Abstract
Chemotherapy, particularly with oxaliplatin, is a key treatment for advanced gastric cancer (GC), and exosomes derived from human bone marrow mesenchymal stem cells (hBM-MSCs) play a vital role in the tumor microenvironment. The study aims to elucidate the previously unexplored role of exosomes derived from hBM-MSCs in GC tumorigenesis, especially under the influence of chemotherapy. We conducted an experimental study, utilizing miRNA sequencing and biological experiments, to analyze the tumorigenicity of exosomal miR-424-3p secreted by hBM-MSCs and its target gene RHOXF2 in GC cell lines. The results were confirmed through experimentation using a xenograft mouse model. This study demonstrated the role of hBM-MSCs in the GC microenvironment, focusing on their epithelial-mesenchymal transition (EMT) facilitation through exosomes, which led to enhanced tumorigenicity in GC cells. Intriguingly, this pro-tumor effect was abrogated when hBM-MSCs were treated with oxaliplatin. Exosomal miRNA sequencing revealed that oxaliplatin can upregulate the levels of miR-424-3p in exosomes secreted by hBM-MSCs, thereby inhibiting the EMT process in GC cells. Furthermore, miR-424-3p was identified to target and downregulate RHOXF2 expression, impeding the malignant behavior of GC cells both in vitro and in the mouse model. These findings uncover a potential hidden mechanism of oxaliplatin's anti-tumor action and propose the delivery of miR-424-3p via exosomes as a promising avenue for anti-tumor therapy.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is a common malignant neoplasm that is ranked as the fifth most prevalent cancer and was the fourth leading cause of cancer-related mortality globally in 20201. Despite medical advancements in both surgical and internal medicine treatments that have elevated survival rates for GC patients, there remains a substantial risk of recurrence or metastasis, affecting approximately 60% of patients2. Presently, oxaliplatin is the favored therapeutic agent for the systemic treatment of GC. As a platinum compound, oxaliplatin exerts its effect by forming covalent bonds with specific helical regions of DNA, leading to DNA cross-linking. This interaction disrupts DNA replication and transcription, resulting in cell cycle arrest and apoptosis, thereby restraining tumor cell proliferation and dissemination. Despite these mechanisms, clinical outcomes continue to be less than ideal. Consequently, an in-depth exploration of the molecular mechanisms governing GC chemotherapy has emerged as an immediate scientific imperative.
Tumor development is a complex and dynamic process, involving various cellular and non-cellular elements that constitute the tumor microenvironment (TME). The interactions between the TME and cancer cells are pivotal in tumor progression and treatment response. The non-malignant cellular composition of the TME includes human bone marrow mesenchymal stem cells (hBM-MSCs), cancer-associated fibroblasts, endothelial cells and immune and inflammatory cells3. Among these, hBM-MSCs represent a subgroup of adult stem cells with the ability to undergo multidirectional differentiation and self-renewal. The role of hBM-MSCs in tumor development is contradictory; they have been shown to possess tumor-suppressive properties4, yet there is also evidence that supporting their involvement in promoting GC cell proliferation, survival, invasion, and metastasis5. Exosomes derived from bone marrow mesenchymal stem cells (hBM-MSC-exos), which serve as the principal mediators of hBM-MSC paracrine actions, are acknowledged for their essential functions in cellular communication and epigenetic control, with potential implications in the regulation of tumor cell growth, migration, and metastasis6. Exosomes contain abundant microRNAs (miRNAs), which can be internalized by target cells7. MiRNAs are small non-coding RNAs that negatively regulate gene expression by binding to the 3' untranslated region of target mRNAs8. Once miRNAs are loaded into exosomes, they can form an intercellular genetic information transport network, leading to transient or sustained phenotypic changes in target cells9. Several studies have demonstrated that MSCs promoted tumor proliferation, angiogenesis, invasion, and drug resistance by transferring miRNAs content to tumor cells via exosomes10,11. Conversely, some research suggests that exosomes secreted by MSCs exert tumor-suppressive effects by delivering specific miRNAs to tumor cells12,13,14. Therefore, the relationship between hBM-MSC-exos and tumor progression remains controversial.
The epithelial-mesenchymal transition (EMT) is a pivotal mechanism in cancer malignancy that is characterized by the loss of epithelial cell polarity and adhesion, and transformation into mesenchymal-like cells with augmented migratory and invasive properties15. Recent studies have shown that exosomes can induce target cells in the EMT by modulating intracellular signaling pathways through carried miRNAs16. However, the specific roles and mechanisms of hBM-MSC-exos on GC cells under chemotherapy remain unclear.
In this study, we revealed the attenuation of the hBM-MSC pro-tumorigenic effect following oxaliplatin treatment. Furthermore, we identified that miR-424-3p was significantly upregulated in the exosomes of hBM-MSCs treated with oxaliplatin, and by targeting and inhibiting the RHOXF2 gene, miR-424-3p impeded the EMT process in GC cells, reducing proliferation, migration, and invasion abilities. This research provides new insights and strategies for the treatment of GC.
Methods
Ethics statement
The animal study at Xinxiang Medical College's First Affiliated Hospital was approved by the Ethics Committee (No: EC-023–094) and strictly followed the principles. In compliance with the guidelines set forth by Guide for the Care and Use of Laboratory Animals, 8th Edition17, we conducted animal experiments while making concerted efforts to reduce the distress experienced by the laboratory subjects. All methods are reported in accordance with ARRIVE guidelines.
Cell culture
Human gastric cancer cell lines SGC-7901 and AGS were obtained from the cell bank of the Chinese Academy of Sciences, Shanghai, China. Human bone marrow-derived mesenchymal stem cells (hBM-MSCs) were purchased from Cyagen Biotechnology Co., Ltd. (Guangzhou, China). hBM-MSCs were resuspended and adjusted to a concentration of 1 × 106 cells/mL (200 μL) with PBS. The hBM-MSC suspension was incubated with 5 μL of different fluorescently labeled monoclonal antibodies (CD105 (ab11414), CD73 (ab202122), CD34 (ab8536), CD45 (ab8216), CD90 (ab23894), and HLA-DR (ab20181)) at 4 °C in the dark for 15 min. Flow cytometry was used to evaluate the hBM-MSCs, as shown in Fig. S1A. The cells were cultured in an incubator (Thermo Fisher Scientific, Waltham, MA, USA) at 37 °C with 5% CO2. SGC-7901 and AGS cells were cultured in DMEM medium (CORNING Inc, NY, USA), both supplemented with 10% fetal bovine serum, while hBM-MSCs were cultured in serum-free culture system (Biological Industries, Israel).
Co-culture of hBM-MSCs with gastric cancer cells
A non-contact Transwell co-culture system was used to co-culture hBM-MSCs with GC cells (1 × 105 cells/cm2). hBM-MSCs were seeded in six-well chambers (1 × 104 cells/cm2) with 0.4-μm permeable polycarbonate membrane pores, which allowed for the passage of exosomes and cytokines secreted by the cells while restricting physical cell transfer. After 48 h of co-culture, the cells were rinsed three times with PBS before being harvested for further analysis.
To collect conditioned medium from hBM-MSCs treated with oxaliplatin (Sanofi, Paris, France), the hBM-MSCs were treated with 0.25 mM (1/2 IC50) oxaliplatin for 24 h and then the culture medium was replaced with fresh exosome-free serum medium. After 24 h of culture, the conditioned medium of oxaliplatin-treated hBM-MSCs was collected and filtered with a 0.22-μm filter. The conditioned medium of untreated hBM-MSCs was collected as a control. These mediums were then ultracentrifuged to remove exosomes according to the experimental requirements. GC cells were cultured in exosome-depleted hBM-MSC-conditioned medium (Ne-Oxa-CM), and exosome-depleted oxaliplatin-treated hBM-MSC-conditioned medium (Ne-Oxa-CM) for 48 h. Afterward, the GC cells were cultured in regular medium for an additional 48 h and harvested for subsequent experiments.
