic” (pH 5.5 ÷ 6.5) according to the Polish Soil Science Society. The moisture of the soils from the both examined fields was approximately equal (Table 2). Also the temperature at each collection site was alike. Altogether, as solely slight differences in pH, moisture, and temperature were recorded between the soils collected from two fields of low and high diseases incidences, it was concluded that these factors alone could not have accounted for the observed differences in the occurrence of blackleg and soft rot symptoms at the studied farmlands.
During the conducted physicochemical analyses of two soils, some deviations were noted in the composition of minerals between the studied fields (Table 2). Three times more Mg and twice as much Mn were determined in the soil collected from the field of low compared to high diseases incidences and these deviations exhibited statistical significance. The previous studies showed that Mn ions inhibit the production of pectin lyases, one of the most important virulence factors of Dickeya and Pectobacterium spp.32. On the other hand, Mg assures greater resistance of the older plant tissues to the necrotrophic pathogens as this constituent of the middle lamella together with Ca boost the resistance of pectic chains to the action of pectolytic enzymes. Moreover, the elevated Mg levels in tubers were discovered to be associated both with the reduced occurrence of soft rot, as observed by McGuire and Kelman33, and with the decreased stem rot incidence, as indicated by Bain et al.34. Anyhow, the impact of Mg on rot prevention was not as pronounced as that of the equivalent amounts of calcium (Ca). These experimental findings align with the results of Dubois et al., who demonstrated that the increased Mg input could effectively delay blackleg disease outbreaks30. Based on this data we can assume that the higher concentration of Mg in the soil acquired from the potato field of low in contrast to high diseases incidence might contribute to the lack of blackleg and soft rot diseases symptoms observed at this location.
Interestingly, the soil obtained from the field showing low diseases incidences contained about twice as much organics as the one collected from the field of high diseases incidences, and the herein-mentioned results are statistically significant. Ficke et al. provided evidence that the enrichment of soil with organic compounds extended the survival of P. atrosepticum10. Thus, it might be hypothesized that even if pectinolytic bacteria survived longer in soil at the Siemysl site in contrast to the Bonin location, their persistence did not lead to the increased diseases incidences due to the impact of some other environmental or biotic factors.
It is worth acknowledging that out of the 17 examined physicochemical features, solely 4 i.e. the contents of Mn, Mg, organic C and organic substance, differed in a statistically significant manner between the soils acquired from two fields of low and high diseases incidences. This finding suggests that it is worth to search for other factors that might have contributed to the observed differences between the investigated two fields of various blackleg and soft rot diseases incidences.
The occurrence of Dickeya spp. and Pectobacterium spp. in the studied potato plantations
Microbiological analysis of soil samples collected from two fields of low and high soft rot and blackleg diseases incidences involved culturing of the serially-diluted plant homogenates and soil filtrates on Crystal Violet Pectate (CVP) media in addition to PCR-based identification to the species level of the so-acquired SRP strains. Importantly, the mother tubers were not subjected to microbiological analysis for the presence of Dickeya spp. and Pectobacterium spp. before planting. Also plants and stems that grew in both fields were not tested for the presence of bacteria from the Pectobacteriaceae family.
As shown in Table 3, four Pectobacterium spp. strains were obtained from the soil collected from the field of high diseases incidences (Bonin), whereas no Dickeya nor Pectobacterium spp. were detected in the case of soil obtained from the field of low diseases incidences (Siemysl). Additionally, 16 strains of Pectobacterium spp. were isolated from young potato tubers harvested from the field showing high soft rot and blackleg diseases incidences. Six out of these strains were initially classified as P. carotovorum/P. parmentieri with the use of multiplex PCR method35. On the other hand, no pectinolytic Pectobacteriaceae were detected in the young potato tubers harvested from the field of low diseases incidences.
Based on the BLAST comparison of the sequences of fragments of dnaX37 and recA38 genes, the isolated pectinolytic bacteria were identified. Three strains of P. brasiliense (IFB5712; IFB5713; IFB5714) and 1 strain of P. betavasculorum (IFB5711) were detected in soil of high disease incidence. Moreover, 6 strains of P. brasiliense (IFB5705; IFB5706; IFB5707; IFB5708; IFB5709; IFB5710), 2 strains of P. betavasculorum (IFB5703; IFB5704) and 8 strains of P. versatile (IFB5695; IFB5696; IFB5697; IFB5698; IFB5699; IFB5700; IFB5701; IFB5702) were identified in young tubers (Table 3). The sequences of dnaX and recA genes of Pectobacterium spp. isolates were submitted to the GenBank database and are available under the following accession numbers: PP869790, PP869791, PP869792, PP869793, PP869794, PP869795, PP869796, PP869797, PP869798, PP869799, PP869800, PP869801, PP869802, PP869803, PP869804, PP869805, PP869806, PP869807, PP869808, PP869809, PP869810, PP869811, PP869812, PP869813, PP869814, PP869815, PP869816, PP869817, PP869818, PP869819, PP869820, PP869821, PP869822, PP869823, PP869824, PP869825, PP869826, PP869827, PP869828, PP869829.The analysis of the obtained sequences allowed to conclude that the same single nucleotide polymorphisms (SNPs) were present in the sequences of recA and dnaX amplified from the DNA of P. brasiliense strains from soil and young tubers. Similar observation on the occurence of identical SNPs among strains of various origin concerned P. betavasculorum.
The genomic profiling with the use of ERIC primers39 (Fig. S1), was applied to examine clonality of the analyzed strains. The acquired ERIC patterns for IFB5705, IFB5706, IFB5707, IFB5708, IFB5709 and IFB5710 strains of P. brasiliense isolated from young tubers and IFB5712 and IFB5713 acquired from soil were the same (Fig. S1). Only P. brasiliense IFB5714 isolated from soil showed a different ERIC profile from the other 8 isolates belonging to this species. Moving to the representatives of the next pectinolytic species P. betavasculorum, these strains were identified both in soil and young tubers. We observed that all three strains IFB5703, IFB5704 (both from young tuber) and IFB5711 (soil) showed the same ERIC profile (Fig. S1). Finally, the P. versatile strains were isolated only from young tubers and according to the results of genomic profiling, all 8 strains were clonal (Fig. S1).