Isolation and identification of exosomes from hBM-MSCs
hBM-MSCs that were exposed to PBS or oxaliplatin, or transfected with miR-424-3p mimics, agomirs, or their controls were cultured in a serum-free culture system for 48 h and then centrifuged at 1 200 g for 25 min at 4 °C to remove the cell debris and dead cells. Exosomes were isolated by using differential centrifugal speeds. The cells were filtered through a 0.2 mm filter, followed by ultracentrifugation at 4° C and 20 000 g for 30 min to remove the large cell blisters. The collected supernatant was filtered through a 0.22-μm filter and centrifuged again at 4 °C and 100 000 g for 90 min. After removing the supernatant, the white sedimentation from the tube bottom was suspended in 500 μL of PBS and stored at − 80 °C for future experiments.
Characterization and application of MSC-secreted exosomes
The final pellet containing exosomes was re-suspended in PBS and the amount and size of exosomes were determined by a Zetasizer Nano ZS instrument (Malvern Instruments, Worcestershire, UK). The protein content of the concentrated exosomes was quantified using BCA Protein Assay (Beyotime, Shanghai, China). To characterize the exosomes, the expression patterns of exosome-specific surface markers CD63, TSG101 and calnexin were analyzed through western blot analysis. Subsequently, an exosome suspension was placed on a carbon-coated copper grid, and the grid was placed in 2% phosphotungstic acid for 1 min, and then rinsed with PBS followed by desiccation. Then the morphology of the exosomes was observed using transmission electron microscopy (TEM) (HT7800, Hitachi, Japan). For in vitro use of exosomes, 1 × 106 cells were incubated with 50 μg of exosomes, and for in vivo use, a total of 1 mg of exosomes were intravenously injected into BALB/c nude mice via the tail vein.
Fluorescent labeling and uptake of exosomes
In order to observe whether GC cells could endocytose exosomes from MSCs, we labeled the extracted exosomes using a PKH-67 fluorescent staining kit (Sigma-Aldrich, MO, USA). Specifically, 200 μL of diluent C solution was added to every 100 μL of the PBS-exosome suspension; then 2 μL of PKH-67 dye and 200 μL of the diluent C solution were taken and added to a 1.5-mL Eppendorf (EP) tube. The two liquids were mixed in a 1.5-mL EP tube and left in the tube for 5 min in the dark, followed by filtration through a 0.22 μm filter and ultracentrifugation at 100,000 g for 90 min. PKH26-labeled exosomes were added to fresh media after the cells had grown to 70% cell confluence of growth in a 12 well plate. After 12 h, the cells were then washed three times with diluted PBS, followed by fixation of SGC-7901 and AGS cells with 4% paraformaldehyde for 20 min and staining of nuclei with 4′,6-diamidino-2-phenylindole (DAPI, Sigma, St. Louis, USA), followed by observation of exosome endocytosis under a confocal fluorescence microscope.
qRT-PCR analysis
Total RNA was extracted utilizing a RNeasy mini kit (Qiagen, Duesseldorf, Germany). The mRNA concentration and quality were subsequently assessed by a Qubit 3.0 fluorometer (Life Technologies, Gaithersburg, USA) and a Nanodrop One spectrophotometer (Thermo Fisher Scientific Inc., Waltham, USA), respectively. The integrity of the total RNA was evaluated using an Agilent 2100 bioanalyzer (Agilent Technologies Inc., Santa Clara, USA), and only samples with RNA integrity number (RIN) values exceeding 7.0 were selected for subsequent experiments. qRT-PCR detection was conducted with primers designed via Primer-BLAST software (NCBI, https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi), and the sequences are listed in Table S1. In the examination of cell and tissue lysates, mRNA levels were normalized to β-actin, and miRNA levels were normalized to hsa-U6.
mRNA sequencing
Paired-end libraries were constructed using the Stranded mRNA-seq Lib Prep Kit for Illumina (ABclonal, Wuhan, China), adhering to the Preparation Guide. In brief, mRNA molecules containing poly-A were isolated with poly-T oligo-attached magnetic beads. After purification, the mRNA was fragmented into small pieces using divalent cations at 94 °C for 10 min. The fragmented RNA was then transcribed into first-strand cDNA using reverse transcriptase and random primers, followed by second-strand cDNA synthesis with DNA Polymerase I and RNase H. The resulting cDNA fragments underwent end repair, a single 'A' base addition, and adapter ligation. The products were purified and enriched via PCR to form the final cDNA library. The purified libraries were quantified by the Qubit 3.0 Fluorometer (Life Technologies, Gaithersburg, USA) and validated by the Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, USA) to verify the insert size and determine the molar concentration. Clusters were generated by cBot, with the library diluted to 10 pM, and sequencing was performed on the Illumina NovaSeq 6000 (Illumina, San Diego, USA). Deferential expressions between two sets of samples were defined according to the criteria of | log2 (fold change) |≥ 1 and p-value < 0.05. Through the utilization of the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, the signal transduction pathways significantly enriched within the Differentially Expressed Genes (DEGs) were identified and confirmed.
miRNA sequencing
For the cDNA library construction, approximately 1 μg of total RNA was used, following the manufacturer's instructions for the TruSeq Small RNA sample prep kits (Illumina, San Diego, USA). The procedure encompassed ligation of the RNA to 3ʹ adapters, and the attachment of 5′ adapters to the other end of the RNA molecules. Subsequently, the adapter-ligated RNAs were subjected to RT-PCR and amplified using a low number of cycles. Post-PCR amplification, the libraries were pooled and purified via a PAGE gel. Finally, 50-bp single-end sequencing was conducted on an Illumina Hiseq 2500 sequencer (LC Bio, Hangzhou, China) following the manufacturer's guidelines, and the resulting data was subsequently analyzed. MiRDeep2 was used to identity known mature miRNAs based on miRBase21 (www.mirBase.org) and to predict novel miRNAs. Expression data were were normalized in Reads Per Million (RPM). The identification of differentially expressed miRNAs was performed using the DESeq2 package within R software (version 4.0, https://www.r-project.org/), employing the default parameters. The differential expression between two sets of samples was defined according to the criteria of |log2 (fold change) |≥ 1 and p-value < 0.05.
Cell transfection
The miR-424-3p mimics, miR-424-3p inhibitors, miR-424-3p agomirs, and negative controls were obtained from Shanghai Sangon Biotech Services Co., Ltd. (Shanghai, China). Lentiviral shRNA vectors targeting human RHOXF2 (sh-RHOXF2) and scrambled non-specific sh-RNA (sh-NC) lentiviral vectors were purchased from Guangzhou Ribobio Co., Ltd. (Guangzhou, China). The sequences are shown in Table S2. The day before transfection, cells were seeded on a cell culture plate with antibiotic-free culture medium, aiming for a cell density of 50% at the time of transfection. The plasmids or lentivirus were transfected into the cells using Lipofectamine 2000 (Invitrogen, Carlsbad, USA) according to the instructions. After 24 h of transfection, the culture medium was changed to medium containing FBS. The cells were harvested 48 h later for subsequent experiments.
For the exosome experiment, hBM-MSCs were treated with a miR-424-3p mimic, inhibitor, agomir, or their respective negative controls (NC mimic, inhibitor, or agomir) for 24 h. The corresponding supernatant was collected and ultracentrifuged to yield exosomes corresponding to Exo-miR-424-3p mimic, Exo-NC mimic, Exo-miR-424-3p inhibitor, Exo-NC inhibitor, Exo-miR-424-3p agomir, and Exo-NC agomir. The expression levels of miR-424-3p and RHOXF2 mRNA were determined by qRT-PCR, and the RHOXF2 protein level was analyzed by western blotting.
Dual luciferase reporter assay
The predicted binding site of miR-424-3p was identified using an online resource (https://www.targetscan.org/vert_72/). A dual luciferase reporter gene assay was employed to verify whether RHOXF2 is a target gene of miR-424-3p. The sequence of RHOXF2 wild type (RHOXF2-wt) containing miR-424-3p binding sites and its mutated sequence (RHOXF2-mut) were integrated into a PGL3 luciferase reporter gene vector. The reporter gene plasmids, along with miR-424-3p mimics and NC mimics, were co-transfected into AGS and SGC-7901 tumor cells for 24 h, followed by a 48-h cultivation period. A Dual Luciferase Reporter assay system (Promega, USA) was used to detect luciferase activity by a Luminoskan Ascent reader system (Thermo Scientific, Waltham, USA). The relative luciferase activity was represented by the ratio of firefly luciferase to Renilla luciferase.
Flow cytometry
A solution was configured with a 100:1 ratio of exosome extract and fluorescent dye, followed by incubation at 37 °C for 30 min. The mixture was filtered with a 0.22-μm filter to remove fluorescent dye residues before infiltrating the cells. To determine the most efficient exosome infiltration concentration and time for this experiment, flow cytometry was used to observe the 12-h and 24-h infiltration rates of SGC-7901 and AGS cells at different exosome concentrations (50 μg/mL and 100 μg/mL).
Cytotoxicity and proliferation assays
Cell cytotoxicity and proliferation were analyzed using the Cell Counting Kit-8 (CCK-8) method (Dojindo, Kumamoto, Japan). For cytotoxicity analysis, hBM-MSCs were plated into 96-well plates and treated with varying concentrations of oxaliplatin (10 μM, 50 μM, 100 μM, 200 μM, 500 μM, 1000 μM). After 24 h, the medium was replaced with 10 μL of CCK-8 solution and 90 μL of DMEM medium for 1 h. Then the optical density (OD) of each well at 450 nm was measured (Thermo Scientific, Waltham, USA), and the half inhibitory concentration (IC50) of oxaliplatin was calculated based on the OD value. Cell cytotoxicity was calculated according to the following formula: inhibition ratio (%) = (OD (drug) − OD (blank)) / (OD (drug control) − OD (blank)) × 100%.
For proliferation analysis, differently treated GC cells were seeded into a 96-well plate at 1 × 103 cells per well. Three duplicate wells for each group were used, and ells were placed in a cell incubator. At different time intervals (every 24 h) of culture, ranging from 0 to 7 days, 10 μL CCK-8 regent was added to each well and incubated for 1 h in the incubator. The optical density (OD) at 450 nm represented the cell proliferation index. The experiment was repeated three times and the mean values (mean ± s.d.) were presented.
Colony formation assay
To determine the colony forming ability of the cells, 500 cells were grown in six-well plates and cultured for 2 weeks. At the end of the culture, the cells were fixed with 4% paraformaldehyde and then stained with 0.5% crystal violet (Solarbio, Beijing, China). The number of cell clones containing more than 50 cells was counted under an optical microscope. Each experiment was replicated three times.
Wound healing assay
GC cells with different treatments were inoculated on 24-well culture plates at a density of 1 × 106 cells/well. When the GC cells were adhered to the plate, they were directly scratched with a 10-μL pipette tip and incubated in serum-free medium for 24 h. All samples were analyzed in triplicate and the data were expressed as mean ± standard deviation.
Transwell invasion assay
Cell invasion experiments were performed on 24-well plates using Matrigel (Sigma Aldrich, St. Louis, UK) Transwell chambers. Matrigel was thawed at 4 °C and then diluted at a 1:8 ratio with culture medium. The diluted Matrigel was evenly coated on the upper membrane of the Transwell chamber and solidified at 37 °C for 4 h. After removing the excess culture medium, add 500 μL of serum-free culture medium was added to the lower chamber and left it for 1 h to hydrate the basement membrane. The upper chamber was filled with 100 μL of a GC cell suspension with different treatments, and the lower chamber was filled with 0.5 mL of DMEM containing 20% fetal bovine serum (FBS). After 24 h of incubation at 37 °C in a 5% CO2 incubator, the remaining cells of the upper chamber were removed, rinsed with PBS and fixed with 4% paraformaldehyde for 30 min. Then, the cells were stained with 0.1% crystal violet for 20 min and rinsed with PBS. Five randomly selected fields of view were photographed and counted under an inverted microscope (TE2000, Nikon, Tokyo, Japan). The average number of cells was regarded as the number of cells passing through the chamber.
Western blotting
To obtain total proteins from different cell samples, cells were centrifuged at 4 °C at 12,000 g for 10 min. RIPA lysis buffer (KeyGen, Nanjing, China) was added to the precipitate after centrifugation in an ice bath for 30 min. The supernatant was transferred to the EP tube, and a portion of the supernatant was used for BCA testing. A reasonable amount of the 5 × loading buffer was added to the remaining supernatant. The samples were stored at − 20 °C after boiling for 5 min. The protein was transferred to a PVDF membrane following SDS-PAGE. The membrane was blocked with 5% skim milk for 1 h and then incubated overnight at 4 °C with primary antibodies. The primary antibodies were as follows: E-cadherin (1:1000, ab40772, Abcam, Cambridge, UK), N-cadherin (1:1000, ab18203, Abcam, Cambridge, UK), vimentin (1:1000, ab92547, Abcam, Cambridge, UK), CD63 (1:1000, ab68418, Abcam, Cambridge, UK), TSG101 (1:1000, ab125011, Abcam, Cambridge, UK), calnexin (1:500, ab92573, Abcam, Cambridge, UK), RHOXF2 (1:1000, YN1233, Immunoway, Plano, USA), and β-actin (1:1000, #8457, Cell Signaling Technology, Danvers, USA). After an overnight incubation at 4 °C, the membrane was washed three times for 5 min with TBST (Solarbio, Beijing, China). The membrane was incubated with a secondary antibody (1:5000) (Cell Signaling Technology, Danvers, USA) for 1 h at room temperature. After washing the membrane with TBST, an ECL chemiluminescence reagent was used for the color reaction.
Immunohistochemistry (IHC)
Tissue samples were dehydrated using an alcohol gradient, fixed with 4% paraformaldehyde, embedded in paraffin, and sectioned. The sections were flattened on a slide spreader at 40℃, transferred onto slides, and baked at 60 °C. The tissue sections underwent an incubation process with normal goat serum (Beijing Boao Sen Biotechnology Co., Beijing, China) for a duration of 1 h. Tissue sections were incubated with primary antibodies (RHOXF2 1:1000, E-cadherin 1:100, N-cadherin 1:200, and vimentin 1:500) at 4 °C overnight. Subsequently, the sections were incubated with a secondary antibody (1:2000) for 1 h at ambient temperature. Post-incubation, the sections were stained with diaminobenzidine and counterstained with hematoxylin. After a washing step to achieve a blue coloration, the sections were examined under a microscope. The results were evaluated using an immunoreactivity scoring (IRS = SI × PP) system, based on the staining intensity (SI) and the percentage of positive cells (PP). SI was defined as: 0 = negative; 1 = weak; 2 = moderate; 3 = strong. PP was defined as: 0 = 0%; 1 = 0–25%; 2 = 25–50%; 3 = 50–75%; 4 = 75–100%.
Tumor xenografts in nude mice
The impact of miR-424-3p on GC cells in vivo was evaluated using a xenograft model in BALB/c nude mice. Female nude mice (n = 20, BALB/c, aged 3–4 weeks, weighing 9–12 g) were procured from the Beijing Vital River Laboratory Animal Technology Center (Beijing, China). The mice were housed in a sterile, temperature-controlled facility with access to lab feed and water. The mice were randomly allocated into four groups, with two groups each implanted with either AGS or SGC-7901 cells, and each group was separately injected with an Exo-miR-424-3p agomir or Exo-NC agomir. Cultured GC cells were suspended in PBS at a density of 5 × 106 cells/mL. A 200-μL cell suspension was subcutaneously injected into the left axillary region of each mouse. Visible tumor nodules were observed five days post-implantation. On days 5, 10, 15, 20, and 25, the mice received tail vein injections of 200 μg of exosomes18. Tumor dimensions were periodically measured and volumes calculated (tumor volume = length x width2 × 0.5). After 28 days of implantation, the mice were euthanized by cervical dislocation. Tumors were removed for subsequent analysis.