The presence of SRP in the Bonin potato field (with a total of 20 acquired Pectobacterium spp. strains) and the absence of such strains in the farmland in Siemysl is in agreement with the data collected during the long-term monitoring of these potato fields performed by our research group14,15. The herein presented data indicate that SRP were detected only in the potato field in Bonin and that a higher number of strains was acquired from potato tubers than from the soil. This observation is most likely associated with a higher survival rate of Dickeya spp. and Pectobacterium spp. cells in association with plant tissues than in the less favorable soil environment17. According to the literature, SRP can be efficiently isolated from post-harvest plant residues that provide a suitable environment for growth and multiplication of bacteria17. The latter remark is in line with the recent data of Toth et al. indicating that SRP populations are often detected after potato harvesting, but over the winter period their numbers decrease to very low levels8. Additionally, ERIC-based genomic profiling showed that in terms of both species, i.e. P. brasiliense and P. betavasculorum, the strains isolated from soil and young tubers collected from the field of high disease incidence were clonal. This observation may indicate that potato plants have been infected by the Pectobacteriaceae present in the soil. However, as the mother tubers had not been tested before planting for the presence of pectinolytic bacteria, we cannot exclude that they formed the source of inoculum. In addition, the presence of pectinolytic bacteria was also not examined in progeny plants. The obtained results are consistent with previous data stating that pectinolytic bacteria of either mother tubers or soil origin are both capable of causing wilting and blackleg disease symptoms on potato under greenhouse and field conditions40.
The numbers of amplicon sequence variants (ASVs) and bacterial diversity in the soil samples collected from two fields of either low or high blackleg and soft rot diseases incidences
Sequencing was performed on 16S rRNA amplified from a library generated from total DNA isolated from five soil samples collected either from the field of low or high soft rot and blackleg diseases incidences. The number of paired raw reads obtained for each sample ranged from 91,124 to 101,293. On average 76% of all the readings passed the quality control threshold (Table S1). After the clustering step, the number of the determined unique ASVs enclosed in the range of 767–1024 ASVs per sample (Table S1). The numbers of ASVs computed for the soil samples originating either from the field of low or high diseases incidences are comparable. The rarefaction curves obtained for the generated ASVs reached a plateau for all the included soil samples (Fig. S2), which indicated an achievement of a satisfactory sequencing depth for studying bacterial diversity of soils acquired from two fields of either low or high blackleg and soft rot diseases incidences.
In order to get a better insight into the bacterial diversity of soils collected from these two fields, alpha- and beta-diversities were calculated. At first, the alpha diversity estimates were computed using Evenness (Pielou) index. No notable deviations were detected in the alpha-diversity of the soils obtained from the field of low in juxtaposition to high blackleg and soft rot diseases incidences (p > 0.05; Kruskal–Wallis test) (Fig. 1A). However, the samples of soil collected from different parts of the field of low diseases incidences varied to a greater extent in terms of complexity of bacterial populations than the replicates obtained from the field of high diseases incidences.
Alpha and beta-diversity indices of the soil samples collected from two fields either of low or high blackleg and soft rot diseases incidences. (A) Pielou’s indices. The differences were not statistically significant based on Kruskal–Wallis tests (p > 0.05). (B) Principal coordinates analysis (PCoA) using the Bray–Curtis distance matrix. PC1 and PC2 represent the two principal components, and the percentage values refer to the contribution of the principal component to the differences between samples.
Subsequently, PCoA and PERMANOVA were used to investigate bacterial beta-diversity in the examined soils. PCoA calculations using the Bray–Curtis distance matrix were performed to determine the relatedness of soil samples acquired from each of two fields of diverse diseases incidences. The PCoA plot revealed separate grouping of soil samples originating from the field of low comparing to high diseases incidences (Fig. 1B), suggesting some structural differences between the two studied fields. PERMANOVA-based analysis pointed to statistical significance of the observed deviations according to pseudo-F at p = 0.01 (Fig. S3). The obtained data suggests that the bacterial communities of the soils collected from two potato fields of either low or high diseases incidences differ. According to the literature a high diversity in soil bacterial community has been associated with inhibition of pathogens23, the herein observed dissimilarities in biodiversity of bacterial populations between the examined fields might have had some reflection in the frequencies of detection of blackleg and soft rot symptoms. In turn, according to the research conducted by Hamed et al.41 covering the impact of algal-based products on the development of brown rot disease caused by R. solanacearum, it was shown that the soil infested by this plant pathogen differs significantly in population biodiversity from the non-infested soil. Moreover, it was demonstrated before that the soil microbial diversity affects the ability of R. solanacearum to induce wilting disease in tomato24,42.
Determination of the bacterial constituents of soils collected from two fields of either low or high blackleg and soft rot diseases incidences
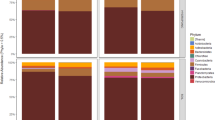
The structural diversity of bacterial communities at the phylum level is shown in Fig. 2. Actinobacteriota, Proteobacteria, Acidobacteriota, Chloroflexi, Firmicutes, and Gemmatimonadota were the six most abundant bacterial phyla and together accounted for more than 85% of all the identified ASVs. The contribution of the most frequently represented phyla to the studied bacterial populations of two investigated fields was comparable in the case of all the analysed soil samples.
Our findings are in agreement with the analysis conducted by Cangioli et al. on the taxonomic composition of bacterial microbiota in four potato farmlands of Italy43. There, Acidobacteriota, Actinobacteriota, Chloroflexi and Proteobacteria turned out to be the four phyla of the highest abundance. The above-listed bacterial taxa were also present among the herein revealed 10 most frequently detected phyla in the two investigated potato fields in Poland. A similar distribution of these phyla was also observed in a study conducted in Japan, in which sweet potato fields were sampled from June to September 2017 across three different geographical locations44.
Going down in the taxonomic rankings of the identified bacterial microbiota in the soils acquired from two fields of either low or high diseases incidences (Fig. 3), a clear sharp clustering of the samples from the aforementioned fields was visible if the focus was turned into bacterial classes. Here, the most abundant classes were Actinobacteria, Alphaproteobacteria, Gammaproteobacteria and Thermoleophilia. It is worth noticing that among these groups, the members of Thermoleophilia were shown to be present in significantly higher numbers in the field of low diseases incidences in contrast to the one of high diseases incidences (p < 0.05). On the contrary, concerning the other classes exhibiting low abundance, no significant differences based on the Wilcoxon rank sum test were found. The presence of Thermoleophilia in soil can influence the overall microbial diversity45. Furthermore, these bacteria might contribute to breaking down organic matter and play a role in nutrient cycling processes, especially in the environments in which the temperatures form a limiting factor for multiplication of other microorganisms46. Intriguingly, Yang et al., observed that soil salinity negatively affects the number of the representatives of Thermoleophilia47. Nonetheless, additional research on the contribution of the members of this class to the natural bacterial populations is needed, given that only a small number of strains belonging to this group has been cultured and characterized thus far48.
The clustered heatmap of ASVs identified according to the class level in the samples of soils collected from two investigated fields either of low or high diseases incidences L: field of low diseases incidences, H: field of high diseases incidences. If the Wilcoxon Rank Sum Test showed statistically significant differences at p < 0.05, they are marked with *.