Statistical analysis
Statistical evaluations were conducted using SPSS 22 software (IBM Corporation, Armonk, USA, https://www.ibm.com/spss), and GraphPad Prism9 (GraphPad Software Inc., La Jolla, USA, https://www.graphpad-prism.cn) was employed for graph creation. Data are represented as mean and standard deviation (SD). Depending on the experimental design, Student t-tests were used for two-group comparisons, while one-way ANOVA with Tukey’s post hoc test was utilized for comparisons among three or more groups. The IC50 of oxaliplatin was determined via normal probability transformation and probit regression analysis. P values less than 0.05 were deemed statistically significant.
Results
hBM-MSC-exos promoted gastric cancer tumorigenicity, and oxaliplatin attenuated this effect
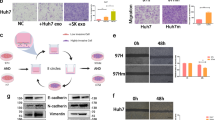
First, we measured the IC50 value of hBM-MSCs for oxaliplatin (Fig. 1A). hBM-MSCs treated with 0.25 mM oxaliplatin were transferred to the upper chamber and co-cultured with SGC-7901 or AGS cells in a Transwell system for 24 h (Fig. 1B). Then, the morphology of the hBM-MSCs was observed under a light microscope (Fig. S1B). Before and after oxaliplatin treatment, we were able to extract exosomes from the MSC culture medium. Examination of exosomes under a transmission electron microscope revealed that they all exhibited a round or oval vesicle morphology (Fig. 1C), with diameters ranging from 30 to 100 nm (Fig. 1D, Fig. S1C). Western blotting showed the absence of the cell-specific marker calnexin in exosomes, but a rich presence of the exosomal marker CD63 and TSG101 (Fig. 1E). To further investigate whether hBM-MSC-derived exosomes were taken up by GC cells in the co-culture system, we co-cultured GC cells with hBM-MSC-exos labeled with PKH-67 for 12 h. As depicted in Fig. 1F, confocal microscopy revealed green fluorescence in GC cells, suggesting successful uptake of exosomes by SGC-7901 and AGS cells. To optimize the use of exosomes in subsequent experiments, flow cytometry determined that a concentration of 50 μg/mL of exosomes achieved optimal uptake rates after 24 h (Fig. S1D). In conclusion, we discovered that exosomes secreted by hBM-MSCs, exhibiting typical exosomal characteristics, were be rapidly internalized by GC cells.
hBM-MSC-exos enhanced gastric cancer tumorigenicity while oxaliplatin diminished this effect. (A) Determination of the IC50 value of oxaliplatin in hBM-MSCs, calculated by non-linear regression to 522 μM (0.522 mM). (B) Schematic representation of the co-culture system after oxaliplatin treatment of hBM-MSCs. (C) Morphological representation of hBM-MSC-exos and Oxa-MSCs-exos (scale bar = 200 nm). (D) Nanoparticle tracking analysis of hBM-MSC-exos. (E) Western blot analysis of the hBM-MSC-exos surface markers calnexin, TSG101 and CD63. (F) Fluorescence microscopy image of exosome endocytosis. PKH-67(green) labeled exosomes internalized by SGC-7901 and AGS cells. DAPI (blue), 4′,6-diamidino-2-phenylindole (scale bar = 25 μm). (G) The proliferation ability of gastric cancer cells is significantly enhanced under co-culture conditions with hBM-MSCs or hBM-MSC-exos. However, no significant differences in proliferation ability were observed among Oxa-MSC, Oxa-MSC-exo, Ne-Oxa-CM, and Ne-MSC-CM. OD at 450 nm represents the cell proliferation index. (H) Analysis of clone formation in gastric cancer cells co-cultured with hBM-MSCs and exosomes under different conditions. Both hBM-MSCs and hBM-MSC-exo significantly enhanced the clonogenic ability of SGC-7901 and AGS cells. (I) The migration ability of gastric cancer cells under different conditions was measured by a wound healing assay (scale bar = 100 μm). The left image shows representative microscopic images, with the red line marking the inner average line of the wound. The right image shows statistical results after 24 h. (J) Transwell invasion assay (scale bar = 50 μm) indicates that gastric cancer cells co-cultured with hBM-MSCs or hBM-MSC-exos have a greater migration ability. The left image shows representative crystal violet-stained microscopic images, and the right image shows statistical results after 24 h. Data are presented as mean ± SD, averaged from three experiments. Data comparison between groups was performed using one-way ANOVA with Tukey’s post hoc test. Ns., not significant. ***p < 0.001.
Morphological changes were detected in SGC-7901 and AGS cells co-cultured with hBM-MSCs and hBM-MSC-exos, characterized by a transition to a rounded or elongated shape, and accompanied by a loosening of the intercellular junctions (Fig. S1E). A CCK-8 assay further substantiated that co-culture with hBM-MSCs increased the proliferation rate of SGC-7901 and AGS cells (Fig. 1G). In comparison to the control group treated with PBS (Mock group), the cells co-cultured with hBM-MSCs demonstrated increased colony numbers (Fig. 1H, Fig. S1F), accelerated wound closure (Fig. 1I), and increased numbers of invaded cells (Fig. 1J), in both SGC-7901 and AGS cells co-cultured with hBM-MSCs, signifying amplified migratory and invasive capacities. Analogous to the hBM-MSCs group, the enhancement effects of hBM-MSC-exos on the clonogenicity, proliferation, migration and invasion of GC cells were observed (Fig. 1G–J, Fig. S1F). Furthermore, compared to the hBM-MSCs group and hBM-MSC-exo group, a significant reduction in colony formation numbers (Fig. 1H, Fig. S1F), wound closure rates (Fig. 1I), and invasion cell counts (Fig. 1J) was noted in SGC-7901 and AGS cells co-cultured with oxaliplatin (Oxa)-MSCs and Oxa-MSC-exos. hBM-MSC-exos were removed from hBM-MSC culture medium via ultracentrifugation to assess the conditioned medium's ability to promote cell proliferation and migration. Consistent with expectations, SGC-7901 and AGS cells cultured with Ne-MSC-CM or Ne-Oxa-CM exhibited no marked enhancement in colony formation (Fig. 1H, Fig. S1F), proliferation, migration or invasion abilities (Fig. 1G–J) relative to the Mock group. These results collectively implied that hBM-MSCs augment the tumorigenic potential of GC cells through exosome secretion.
hBM-MSC-exos promote the EMT in GC cells
To understand the gene profile changes and enhanced tumorigenicity of GC cells in an hBM-MSC co-culture environment, we proceeded to assess the transcriptional changes in AGS cells induced by hBM-MSCs. We determined that hBM-MSCs induced an EMT state in AGS cells, marked by the upregulation of numerous genes (Fig. 2A). Hierarchical clustering revealed the upregulation of genes associated with cellular glucose metabolism, hypoxia, and the EMT in AGS cells co-cultured with hBM-MSCs (Fig. 2B). In the study, KEGG analysis pinpointed a minimum of 10 signaling pathways with established roles in tumorigenesis. Alongside the detected morphological alterations in GC cells (Fig. S1E), this led to the hypothesis that hBM-MSCs might facilitate the EMT process within GC cells. To validate this phenotype, the expression of EMT markers in SGC-7901 and AGS cells under diverse culture and treatment conditions was assessed by western blotting and mRNA analysis. In both the hBM-MSCs and hBM-MSC-exo groups, our data demonstrated a marked reduction in the expression of the EMT epithelial marker E-cadherin, coupled with an elevation in the mesenchymal markers N-cadherin and vimentin relative to the Mock group (Fig. 2C–F). However, after co-culturing with Oxa-MSCs or Oxa-MSC-exos, significant reversed findings were identified when compared to the hBM-MSC and hBM-MSC-exo group (Fig. 2C–F). These results clearly indicate that hBM-MSCs enhanced the tumorigenic potential of GC cells by inducing EMT by delivering exosomes.
Transcriptome analysis of GC cells with hBM-MSCs. (A) Volcano plot demonstration of gene expression changes in AGS cells co-cultured with hBM-MSCs compared to control AGS cells. (B) Enhancement of pathways related to cancer phenotypes in AGS cells subjected to hBM-MSCs. (C, D) Western blot detection of E-cadherin, N-cadherin and Vimentin proteins in GC cevlls normalized to β-actin. (E, F) Expression of gene encoding E-cadherin, N-cadherin and vimentin genes by qRT-PCR analysis. β-actin was used as an internal control. The data in the figure were measurement data. Data comparison between groups was performed using one-way ANOVA with Tukey’s post hoc test. All statistical data were obtained from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Exosomal miR-424-3p of hBM-MSCs upregulated by oxaliplatin inhibited the EMT in GC cells
To understand what changes in the exosomal content led to the observed phenotypic alterations, we conducted miRNA sequencing on the exosomes secreted by hBM-MSCs. This analysis identified two miRNAs, miR-424-3p and miR-127-3p, that were differentially expressed between the Oxa-MSC-exo and hBM-MSC-exo groups (Fig. 3A). In a particular investigation, miR-424-3p was found to significantly inhibit the proliferation, migration, and invasion of non-small cell lung cancer cells19. This result aligned with our sequencing data, suggesting that miR-424-3p might be linked to potential tumor-suppressing effects in GC cells. To further explore the expression trend of miR-424-3p, we extracted total RNA from Oxa-MSC-exos and hBM-MSC-exos. The qRT-PCR results demonstrated a significant upregulation of miR-424-3p in Oxa-MSC-exos (Fig. 3B), consistent with the trend observed in the screening study.
Exosomal miR-424-3p suppresses EMT in gastric cancer cells. hBM-MSCs or GC cells were transfected with miR-424-3p mimics, inhibitors, and their controls (NC mimics or NC inhibitors). (A) A volcano plot illustrates the differential expression of miRNAs in Oxa-MSC-exo and hBM-MSC-exo, as determined by high-throughput sequencing analysis of exosomal miRNA. (B) qRT-PCR quantification validates miR-424-3p in Oxa-MSC-exo and hBM-MSC-exo. (C) qRT-PCR analysis evaluating the expression levels of miR-424-3p after transfection with miR-424-3p mimics, inhibitors, and their controls in hBM-MSCs and their exomes (Exo-miR-424-3p mimics, Exo-miR-424-3p inhibitors, Exo-NC mimics and Exo-NC inhibitors). (D) miR-424-3p expression in GC cells treated with MSC-derived exosomes by qRT-PCR. (E) Colony formation experiments in GC cells treated with PBS, Exo-miR-424-3p mimics, Exo-miR-424-3p inhibitors, Exo-NC mimics and Exo-NC inhibitors. (F) Cell proliferation experiments in GC cells treated with PBS, Exo-miR-424-3p mimics, Exo-miR-424-3p inhibitors, Exo-NC mimics and Exo-NC inhibitors. The proliferation index (OD 450 nm) in the Exo-miR-424-3p mimic group was significantly lower compared to the Exo-NC mimic group, while the rate in the Exo-miR-424-3p inhibitor group was significantly higher than in the Exo-NC inhibitor group. (G) Wound healing assays (scale bar = 100 μm) in GC cells treated with PBS, Exo-miR-424-3p mimics, Exo-miR-424-3p inhibitor, Exo-NC mimics and Exo-NC inhibitors. (H) Transwell assays (scale bar = 50 μm) in GC cells treated PBS, Exo-miR-424-3p mimics, Exo-miR-424-3p inhibitors, Exo-NC mimics and Exo-NC inhibitors. (I) Western blotting detects the expression of E-cadherin, N-cadherin, and Vimentin proteins in gastric cancer cells under different treatment, normalized to β-actin. (J, K) qRT-PCR detects the mRNA expression of E-cadherin, N-cadherin, and Vimentin in gastric cancer cells under different treatment, normalized to β-actin. Unpaired t-test were used for comparisons of data between two groups. One-way ANOVA and Tukey's post-hoc test was used for comparisons between groups. All statistical data were obtained from three independent experiments, with results presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001.
Subsequently, we successfully manipulated the expression of miR-424-3p in hBM-MSCs and their exosomes using miR-424-3p mimics or inhibitors (Fig. 3C). qRT-PCR revealed that altering the expression of miR-424-3p in hBM-MSCs led to corresponding changes in its exosomes (Fig. 3C). To investigate the impact of miR-424-3p delivered via exosomes, we treated GC cells with these exosomes. Treatment with Exo-miR-424-3p mimics increased miR-424-3p expression in SGC-7901 and AGS cells, while treatment with an Exo-miR-424-3p inhibitor showed the opposite trend (Fig. 3D). Compared to the PBS-treated control group (Mock group), Exo-miR-424-3p mimics reduced the clonogenic ability (Fig. 3E, Fig. S2), decreased cell proliferation (Fig. 3F), and inhibited the migration (Fig. 3G) and invasion (Fig. 3H) abilities of GC cells. The analyses also showed an increase in the EMT-related molecular markers N-cadherin and vimentin, and a decrease in E-cadherin (Fig. 3I–K). Conversely, the Exo-miR-424-3p inhibitor group exhibited the opposite effects (Fig. 3I–K). These findings underscore the significant influence of hBM-MSC-exos containing miR-424-3p on the behavior of GC cells.
miR-424-3p inhibits RHOXF2 in GC cells
To explore the molecular mechanisms of miR-424-3p in the malignant progression of GC cells, we used the TargetScan database to identify potential target genes of miR-424-3p (Fig. 4A). Among these, RHOXF2 was identified as a putative target of miR-424-3p. To validate this, we co-transfected SGC-7901 and AGS cells with the RHOXF2-wt or RHOXF2-mut together with miR-424-3p mimic or NC mimic. Luciferase activity assays revealed a significant reduction in the activity of the RHOXF2 reporter vector in these cells, while the mutant form was unaffected by miR-424-3p (Fig. 4B, C). When transfected with miR-424-3p mimics, RHOXF2 mRNA levels were significantly suppressed; conversely, when transfected with miR-424-3p inhibitor, miR-424-3p expression was significantly inhibited, leading to a significant increase in RHOXF2 mRNA levels (Fig. 4D, E). Western blot analysis further confirmed that the miR-424-3p inhibitors significantly increased RHOXF2 protein expression, whereas the mimics significantly decreased it (Fig. 4F). These results indicated that miR-424-3p negatively regulates RHOXF2.