Upon transitioning to the order level, we identified statistically significant differences (Wilcoxon Rank Sum; p < 0.05) between the investigated soils concerning the bacteria from Ktedonobacterales, Clostridiales, Saccharimonadales, Micropepsales, Frankiales, Acidobacteriales, Acetobacterales, Solirubrobacterales, Solibacterales, Chthoniobacterales, Streptomycetales, and Xanthomonadales orders (Fig. 4). It is also interesting to acknowledge the presence of Acidobacteriales, Acetobacterales, Streptomycetales, and Bacillales among the 30 most differently abundant orders basing on log2fold change considering that the members of these taxa have been recognized before for their effectiveness in inhibiting the growth of pathogenic bacteria25. Different representation of Streptomycetales in the soils from two fields of contrasting soft rot and blackleg diseases incidences finds confirmation in the former data on decreases in the numbers of three Actinomycetes families, including Streptomycetaceae, post subjection of the soil suppressing the fungal root pathogen Rhizoctonia solani to selective heat (80 °C; 1 h), as the latter treatment led to total elimination of the inhibitory effect of that soil26. Furthermore, our analysis revealed 12 orders of bacteria of significantly higher abundance in the field characterized by low diseases incidences compared to the one of high diseases incidences. Noteworthily, there are among them Gaiellales and Solirubrobacterales orders, members of the Thermoleophilia class that also exhibited statistically significant differences in abundance between the herein investigated two potato fields.
The log2fold change among 30 most differentially represented bacterial orders between the field of low and high diseases incidences. Values above zero indicate that a stated order is more abundant in the soil collected from the field of low diseases incidences, whereas log2fold change below zero points to higher abundance of a certain bacterial order in the soil acquired from the field of high diseases incidences. Taxa, which are according to the literature known for involvement in the control of plant pathogens, are displayed in bold49. If the Wilcoxon Rank Sum Test showed statistically significant differences at p < 0.05, they are marked with *.
Finally, the comparison of the normalized counts for the four most divergently represented genera between two fields of interest, led to identification of Bacillus, Rummeliibacillus, Acidobacterium and Gaiella as the genera present in high numbers in the field of low soft rot and blackleg diseases incidences in contrast to their total absence in the field of high occurrence of SRP-triggered disease symptoms (Fig. 5). In the case of Gaiella spp., which belongs to the Thermoleophilia class, we noted an overlap with the results obtained from the insight into the previously examined bacterial taxonomic levels (classes and orders) (Figs. 3 and 4). The total numbers of the occurrences of ASVs, assigned to the genera Bacillus and Rummeliibacillus in the soil acquired from the field of low diseases incidences, were 209 and 135, respectively, while in the soil obtained from the field of high diseases incidences these ASVs were not found at all. In turn, the counts of ASVs corresponding to the Gaiella and Acidobacterium genera in the soil samples from the field of low diseases incidences were 188 and 133, respectively. As in the case of Bacillus and Rummeliibacillus, the ASVs assigned to Gaiella and Acidobacterium did not occur in the soil samples collected from the field of high diseases incidences. These results point to interesting differences in the presence of bacterial taxa between the two studied fields.
Noteworthily, the representatives of the genus Bacillus were identified more frequently in the soil collected from the field showing low in contrast to high incidence of blackleg and soft rot, which is in accordance with the fact that Bacillus spp. belongs to plant growth promoting bacteria (PGPR), well-known for improving plant health and productivity50. PGPR act by three diverse mechanisms: production of antagonistic biocidals, releasing substances promoting plant growth and/or induction of plant defense systems49. Furthermore, the use of bioformulations with different strains of Bacillus spp. was stated to restore the biological balance and self-regulating capacity of the soil49. For example, Bacillus amyloliquefaciens subsp. plantarum51 and Myxococcus sp.52 isolated from soil have been identified as potential biocontrol agents that could efficiently inhibit development of soft rot diseases. Moreover, Weinert et al.53, observed, while studying microbial diversity on the surface of the soil-grown potato tubers, that the occurrence of Bacillus spp. and Streptomyces spp. was responsible for suppressing disease symptoms caused by potato pathogens Rhizoctonia solani, Verticillium dahliae or Phytophthora infestans. It was also suggested that Bacillus spp. can improve the fertilization efficacy and provide plants with essential elements, e.g. phosphorus, nitrogen, potassium, manganese and iron, from the limited soil resources54. Additionally, bacteria from this genus support plants in acquisition of other nutrients or production of phytohormones, exhibit antagonistic activity against phytopathogens and/or improve crops protection against abiotic stresses55.
In turn, Gaiella spp. is one of the representatives of the Thermoleophilia class, whose members have been so far isolated from the aquatic and soil environments. Zhao et al. showed positive relationship between the abundance of bacteria from the Gaiella group and physicochemical properties of the soil (pH, NH4+, NO3, soil organic matter and dissolved organic carbon)56. A notable linkage between physicochemical properties of the soil and the presence of Gaiella spp. was also demonstrated during the analyses of sugarcane fields57.
Moving to Acidobacteria, a significant dominance of bacterial phyla Acidobacteria, Actinobacteria and Firmicutes was demonstrated by Xiong et al. in soil suppressing development of Fusarium wilt disease in vanilla58. Among others, Acidobacteria participate in biogeochemical cycles, involving carbon, nitrogen or sulphur, by these means contributing to the balance in the plant-soil ecosystem. These microorganisms not only play a critical role in recycling organic matter by increasing the organics and nitrogen contents in soil along with boosting the availability of the essential macro and microelements, but also Acidobacteria secrete exopolysaccharides, which participate in the formation of the soil matrix and contribute to plant growth promotion by facilitating the water and nutrients uptake59. In addition, some species of Acidobacteria produce a phytohormone indole-3-acetic acid (IAA) and siderophores, molecules showing activities that significantly improve plant growth parameters60.
Previous research demonstrated that different microorganisms are antagonistic to Dickeya spp. and Pectobacterium spp. The so-far obtained strains showing biocontrol potential belong mostly to the genera Bacillus, Pseudomonas or Serratia61,62,63,64. Gerayeli et al.65 revealed antagonistic action of Bacillus subtilis, Bacillus pumilus, Bacillus amyloliquefaciens, and Bacillus thuringiensis strains against P. brasiliense. Also, Des Essarts et al.66 isolated Bacillus simplex and Pseudomonas brassicacearum strains, which inhibited the development of P. atrosepticum-induced symptoms on plants besides acquisition of Pseudomonas fluorescens and P. putida strains that exhibited high antagonistic action against D. dianthicola. In the latter study, the noted inhibitory effects, observed both under laboratory and greenhouse conditions, have been attributed to the presence in the genomes of antagonistic bacteria of specific clusters responsible for the production of bacteriocins, adhesins and siderophores67.
Determination of the constituents of bacterial microbiota of the soil may in the future serve as one of predictive factors contributing to assessment of risks of blackleg and soft rot diseases development at a stated location. The prognostic value of soil microbial inhabitants might have been exploited during search for optimal fields for cultivating elite seed potatoes. In addition, this research points to bacterial taxa enclosing considerable candidates for development of sophisticated and efficient biological formulations that could potentially serve for plant protection purposes in the modern sustainable agriculture biocontrol schemes.