The predicted binding gene RHOXF2 of miR-424-3p is involved in regulating the EMT in gastric cancer cells. (A) Prediction of miR-424-3p binding sites in the RHOXF2 3′-UTR. (B, C) Luciferase reporter assays confirmed that miR-424-3p directly targets the RHOXF2 3′-UTR in cells co-transfected with RHOXF2-wt or -mut luciferase reporter vector and NC mimics or miR-424-3p in SGC-7901 (B) and AGS cells (C). (D, E) qRT-PCR assessed miR-424-3p and RHOXF2 mRNA expression levels after overexpression or inhibition in SGC-7901 cells (D) and AGS cells (E). (F) Western blot analysis reveals RHOXF2 expression levels after overexpression or inhibition miR-424-3p in SGC-7901 and AGS cells. (G) qRT-PCR analysis shows RHOXF2 mRNA expression levels in SGC-7901 and AGS cells transfected with sh-NC or sh-RHOXF2. (H) Western blot analysis assessed RHOXF2 protein levels after transfection in GC cells. (I) Colony formation assays evaluated the tumorigenic ability in GC cells treated with sh-NC or sh-RHOXF2. (J) A CCK-8 proliferation assay measured the proliferation index in GC cells treated with sh-NC or sh-RHOXF2. (K) Wound healing assays (scale bar = 100 μm) examined migration in GC cells treated with sh-NC or sh-RHOXF2. (L) Transwell assays (scale bar = 50 μm) investigated invasion in GC cells treated with sh-NC or sh-RHOXF2. (M) Western blot analysis revealed the E-cadherin, N-cadherin, and vimentin protein expression levels in SGC-7901 and AGS cells transfected with sh-NC or sh-RHOXF2. (N) qRT-PCR measured mRNA expression of E-cadherin, N-cadherin, and vimentin in GC cells transfected with sh-NC or sh-RHOXF2. Comparisons between two groups were made using an unpaired t-test; comparisons between multiple groups were analyzed by one-way ANOVA, followed by Tukey’s multiple comparison test. All statistical data were obtained from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Silencing RHOXF2 inhibits the EMT in GC cell lines
To further investigate the pro-tumorigenic role of RHOXF2 in GC cells, we synthesized short hairpin RNA specific for RHOXF2 (sh-RHOXF2) (Table S2) together with a non-targeting control (sh-NC) and transfected these constructs into SGC-7901 and AGS cells. Following transfection with sh-RHOXF2, we observed a significant decrease in both mRNA and protein levels of RHOXF2 (Fig. 4G, H). In addition, clonogenic assays, CCK-8 proliferation assays, wound healing assays and Transwell invasion experiments all showed a reduction in clonogenic ability (Fig. 4I, Fig. S3), cell proliferation (Fig. 4J), migration (Fig. 4K), and invasion (Fig. 4L) abilities with RHOXF2 silencing. We then examined the expression patterns of EMT-related factors (N-cadherin, E-cadherin, vimentin) by Western blot analysis and qRT-PCR and found that RHOXF2 knockdown resulted in decreased expression of N-cadherin and vimentin and increased expression of E-cadherin (Fig. 4M, N). Collectively, these results suggest that RHOXF2 silencing can inhibit the proliferation, migration and invasion capabilities of GC cells.
miR-424-3p prevents EMT by inhibiting RHOXF2 in vitro
Exosomal miR-424-3p, consistent with the silencing of the RHOXF2 gene, was demonstrated to inhibit the EMT in GC cells, resulting in the suppression of GC cell progression. To investigate whether RHOXF2 is a critical gene under negative regulation by miR-424-3p, contributing to its tumorigenic effects, we performed a series of experiments. We transfected or co-transfected SGC-7901 and AGS cells with miR-424-3p mimics, miR-424-3p inhibitors and sh-RHOXF2, including appropriate controls, and monitored the phenotypic changes. By modulating the expression of miR-424-3p and RHOXF2, we were able to inhibit the growth, migration and invasion of SGC7901 and AGS cells (Fig. 5A–F, Fig. S4). Furthermore, downregulation of miR-424-3p in these cells enhanced tumorigenicity, which was abolished by RHOXF2 silencing (Fig. 5A–F). In addition, analyses of EMT-related markers revealed an increase in N-cadherin and vimentin expression when miR-424-3p was downregulated, and a loss of E-cadherin expression, which was subsequently restored by RHOXF2 silencing (Fig. 5G–I). Taken together, these findings strongly support the role of miR-424-3p in inhibiting the EMT in GC cells through the inhibition of RHOXF2.
miR-424-3p upregulation inhibits the EMT in gastric cancer by targeting RHOXF2. (A, B) Clonogenic assay assessing the tumorigenic potential of gastric cancer cells treated with miR-424-3p mimics, miR-424-3p inhibitors, or miR-424-3p inhibitors combined with sh-RHOXF2 or sh-NC. (C, D) A CCK-8 cell assay evaluated the growth of gastric cancer cells treated with miR-424-3p mimics, miR-424-3p inhibitors, or miR-424-3p inhibitors combined with sh-RHOXF2 or sh-NC. (E) Wound healing assays (scale bar = 100 μm) examined the migration of gastric cancer cells treated with miR-424-3p mimics, miR-424-3p inhibitors, or miR-424-3p inhibitors combined with sh-RHOXF2 or sh-NC. (F) Transwell invasion assays (scale bar = 50 μm) investigated the invasion of gastric cancer cells treated with miR-424-3p mimics, miR-424-3p inhibitors, or miR-424-3p inhibitors combined with sh-RHOXF2 or sh-NC. (G) Western blot analysis of E-cadherin, N-cadherin, and vimentin protein expression in gastric cancer cells treated with miR-424-3p mimics, miR-424-3p inhibitors, or miR-424-3p inhibitors combined with sh-RHOXF2 or sh-NC. (H, I) qRT-PCR analysis of E-cadherin, N-cadherin, and vimentin mRNA levels in gastric cancer cells treated with miR-424-3p mimics, miR-424-3p inhibitors, or miR-424-3p inhibitors combined with sh-RHOXF2 or sh-NC. Measurements are expressed as mean ± standard deviation. Comparisons between multiple groups were analyzed by one-way ANOVA, followed by Tukey’s multiple comparison test. All statistical data were obtained from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
miR-424-3p exhibited anti-tumor characteristics in vivo
Subsequently, we isolated exosomes from hBM-MSCs transfected with miR-424-3p agomir and NC agomirs, and administered them into nude mice via a tail vein injection. To confirm the successful delivery of miR-424-3p to the tumor tissue, we assessed its expression levels. The results revealed a significant increase in miR-424-3p expression following the injection of Exo-miR-424-3p agomirs (Fig. 6A). Upon final tumor extraction and weighing, the tumors treated with Exo-miR-424-3p agomirs exhibited a significant reduction in final weight (Fig. 6B). Furthermore, we observed a marked reduction in tumor weight and growth rate in mice receiving hBM-MSC-derived exosomes overexpressing miR-424-3p (Fig. 6C, D). We then collected tumor tissues from the nude mice (Fig. 6E) and evaluated the expression patterns of RHOXF2 and EMT-related markers. The findings demonstrated that upregulation of miR-424-3p via hBM-MSC-derived exosomes significantly decreased the expression of RHOXF2, N-cadherin, and vimentin in the tumor tissues (Fig. 6F-H). Further IHC staining of tumor sections corroborated these results, showing an increase in E-cadherin-positive cells and a decrease in RHOXF2, N-cadherin, and vimentin-positive cells, with a corresponding decline in IRS immunoreactivity scores relative to the Exo-NC agomir group (Fig. 6I). In summary, the data convincingly demonstrated that miR-424-3p, contained within hBM-MSC-derived exosomes, was effectively transferred to GC cells. By downregulating RHOXF2, miR-424-3p disrupted the EMT process, leading to the suppression of heterotransplanted tumors in nude mice.