Conclusions
In this work, two potato fields of various incidences of blackleg and soft rot diseases were determined to share 17 physicochemical features and to differ in Mg, Mn, organic C and organic substance contents. Relying on classical culturing methods, 20 Pectobacterium spp. strains were isolated from soil and tubers growing on the field of high disease incidences. Molecular diagnostics methods allowed for identification of P. brasiliense (6 strains), P. betavasculorum (3 strains) and P. versatile (8 strains). In the presented case study, the mother tubers were not tested before planting for the presence of pectinolytic bacteria. Based on this, we cannot exclude that they formed the source of inoculum. By applying sequencing of 16S rRNA followed by bioinformatic analysis, the two studied fields showed similar alpha-diversity, but contrasting beta-diversity values. Examination of the abundances of various bacterial taxa, revealed that the members of Thermoleophilia class were present in higher numbers in the field of low in juxtaposition to high diseases incidences. Also, the members of Bacillus, Rumeliibacillus, Acidobacterium, and Gaiella genera were more commonly found in soil samples collected from the field showing low soft rot and blackleg diseases incidences, as opposed to the field of high diseases incidences. The herein presented case study demonstrated, for the first time, considerable differences in the occurrences of specific bacterial taxa between two fields of various soft rot and blackleg diseases incidences.
Materials and methods
Sampling of potato fields and physicochemical analysis of the soil
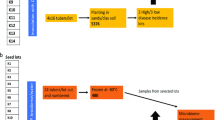
The soil samples were collected in July 2021 from two potato fields, showing either low or high soft rot and blackleg diseases incidences. Five soil samples were picked up from each of these two fields. The fields selected for this case study are located in Siemysl (54° 38′ E, 18° 58′ N) and Bonin (54° 15′ E, 16° 24′ N) in Poland, respectively. At the time of sampling and in the previous week, it was sunny and the air temperature approximated 30 °C at 12.00 PM. The mineral formations of the collected soil samples were assigned on the basis of the soil classification system of the Polish Society of Soil Science by relying on the grain size. From each field, five soil samples were collected with a sterile metal spatula and placed directly into sterile plastic string bags. The soil was collected exactly from beneath the potato plants at a depth of 20 cm. In each field, the sampling points were located at 5 m distance one from another. The soil samples intended for DNA isolation were immediately frozen and kept on dry ice. The samples designated for microbiological and chemical analyses were transported to the Laboratory of Plant Protection and Biotechnology at the Faculty of Biotechnology University of Gdansk and Medical University of Gdansk (IFB UG & MUG) at an ambient temperature and then stored one week at 4 °C. In addition, soil samples and potato tubers were collected during harvest in August and sent to IFB UG & MUG for analyses involving detection and identification of pectinolytic bacteria.
The physicochemical composition of the soil samples was determined by the District Chemical and Agricultural Station in Gdansk, Poland. Three samples from each field were analysed in terms of their pH, salinity and the contents of microelements and macroelements such as: N–NO3, P, K, Mg, Ca, Cl, Na, Zn, Cu, Mn, Fe, B, organic C and organic substance. This analysis was performed in accordance with the research procedure including the scope of accreditation for the testing laboratory for horticultural soil (No AB 787). Additionally, the moisture of the collected soil samples was examined by a gravimetric method68. In the latter case, the percentage average moisture content (weight; W) was calculated from the ratio of the water mass present in the soil (Mw) to the mass of the solid components of the soil post drying (mass after drying; Ms) according to the following formula \(W=\frac{Mw}{Ms}x100\%\). In order to compare physicochemical properties of the soils acquired from two fields of either high or low blackleg and soft rot diseases incidences, individual Student’s t-tests were performed for each feature examined in terms of the statistical significance of differences between means.
Isolation and identification of Dickeya spp. and Pectobacterium spp. strains by culturing and molecular diagnostics approaches
The soil and young tuber samples collected from two studied fields were tested for the presence of pectinolytic bacteria from the genera Dickeya and Pectobacterium by using a slightly modified methodology of Potrykus et al.15. Molecular detection of pectinolytic bacteria was not conducted on mother tubers prior to planting. Furthermore, the presence of pectinolytic bacteria was not tested in progeny stems either. In terms of young tubers samples, 1 g of this material was added to 10 ml of 50 mM phosphate buffer pH 7.2, mixed and manually homogenized with a Bioreba device (Bioreba, Switzerland). Regarding soil samples, 10 g of soil were suspended in 50 ml of pectate enrichment medium (PEM)69 and incubated for 48 h at 28 °C with 140 rpm shaking. Afterwards, soil suspensions were filtered through sterile paper discs. The resulting plant homogenates and soil filtrates were serially diluted and plated on CVP media70 prior to incubation for 48 h at 28 °C. The pectinolytic bacterial colonies forming characteristic cavities on CVP were further replated on CVP, and then TSA, until the axenic culture state was reached.
Bacterial cells were lysed by suspending a single colony in 1 ml of distilled water and providing cold shock conditions for 1 h at -20 °C. Subsequently, these suspensions were heated up for 10 min at 99 °C prior to transferring 100 µl of the resultant bacterial lysate to 400 ml of sterile distilled water. Multiplex PCR and a single PCR with Y1/Y2 primers were performed to identify pectinolytic strains to the species level as described previously35,36. The applied multiplex PCR reaction included Df and Dr primers (for identification of the Dickeya genus71), Y45 and Y46 primers (targeting P. atrosepticum)72 in addition to ExpccF and ExpccR primers (for bacteria formerly classified as P. carotovorum)73.
The 20 isolated pectinolytic strains were cultured overnight in tryptone soya broth (TSB) (BTL, Łodz, Poland) at 28 °C prior to isolation of genomic DNA using the Genomic Mini AX Bacterial Kit (A&A Biotechnology, Łodz, Poland). The quality and concentration of the obtained DNA were evaluated with a NanoDrop ND-1000 (Thermo Fisher Scientific, Minneapolis, MN, USA). Single PCR reactions were performed according to the formerly published protocols37,38 to amplify the fragments (535 and 730 bp, respectively) of dnaX and recA housekeeping genes. The acquired amplicons underwent sequencing from both ends, which was commissioned as a commercial service to the Genomed company (Warsaw, Poland). The chromatograms were manually edited and aligned utilizing the MEGA Software (Auckland, New Zealand) by employing the default parameters of ClustalW algorithm. The generated consensus sequences of the dnaX and recA gene fragments originating from the herein isolated pectinolytic strains were compared with the corresponding gene fragments deposited in the GenBank database (accessed: June 4, 2024), which enabled to identify these strains to the species level.
Genomic profiles of 20 pectinolytic isolates were generated using repetitive sequence-based (REP) PCR with ERIC1F and ERIC2R primers, following the method described previously39. The resulting amplicons were separated by electrophoresis in 1% agarose (Basica) gel suspended in 0.5 × Tris–borate-EDTA buffer. The separation lasted 4 h at 50 V. Then the gel was stained in 5 mg mL−1 ethidium bromide, washed in distilled water and visualized with the Gel Doc imaging system (Bio Rad Laboratories Inc.) using Quantity One Software (Bio Rad Laboratories Inc.).
Extraction of the total DNA from soil and sequencing of the 16S rRNA library
Whole DNA from five soil samples per each of the two investigated fields was isolated with the use of a commercially available NucleoSpin Soil Mini kit (Macherey–Nagel, Duren, Germany) following the manufacturer’s guidelines. During the isolation procedure, 300–400 mg of soil per sample and the SL1 lysis buffer were utilized. Purification of the isolated DNA was accomplished with NucleoSpin gDNA Clean‑up Mini kit (Macherey–Nagel, Duren, Germany). The quality and concentration of the isolated DNA were assessed spectrophotometrically with NanoDrop ND-1000 (Thermo Fisher Scientific, Minneapolis, MN, USA). At least 3 μg of DNA (OD260/280 1.8–2.0) per soil sample were sent to Genomed (Warsaw, Poland) for preparation of libraries and sequencing of amplicons of the hypervariable V3-V4 region of the 16S rRNA gene. In more detail, the 341F (5'-CCT ACG GGN GGC WGC AG-3') and 785R (5'-GAC TAC HVG GGT ATC TAA TCC-3') primers were used to amplify the selected 16S rRNA region. The preparation of the library was based on performing PCR reactions with the KAPA HiFi HotStart ReadyMix, attaching dual indices and Illumina sequencing adapters using the Nextera XT Index Kit. PCR reaction was performed with the Q5 Hot Start High-Fidelity 2 × Master Mix (New England Biolabs, Ipswich, USA). The DNA was sequenced on the Illumina MiSeq instrument post utilization of the Illumina v3 kit (Illumina, San Diego, USA) in order to generate 300 bp paired-end reads. An automatic preliminary report was generated on the MiSeq sequencer using the MiSeq Reporter (MSR) v2.6 software (Illumina, San Diego, USA).
Processing of the raw data, clustering and taxonomic assignment of the reads
Initial quality control of the reads, including investigation of the error profile of individual samples and generation of the dynamic parameters for quality control, was conducted with the FIGARO tool74 by Genomed (Warsaw, Poland). Subsequently, pre-processing of the data relied on the removal of the adapter sequences and rejection of the short reads (< 30 nt) with the Cutadapt tool75. Final quality assessment of the filtered data was performed with FASTQC76.
Bioinformatic analysis, which allowed for taxonomic classification of the soil-inhabiting microbes basing on 16S rRNA reads, was carried out with QIIME 277. The obtained reads were denoised and unique sequences of biological origin, i.e. ASVs were assigned to them by the DADA2 pipeline78. In the next step, a self-trained naive Bayes classifier and reference data from the Silva 138 nr_v138 database were implemented to assign ASVs to the specific bacterial taxa79. The annotated ASVs count tables were processed using the ‘Phyloseq’ R package (RStudio version 2021.09.0)80.
Downstream analysis of the 16S rRNA gene amplicon sequencing data
Evaluation of diversifications between bacterial populations involved providing an insight into alpha and beta diversities. In terms of alpha diversity, Evenness (Pielou) indices for the soil samples, collected either from the field of high or low diseases incidences, were calculated. Beta diversity was analyzed by a non-parametric, multivariate statistical test Permutation Based Analysis of Variance (PERMANOVA) in addition to Principal coordinates analysis (PCoA) based on Bray–Curtis distances. Finally, rarefaction curves were generated. The conducted statistical analysis was based on Kruskal–Wallis test (p < 0.05) for alpha-diversity and pseudo-F test (p < 0.05) for beta-diversity.
Visualization of the results and statistical analyses were performed in R. In the case of the clustered heatmap of ASVs, the relative frequencies were normalized as follows 1/(abs(log10(aggregated_data_phylum + 0.000001))). For statistical evaluation, Wilcoxon Rank Sum Test was applied (p < 0.05). Differential abundance analysis was carried out using the R package DeSeq2 by juxtaposition with the reference sequences deposited in two databases, i.e. NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Sequence Match-RDP (https://www.lcsciences.com/documents/sample_data/16S_sequencing/src/html/top1.html) in order to identify the ASVs/taxa differing in a statistically significant manner between the studied two fields81. The relative frequencies were normalized as follows 1/(abs(log10(relative frequency + 0.000001))), where 0.000001 represents a pseudo-count introduced here to avoid mathematical inconsistency.
Data availability
The 16S rRNA raw sequencing data are publicly available in the Sequence Read Archive repository at https://www.ncbi.nlm.nih.gov/sra/PRJNA902273. All data generated or analyzed during this study are included in this published article (or in its supplementary information files) or will be provided by the corresponding author after a reasonable request. The sequences of housekeeping genes dnaX and recA of 9 strains of P. brasiliense (IFB5712; IFB5713; IFB5714; IFB5705; IFB5706; IFB5707; IFB5708; IFB5709; IFB5710), 3 strains of P. betavasculorum (IFB5711; IFB5703; IFB5704) and 8 strains of P. versatile (IFB5695; IFB5696; IFB5697; IFB5698; IFB5699; IFB5700; IFB5701; IFB5702) were deposited in the GenBank database under the following accession numbers: PP869790, PP869791, PP869792, PP869793, PP869794, PP869795, PP869796, PP869797, PP869798, PP869799, PP869800, PP869801, PP869802, PP869803, PP869804, PP869805, PP869806, PP869807, PP869808, PP869809, PP869810, PP869811, PP869812, PP869813, PP869814, PP869815, PP869816, PP869817, PP869818, PP869819, PP869820, PP869821, PP869822, PP869823, PP869824, PP869825, PP869826, PP869827, PP869828, PP869829.
References
Ben Moussa, H., Pédron, J., Bertrand, C., Hecquet, A., Barny, M. A. Pectobacterium quasiaquaticum sp. nov., isolated from waterways. Int. J. Syst. Evol. Microbiol. 71. https://doi.org/10.1099/ijsem.0.005042 (2021).
Toth, I. K. et al. Pectobacterium and Dickeya: Taxonomy and Evolution. In: Van Gijsegem, F., van der Wolf, J.M., Toth, I.K. (eds). Plant Diseases Caused by Dickeya and Pectobacterium Species. Springer, Cham. 13–37. https://doi.org/10.1007/978-3-030-61459-1_2 (2021).
Zhou, J. et al. Isolation and genome analysis of Pectobacterium colocasium sp. nov. and Pectobacterium aroidearum, two new pathogens of taro. Front. Plant Sci. 13, 1141. https://doi.org/10.3389/fpls.2022.852750 (2022).