Exosomal miR-424-3p derived from mesenchymal stem cells inhibits the growth of gastric cancer in nude mice. (A) Expression of miR-424-3p in tumor tissue as determined by qRT-PCR. (B) Weight of transplanted tumors in nude mice, n = 5. (C, D) Tumor volume changes in SGC-7901 and AGS tumor-transplanted nude mice, n = 5. (E) Representative images of transplanted tumors in nude mice, n = 5. (F) Protein levels of RHOXF2, E-cadherin, N-cadherin, and vimentin in GC tumors detected by Western blot. (G, H) mRNA expression levels of RHOXF2, E-cadherin, N-cadherin, and vimentin in SGC-7901 and AGS tumor tissues as determined by qRT-PCR. (I) Immunohistochemical staining of the target protein in transplanted tumor tissues (scale bar = 100 μm); the left image shows representative microscopic images, and the right image shows statistical results of the IRS score. IRS = SI × PP. SI was defined as: 0 = negative; 1 = weak; 2 = moderate; 3 = strong. PP was defined as: 0 = 0%; 1 = 0–25%; 2 = 25–50%; 3 = 50–75%; 4 = 75–100%. (J). Schematic diagram of the mechanism by which hBM-MSC-derived exosomes containing miR-424-3p affect gastric cancer cell activity. After treatment with oxaliplatin, hBM-MSCs transfer the upregulated miR-424-3p to gastric cancer cells via exosomes, thereby downregulating the expression of RHOXF2, inhibiting the EMT, and subsequently reducing the proliferation, migration, and invasion of gastric cancer cells, ultimately slowing the progression of gastric cancer. By Figdraw. Measurement data are presented as mean ± standard deviation; comparisons between two groups were made using an unpaired t test; comparisons between multiple groups were analyzed by one-way ANOVA, followed by Tukey’s multiple comparison test. All statistical data were obtained from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
Here, we showed that through exosome secretion, human bone hBM-MSCs exert a pro-tumor effect within an hBM-MSC and GC co-culture system. Notably, oxaliplatin treatment of hBM-MSCs upregulates miR-424-3p levels in these exosomes, which in turn potentially constrained the EMT by suppressing RHOXF2, which subsequently inhibited GC cell proliferation, migration, and invasion.
hBM-MSCs have been identified as a critical component of the TME and play an important role in interacting with tumor cells through various mechanisms3. In gastric carcinogenesis and development, hBM-MSCs have been shown to play diverse and complex roles. While some studies have established that hBM-MSCs enhanced the EMT process in GC cells, as demonstrated by increased Transwell and migration, including upregulation of proteins such as β-catenin, snail and twist20, other studies have reported that hBM-MSCs could have a tumor suppressive effect in some cases21,22. This tumor suppressive effect could be related to the origin of hBM-MSCs, their differentiation state and their interaction with tumor cells. To create a clinically relevant scenario, we established a co-culture model of hBM-MSCs with SGC-7901 and AGS cells. Our results showed that hBM-MSCs exhibited pro-tumor properties in the co-culture system (Fig. 1). Furthermore, we confirmed the occurrence of the EMT by cellular pathway enrichment analysis of mRNA sequencing of co-cultured GC cells, which was confirmed by western blotting and qRT-PCR analyses. hBM-MSCs co-cultured with SGC-7901 and AGS cells showed a lack of expression of the epithelial marker E-cadherin and an increase in expression of the mesenchymal cell markers N-cadherin and vimentin (Fig. 2). EMT is a pivotal process in epithelial tumor progression, rendering cancer cells more migratory and invasive15. Our findings revealed that hBM-MSCs facilitated the EMT in SGC-7901 and AGS, thereby stimulating cancer cell proliferation and enhancing migratory and invasive capabilities. Intriguingly, this effect was nullified when hBM-MSCs were treated with oxaliplatin. To the best of our knowledge, this study represents the first exploration of the impact of hBM-MSCs on GC cells following oxaliplatin treatment.
Furthermore, this study characterized hBM-MSC-exos and examined their uptake by SGC-7901 and AGS cells using immunofluorescence staining, confirming a direct interaction between hBM-MSC-exos and these GC cells (Fig. 1). It is noteworthy that exosomes derived from hBM-MSCs exert a substantial influence on tumor progression. These exosomes, which vary in size between 40–160 nm and are composed of nanoscale membrane vesicles, facilitate the transport of an extensive array of biomolecules, encompassing mRNAs, miRNAs, and proteins. Consequently, exosomes have gained recognition as vital mediators in intercellular communication23. For example, studies have demonstrated that hBM-MSC-derived exosomes can trigger the CaM-Ks/Raf/MEK/ERK pathway, leading to drug resistance in GC cells24. In a previous study, exosomes extracted from supernatants of BM-MSCs transfected with miR-1231 oligonucleotides inhibited proliferation, migration, invasion and matrix adhesion of pancreatic cancer cells BxPC-3 and PANC-118. Additionally, the downregulation of miRNAs in gastric cancer often results in the augmented expression and functionality of chemokines, receptors, and proteins linked to tumor cell invasion and metastasis25. Simultaneously, exosomes are instrumental in either enhancing or inhibiting tumor cell invasion and metastasis by conveying specific miRNAs. One study showed that miR-155-5p from exosomes of paclitaxel-resistant GC cells fostered the EMT and chemoresistance in paclitaxel-sensitive cells26. In this study, the absence of exosomes in the hBM-MSC culture medium negated any notable tumor-promoting effect on GC cells, strongly suggesting that hBM-MSCs promote the EMT process in these cells through paracrine exosomes. Following treatment with oxaliplatin, the miRNA expression profiles in hBM-MSC-exos were sequenced and analyzed (Fig. 4). Interestingly, our results revealed that oxaliplatin could alter the exosomes created by hBM-MSCs. This alteration hindered the EMT process in SGC-7901 and AGS cells by upregulating miR-424-3p, thereby inhibiting their proliferation, invasion, and migration (Fig. 4). We hypothesized that this may constitute one of the underlying mechanisms by which oxaliplatin operates. If the mechanisms of oxaliplatin's action within the tumor microenvironment can be fully elucidated, it could pave the way for the development of optimized anti-tumor treatment strategies, potentially benefiting numerous patients.
miR-424-3p, belonging to the miR-15/107 family, has a role in tumor progression that remains a subject of debate. Y. Li's analysis27 suggested that miR-424-3p regulates chemotherapy resistance, proliferation, invasion, and migration of GC cells through ABCC2. C. Lu et al.28 found that miR-424-3p's inhibition of PTEN/PI3K/Akt led to reduced proliferation, increased apoptosis, and heightened cisplatin sensitivity in small cell lung cancer cells. Conversely, J. Xu's research29 posited that miR-424 plays a tumor-suppressive role in cervical cancer by enhancing Chk1 and p-Chk1 expression. In both lung cancer19 and oral squamous carcinoma30, miR-424-3p downregulation may contribute to chemoresistance and tumorigenesis. Additionally, miR-424-3p reduces JAG1 expression, slowing the EMT progression in pituitary adenoma cells31. To explore its function, we augmented miR-424-3p expression in hBM-MSC-exos via hBM-MSC transfection, and successfully inhibited the EMT and tumor growth in GC cells in both in vitro and in vivo experiments (Fig. 5, 6). These results suggest that miR-424-3p could be a potential target to improve chemotherapy efficacy. A slight increase in miR-127-3p was also noted, and further investigation into its role in GC is currently underway. It is noteworthy that exosomes harbor a wide array of miRNAs capable of regulating various signaling pathways, thereby affecting recipient cells broadly. Moreover, tumor development is a complex interplay between cancer cells and the TME, and our co-culture model merely simulated the situation in which hBM-MSCs were unilaterally exposed to oxaliplatin. The changes in cancer cell phenotype may involve the participation of exosomes from various cells, necessitating more sophisticated techniques to enrich the miRNA-rich exosome subpopulation for comprehensive studies.
RHOXF2, also known as PEPP2, has been found to be highly expressed in various tumor cell lines, including GC cell lines. In the work of F. Shibata-Minoshima, the knockdown of RHOXF2 weakened the growth of GC cell lines, while overexpression of RHOXF2 rapidly induced autonomous growth and leukemia onset in transplanted bone marrow monocyte line HF6 cells in mice32. Recent research by L. Zhao also discovered that RHOXF2 is highly expressed in lung cancer tissues, and that downregulating RHOXF2 can inhibit lung cancer cell proliferation, migration, and invasion33. We selected RHOXF2 for further analysis using TargetScan, which provides a comprehensive scoring system. Candidate genes with high scores were considered, and RHOXF2 was chosen based on both its high score and its biological relevance. Our data indicated that high expression of miR-424-3p in GC cells can suppress RHOXF2 expression, and the EMT-promoting effects of miR-424-3p inhibitors were reversed by sh-RHOXF2. This evidence supports that miR-424-3p can halt the EMT process in GC cells by inhibiting RHOXF2 (Fig. 5, 6). Considering that miR-424-3p may have multiple targets in regulating the EMT, and our work has only clarified one target, and further experimental validation is required.