Motyka, A., Zoledowska, S., Sledz, W. & Lojkowska, E. Molecular methods as tools to control plant diseases caused by Dickeya and Pectobacterium spp: A minireview. N. Biotechnol. 39, 181–189. https://doi.org/10.1016/J.NBT.2017.08.010 (2017).
Birch, P. R. J. et al. Crops that feed the world 8: Potato: Are the trends of increased global production sustainable? Food Secur. 4, 477–508. https://doi.org/10.1007/S12571-012-0220-1 (2012).
Van der Wolf, J. M. et al. Diseases caused by Pectobacterium and Dickeya Species Around the World. In: Van Gijsegem, F., van der Wolf, J.M., Toth, I.K. (eds). Plant Diseases Caused by Dickeya and Pectobacterium Species. Springer, Cham. 215–261. https://doi.org/10.1007/978-3-030-61459-1_7 (2021).
Dupuis, B., Nkuriyingoma, P., Van Gijsegem, F. Economic Impact of Pectobacterium and Dickeya species on potato crops: A Review and Case study. In: Van Gijsegem, F., van der Wolf, J.M., Toth, I.K. (eds). Plant Diseases Caused by Dickeya and Pectobacterium Species. Springer, Cham. 263–282. https://doi.org/10.1007/978-3-030-61459-1_8 (2021).
Toth, I. K. et al. Pectobacterium and Dickeya: Environment to Disease Development. In: Van Gijsegem, F., van der Wolf, J.M., Toth, I.K. (eds). Plant Diseases Caused by Dickeya and Pectobacterium Species. Springer, Cham. 39-84; https://doi.org/10.1007/978-3-030-61459-1_3 (2021).
Ge, T., Ekbataniamiri, F., Johnson, S. B., Larkin, R. P. & Hao, J. Interaction between Dickeya dianthicola and Pectobacterium parmentieri in potato infection under field conditions. Microorganisms. 9, 1–10. https://doi.org/10.3390/MICROORGANISMS9020316 (2021).
Ficke, W., Naumann K., Skadow, K., Müller, H., Zielke, R. Die Lebensdauer von Pectobacterium carotovorum var. atrosepticum (van Hall) Dowson auf dem Pflanzgut und im Bodem. Arch. Phytopathol. Plant Protect. 9, 281–293 (1973).
Van der Wolf, J., Czajkowski, R. & Velvis, H. Effectieve kolonisatie van aardappelplanten door Dickeya soorten (Erwinia chrysanthemi). Gewasbescherming Jaargang. 4, 169–171 (2009).
Czajkowski, R. et al. Detection, identification and differentiation of Pectobacterium and Dickeya species causing potato blackleg and tuber soft rot: A review. Ann. Appl. Biol. 166, 18–38; https://doi.org/10.1111/AAB.12166 (2015).
Peltzer, S., Sivasithamparam, K. Sero-groups of Erwinia carotovora associated with water, soil, tuber, and stems of potato plants in Western Australia. New Zealand J. Agric. 16, 265–270. https://doi.org/10.1080/03015521.1988.10425649 (1988).
Motyka-Pomagruk, A., Zoledowska, S., Sledz, W. & Lojkowska, E. The occurrence of bacteria from different species of Pectobacteriaceae on seed potato plantations in Poland. Eur. J. Plant Pathol. 159, 309–325. https://doi.org/10.1007/S10658-020-02163-X/FIGURES/6 (2021).
Potrykus, M. et al. Biodiversity of Dickeya spp. isolated from potato plants and water sources in temperate climate. Plant Dis. 100, 408–417. https://doi.org/10.1094/PDIS-04-15-0439-RE (2016).
Pérombelon, M., & Hyman, L. Survival of soft rot coliforms, Erwinia carotovora subsp. carotovora and E. carotovora subsp. atroseptica in soil in Scotland. J. Appl. Bacteriol. 66, 95–106. https://doi.org/10.1111/j.1365-2672.1989.tb02459.x (1989).
Burr, T. J., & Schroth, M. N. Occurrence of soft rot Erwinia spp. in soil and plant material. Phytopathology. 67, 1382–1387. https://doi.org/10.1094/Phyto-67-1382 (1977).
Gudmestad, N. C. & Secor, G. A. The bionomics of Erwinia carotovora in North Dakota. Am. Potato J. 60, 759–771. https://doi.org/10.1007/BF02856895 (1983).
McCarter-Zorner, N. J. et al. Soft rot Erwinia bacteria in the rhizosphere of weeds and crop plants in Colorado USA and Scotland UK. J. Appl. Bacteriol. 59, 357–368. https://doi.org/10.1111/j.1365-2672.1985.tb03331.x (1985).
De Corato, U. Governance of soil amendment to enhance suppression to soil-borne plant pathogens from a long-term perspective. Appl. Soil Ecol. 182. https://doi.org/10.1016/j.apsoil.2022.104721 (2023).
Schroth, M. N. & Hancock, J. G. Disease-suppressive soil and root-colonizing bacteria. Science. 216. https://doi.org/10.1126/science.216.4553.1376 (1982).
Wang, Q. et al. Investigating the responses of microbial communities to banana Fusarium Wilt in suppressive and conducive soils based on soil particle-size differentiation. Agronomy. 12, 229. https://doi.org/10.3390/agronomy12020229 (2022).
Deng, X. et al. Soil microbiome manipulation triggers direct and possible indirect suppression against Ralstonia solanacearum and Fusarium oxysporum. npj Biofilms Microbiomes. 7, 33. https://doi.org/10.1038/s41522-021-00204-9 (2021).
Messiha, N. A. S. et al. Enhancement of soil suppressive potential to bacterial wilt disease caused by Ralstonia solanacearum. Arch. Phytopathol. Pflanzenschutz. 56. 1127–1165, https://doi.org/10.1080/03235408.2023.2267668 (2023).
Weller, D. M., Raaijmakers, J. M., McSpadden Gardener, B. B. & Thomashow, L. S. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348. https://doi.org/10.1146/ANNUREV.PHYTO.40.030402.110010 (2002).
Ossowicki, A. et al. Microbial and volatile profiling of soils suppressive to Fusarium culmorum of wheat. Proc. R. Soc. B. 287. https://doi.org/10.1098/RSPB.2019.2527 (2020).
Sraphet, S. & Javadi, B. Unraveling techniques for plant microbiome structure analysis. Divers. 14, 206. https://doi.org/10.3390/D14030206 (2022).
Huber, D. M. Effect of organic amendment on soil-borne plant pathogens. Phytopathology. 60, 22. https://doi.org/10.1094/PHYTO-60-22 (1970).
Sledz, W., Zoledowska, S., Motyka, A., Kadzinski, L. & Banecki, B. Growth of bacterial phytopathogens in animal manures. Acta Biochim. Pol. 64. https://doi.org/10.18388/abp.2016_1389 (2017).
Dubois, G. E., Schaerer, S. & Dupuis, B. Factors impacting blackleg development caused by Dickeya spp. in the field. Eur. J. Plant. Pathol. 140, 317–327. https://doi.org/10.1007/s10658-014-0465-y (2014).
Czajkowski, R., Pérombelon, M. C. M., van Veen, J. A. & van der Wolf, J. M. Control of blackleg and tuber soft rot of potato caused by Pectobacterium and Dickeya species: A review. Plant Pathol. 60(6), 999–1013. https://doi.org/10.1111/AAB.12166 (2011).
Ouattara, H. G. et al. Implication of Bacillus sp. in the production of pectinolytic enzymes during cocoa fermentation. World J. Microbiol. Biotechnol. 24, 1753–1760. https://doi.org/10.1007/s11274-008-9683-9 (2008).
McGuire, R. G. & Kelman, A. Calcium in potato tuber cell walls in relation to tissue maceration by Erwinia carotovora pathovar atroseptica. Phytopathology. 76(4), 401–406 (1986).
Bain, R. A., Millard, P. & Perombelon, M. C. M. The resistance of potato plants to Erwinia carotovora subsp. atroseptica in relation to their calcium and magnesium content. Potato Res. 39(1), 185–193 (1996).
Potrykus, M. et al. Simultaneous detection of major blackleg and soft rot bacterial pathogens in potato by multiplex polymerase chain reaction. Ann. Appl. Biol. 165, 474–487. https://doi.org/10.1111/aab.12156 (2014).
Darrasse, A., Priou, S., Kotoujansky, A., Bertheau, Y. PCR and restriction fragment length polymorphism of a pel gene as a tool to identify Erwinia carotovora in relation to potato diseases. Appl Environ Microbiol. 60; https://doi.org/10.1128/aem.60.5.1437-1443 (1994).
Slawiak, M. et al. Biochemical and genetical analysis reveal a new clade of biovar 3 Dickeya spp. strains isolated from potato in Europe. Eur. J. Plant Pathol. 125, 245–261. https://doi.org/10.1007/s10658-009-9479-2 (2009).
Waleron, M., Waleron, K., Podhajska, A. J. & Lojkowska, E. Genotyping of bacteria belonging to the former Erwinia genus by PCR-RFLP analysis of a recA gene fragment. Microbiology. 148, 583–595. https://doi.org/10.1099/00221287-148-2-583 (2002).
Versalovic, J., Schneider, M., & Bruijn, F. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. (1994).
Ansermet, M., Schaerer, S., Kellenberger, I., Tallant, M. & Dupuis, B. Influence of seed-borne and soil-carried inocula of Dickeya spp. on potato plant transpiration and symptom expression. Eur. J. Plant Pathol. 145, 459–467. https://doi.org/10.1007/s10658-016-0859-0 (2016).
Hamed, S. M., Kamal, M., Messiha, N. A. S. Potential of algal-based products for the management of potato brown rot disease. Bot Stud. 64, 29; https://doi.org/10.1186/s40529-023-00402-y (2023).
Irikiin, Y., Nishiyama, M., Otsuka, S. & Senoo, K. Rhizobacterial community-level, sole carbon source utilization pattern affects the delay in the bacterial wilt of tomato grown in rhizobacterial community model system. Appl. Soil Ecol. 34, 27–32. https://doi.org/10.1016/j.apsoil.2005.12.003 (2006).
Cangioli, L. et al. Effect of site and phenological status on the potato bacterial rhizomicrobiota. Microorganisms. 10, 1. https://doi.org/10.3390/microorganisms10091743 (2022).
Puri, R. R. et al. Metagenomic study of endophytic bacterial community of sweet potato (Ipomoea batatas) cultivated in different soil and climatic conditions. World J. Microbiol. Biotechnol. 35, 1. https://doi.org/10.1007/S11274-019-2754-2 (2019).
Shivlata, L. & Satyanarayana, T. Thermophilic and alkaliphilic Actinobacteria: biology and potential applications. Front. Microbiol. 1, 1014. https://doi.org/10.3389/fmicb.2015.01014 (2015).
Li, H. Y. et al. The chemodiversity of paddy soil dissolved organic matter correlates with microbial community at continental scales. Microbiome. 187, 1. https://doi.org/10.1186/s40168-018-0561-x (2018).
Yang, C. et al. Soil salinity regulation of soil microbial carbon metabolic function in the Yellow River Delta. Sci. Total Environ. 790, 1. https://doi.org/10.1016/j.scitotenv.2021.148258 (2021).
Hu, D., Zang, Y. & Mao, Y. Identification of molecular markers that are specific to the class Thermoleophilia. Front. Microbiol. 10, 1. https://doi.org/10.3389/fmicb.2019.01185 (2019).
Kumar, S., Diksha, S. S. & Kumar, R. Biofertilizers: An ecofriendly technology for nutrient recycling and environmental sustainability. Curr. Res. Microb. Sci. 3, 100094. https://doi.org/10.1016/J.CRMICR.2021.100094 (2022).
Wang, Y. et al. Bacillus subtilis DSM29784 alleviates negative effects on growth performance in broilers by improving the intestinal health under necrotic enteritis challenge. Front. Microbiol. 12, 1. https://doi.org/10.3389/FMICB.2021.723187 (2021).
Zhao, Y. et al. Control of postharvest soft rot caused by Erwinia carotovora of vegetables by a strain of Bacillus amyloliquefaciens and its potential modes of action. World J. Microbiol. Biotechnol. 29, 411–420. https://doi.org/10.1007/s11274-012-1193-0 (2013).
Li, Z. et al. Biocontrol potential of Myxococcus sp. strain BS against bacterial soft rot of calla lily caused by Pectobacterium carotovorum. Biol. Control. 126, 36–44; https://doi.org/10.1016/J.BIOCONTROL.2018.07.004 (2018).
Weinert, N. et al. Bacterial diversity on the surface of potato tubers in soil and the influence of the plant genotype. FEMS Microbiol. Ecol. 74, 114–123. https://doi.org/10.1111/J.1574-6941.2010.00936.X (2010).
Radhakrishnan, R., Hashem, A., Abd Allah, E.F. Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 8, 667; https://doi.org/10.3389/FPHYS.2017.00667/BIBTEX (2017).
Saxena, A. K., Kumar, M., Chakdar, H., Anuroopa, N. & Bagyaraj, D. J. Bacillus species in soil as a natural resource for plant health and nutrition. J. Appl. Microbiol. 128, 1583–1594. https://doi.org/10.1111/JAM.14506 (2020).
Zhao, F. et al. Vermicompost can suppress Fusarium oxysporum f. sp. lycopersici via generation of beneficial bacteria in a long-term tomato monoculture soil. Plant Soil. 440, 491–505. https://doi.org/10.1007/s11104-019-04104-y (2019).
Chungopast, S. et al. Correlation of soil physiochemical properties, microorganism numbers, and bacterial communities following unburned and burned sugarcane harvest. Appl. Environ. Soil Sci. 1, 1687–7667. https://doi.org/10.1155/2023/9618349 (2023).
Xiong, W. et al. Distinct roles for soil fungal and bacterial communities associated with the suppression of vanilla Fusarium wilt disease. Soil Biol. Biochem. 107, 198–207. https://doi.org/10.1016/J.SOILBIO.2017.01.010 (2017).
Lacey, J. Ecology of Actinomycetes in fodders and related substrates. Zentralblatt fur Bakteriol. Parasitenkunde, Infekt. und Hyg (1978).
Kalam, S., Basu, A. & Podile, A. R. Functional and molecular characterization of plant growth promoting Bacillus isolates from tomato rhizosphere. Heliyon. 6, 1. https://doi.org/10.1016/J.HELIYON.2020.E04734 (2020).
Beneduzi, A., Ambrosini, A. & Passaglia, L. M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. https://doi.org/10.1590/s1415-47572012000600020 (2012).
Matilla, M. A. & Krell, T. Plant growth promotion and biocontrol mediated by plant-associated bacteria. In: Plant microbiome: Stress response. Springer. 5, 45–80. https://doi.org/10.1007/978-981-10-5514-0_3 (2018).
Jafra, S., Przysowa, J., Gwizdek-Wisniewska, A. & van der Wolf, J. M. Potential of bulb-associated bacteria for biocontrol of hyacinth soft rot caused by Dickeya zeae. J. Appl. Microbiol. 106, 268–277. https://doi.org/10.1111/j.1365-2672.2008.04000.x (2009).
Ossowicki, A., Jafra, S. & Garbeva, P. The antimicrobial volatile power of the rhizospheric isolate Pseudomonas donghuensis P482. PLoS ONE. 12, 1. https://doi.org/10.1371/journal.pone.0174362 (2017).
Gerayeli, N., Baghaee-Ravari, S. & Tarighi, S. Evaluation of the antagonistic potential of Bacillus strains against Pectobacterium carotovorum subsp. carotovorum and their role in the induction of resistance to potato soft rot infection. Eur. J. Plant Pathol. 150, 1049–1063. https://doi.org/10.1007/s10658-017-1344-0 (2018).
Des Essarts, Y. R. et al. Biocontrol of the potato blackleg and soft rot diseases caused by Dickeya dianthicola. Appl. Environ. Microbiol. 82, 268–278. https://doi.org/10.1128/AEM.02525-15 (2016).
Sansinenea, E. Bacillus spp.: As Plant Growth-Promoting Bacteria. In: Singh, H., Keswani, C., Reddy, M., Sansinenea, E., García-Estrada, C. (eds) Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms. Springer. 225–237; https://doi.org/10.1007/978-981-13-5862-3_11 (2019).
Reynolds, S. G. The gravimetric method of soil moisture determination Part I A study of equipment, and methodological problems. J. Hydrol. 11, 3. https://doi.org/10.1016/0022-1694(70)90066-1 (1970).
Meneley, J. C. Isolation of Soft-Rot Erwinia spp. from agricultural soils using an enrichment technique. Phytopathology. 66, 367. https://doi.org/10.1094/PHYTO-66-367 (1976).
Hélias, V., Hamon, P., Huchet, E., Wolf, J. V. D. & Andrivon, D. Two new effective semiselective crystal violet pectate media for isolation of Pectobacterium and Dickeya. Plant Pathol. 61, 339–345. https://doi.org/10.1111/J.1365-3059.2011.02508.X (2012).
Laurila, J. et al. Symptoms and yield reduction caused by Dickeya spp. strains isolated from potato and river water in Finland. Eur. J. Plant Pathol. 126, 249–262. https://doi.org/10.1007/s10658-009-9537-9 (2010).
Frechon, D. et al. Evaluation of a PCR kit for the detection of Erwinia carotovora subsp. atroseptica on potato tubers. Potato Res. 41, 163–173. https://doi.org/10.1007/BF02358439 (1998).
Kang, H. W., Kwon, S. W. & Go, S. J. PCR-based specific and sensitive detection of Pectobacterium carotovorum ssp. carotovorum by primers generated from a URP-PCR fingerprinting-derived polymorphic band. Plant Pathol. 52, 127–133. https://doi.org/10.1046/j.1365-3059.2003.00822.x (2003).
Sasada, R., Weinstein, M., Prem, A., Jin, M. & Bhasin, J. FIGARO: An efficient and objective tool for optimizing microbiome rRNA gene trimming parameters. J. Biomol. Tech. 31, 1 (2020).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. https://doi.org/10.14806/EJ.17.1.200 (2011).
Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. https://doi.org/10.1038/S41587-019-0209-9 (2019).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 13, 581–583. https://doi.org/10.1038/NMETH.3869 (2016).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 1. https://doi.org/10.1093/NAR/GKS1219 (2013).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 8, 1. https://doi.org/10.1371/JOURNAL.PONE.0061217 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Acknowledgements
This work was supported by the Ministry of Science and Higher Education (grant number 531-N107-D801-21) attributed to Ewa Lojkowska and National Science Centre in Poland via project Preludium 21 (grant number UMO-2022/45/N/NZ9/01923) attributed to Dr. Weronika Babinska-Wensierska. Dr. Agata Motyka-Pomagruk got support from Ministry of Education and Science in Poland via outstanding young scientists scholarship (SMN/18/0019/2022). We would like to thank Professor George C. DiCenzo from Queen's University for his discussion on the final version of the manuscript. In addition, we are grateful to MSc Christopher Riccardi from University of Florence for advice on the deposition of 16S rRNA raw reads in the Sequence Read Archive repository. Finally, we would like to thank our collaborators for enabling sampling of soil from two selected potato fields and providing all the necessary information on the conducted agrotechnical treatments.
Author information
Authors and Affiliations
Contributions
E. L., A. M-P and W. B-W. planned and designed all experiments. W. B-W. collected soil samples, performed all microbiological and molecular diagnostics experiments, isolated total DNA from the collected soil samples and performed bioinformatic analysis. W. B-W, M. F., A. M., A. E. M. took part in discussion on the selection of appropriate bioinformatics tools for visualization of 16S rDNA sequencing data. W. B-W., M. F. visualized the collected data. W. B-W, A. M-P., A. M., E. L. took part in the discussion on the collected data. W. B-W. wrote the first version of this manuscript. W. B-W, A. M-P., A. M., E. L. prepared the final version of this manuscript that has been accepted by all the other authors. W. B-W, A. M-P., E. L. corrected the manuscript according to the Reviewers’ comments. W. B-W. and E. L. acquired funding for this study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Babinska-Wensierska, W., Motyka-Pomagruk, A., Fondi, M. et al. Differences in the constituents of bacterial microbiota of soils collected from two fields of diverse potato blackleg and soft rot diseases incidences, a case study. Sci Rep 14, 18802 (2024). https://doi.org/10.1038/s41598-024-69213-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-69213-w
- Springer Nature Limited