Conclusion
In summary, this study confirmed that hBM-MSCs primarily exert a pro-tumor effect in the GC microenvironment through paracrine exosomes, and that oxaliplatin attenuated this effect. We believe that we have uncovered a hidden mechanism through which oxaliplatin works. The upregulation of miR-424-3p in exosomes released by hBM-MSCs treated with oxaliplatin may inhibit the occurrence of EMT by suppressing RHOXF2, thereby inhibiting the proliferation, migration, and invasion of GC cells. Hence, delivering miR-424-3p through exosomes represents a promising anti-tumor treatment approach. However, our research has limitations regarding the mechanisms involved in this study, and further in-depth research is necessary.
Data Availability Statement
All exosomal miRNA sequencing data sets in this study have been deposited in the Gene expression Omnibus (GEO) database (GSE248357).
Abbreviations
- GC:
-
Gastric cancer
- Oxa:
-
Oxaliplatin
- TME:
-
Tumor microenvironment
- hBM-MSCs:
-
Human bone marrow-derived mesenchymal stem cells
- Oxa-MSCs:
-
Human bone marrow-derived mesenchymal stem cells treated with oxaliplatin
- hBM-MSC-exos:
-
Exosomes derived from human bone marrow mesenchymal stem cell
- Oxa-MSC-exos:
-
Exosomes derived from human bone marrow mesenchymal stem cell treated with oxaliplatin
- EMT:
-
Epithelial mesenchymal transition
- miR:
-
MicroRNA
- CCK-8:
-
Cell counting kit-8
- qRT-PCR:
-
Quantitative reverse transcription-polymerase chain reaction
- PBS:
-
Phosphate-buffered saline
- FBS:
-
Fetal bovine serum
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- PVDF:
-
Polyvinylidene difluoride
- SDS-PAGE:
-
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- TEM:
-
Transmission electron microscopy
- DAPI:
-
4',6-Diamidino-2-phenylindole
- RIPA:
-
Radioimmunoprecipitation assay buffer
- TBST:
-
Tris-Buffered Saline and Tween 20
- cDNA:
-
Complementary DNA
- SI:
-
Staining intensity
- PP:
-
Percentage of positive cells
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- DEGs:
-
Differentially expressed genes
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Liu, D. et al. The patterns and timing of recurrence after curative resection for gastric cancer in China. World J. Surg. Oncol. 14, 305. https://doi.org/10.1186/s12957-016-1042-y (2016).
Belli, C. et al. Targeting the microenvironment in solid tumors. Cancer Treat. Rev. 65, 22–32. https://doi.org/10.1016/j.ctrv.2018.02.004 (2018).
Furusaka, Y. et al. Potent antitumor effects of the conditioned medium of bone marrow-derived mesenchymal stem cells via IGFBP-4. Cancer Sci. 114, 2499–2514. https://doi.org/10.1111/cas.15789 (2023).
Ikeda, T. et al. Mesenchymal stem cell-derived CXCL16 promotes progression of gastric cancer cells by STAT3-mediated expression of Ror1. Cancer Sci. 111, 1254–1265. https://doi.org/10.1111/cas.14339 (2020).
Azmi, A. S., Bao, B. & Sarkar, F. H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 32, 623–642. https://doi.org/10.1007/s10555-013-9441-9 (2013).
Huang, X. et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 14, 319. https://doi.org/10.1186/1471-2164-14-319 (2013).
Marques, T. M. & Gama-Carvalho, M. Network approaches to study endogenous RNA competition and its impact on tissue-specific microRNA functions. Biomolecules https://doi.org/10.3390/biom12020332 (2022).
Mittelbrunn, M. et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282. https://doi.org/10.1038/ncomms1285 (2011).
Zhang, F. et al. Mesenchymal stem cell-derived exosome: A tumor regulator and carrier for targeted tumor therapy. Cancer Lett. 526, 29–40. https://doi.org/10.1016/j.canlet.2021.11.015 (2022).
Zhang, X. et al. Exosomes from bone marrow microenvironment-derived mesenchymal stem cells affect CML cells growth and promote drug resistance to tyrosine kinase inhibitors. Stem Cells Int. 2020, 8890201. https://doi.org/10.1155/2020/8890201 (2020).
Choi, D.-W. et al. Extracellular vesicles from tonsil-derived mesenchymal stromal cells show anti-tumor effect via miR-199a-3p. Int. J. Mol. Med. https://doi.org/10.3892/ijmm.2021.5054 (2021).
Xu, H. et al. Mesenchymal stem cell-derived exosomal microRNA-133b suppresses glioma progression via Wnt/β-catenin signaling pathway by targeting EZH2. Stem. Cell. Res. Ther. 10, 381. https://doi.org/10.1186/s13287-019-1446-z (2019).
Kimura, Y. et al. Intraperitoneal transfer of microRNA-29b-containing small extracellular vesicles can suppress peritoneal metastases of gastric cancer. Cancer Sci. 114, 2939–2950. https://doi.org/10.1111/cas.15793 (2023).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15, 178–196. https://doi.org/10.1038/nrm3758 (2014).
Malyla, V. et al. Extracellular vesicles released from cancer cells promote tumorigenesis by inducing epithelial to mesenchymal transition via β-catenin signaling. Int. J. Mol. Sci. https://doi.org/10.3390/ijms24043500 (2023).
National Research Council Committee for the Update of the Guide for the, C. & Use of Laboratory, A. in Guide for the Care and Use of Laboratory Animals (National Academies Press (US) Copyright © 2011, National Academy of Sciences, 2011).
Shang, S. et al. Exosomal miRNA-1231 derived from bone marrow mesenchymal stem cells inhibits the activity of pancreatic cancer. Cancer Med. 8, 7728–7740 (2019).
Zhang, M., Zeng, J., Zhao, Z. & Liu, Z. Loss of MiR-424-3p, not miR-424-5p, confers chemoresistance through targeting YAP1 in non-small cell lung cancer. Mol. Carcinog. 56, 821–832. https://doi.org/10.1002/mc.22536 (2017).
Xue, Z. et al. Mesenchymal stem cells promote epithelial to mesenchymal transition and metastasis in gastric cancer though paracrine cues and close physical contact. J. Cell Biochem. 116, 618–627. https://doi.org/10.1002/jcb.25013 (2015).
Li, W. et al. Gastric cancer-derived mesenchymal stem cells prompt gastric cancer progression through secretion of interleukin-8. J Exp. Clin. Cancer Res. 34, 52. https://doi.org/10.1186/s13046-015-0172-3 (2015).
Chaturvedi, P. et al. Hypoxia-inducible factor-dependent breast cancer-mesenchymal stem cell bidirectional signaling promotes metastasis. J. Clin. Invest 123, 189–205. https://doi.org/10.1172/JCI64993 (2013).
Lin, Z. et al. Mesenchymal stem cell-derived exosomes in cancer therapy resistance: recent advances and therapeutic potential. Mol. Cancer 21, 179. https://doi.org/10.1186/s12943-022-01650-5 (2022).
Ji, R. et al. Exosomes derived from human mesenchymal stem cells confer drug resistance in gastric cancer. Cell Cycle 14, 2473–2483. https://doi.org/10.1080/15384101.2015.1005530 (2015).
Wei, L. et al. Noncoding RNAs in gastric cancer: implications for drug resistance. Mol. Cancer 19, 62. https://doi.org/10.1186/s12943-020-01185-7 (2020).
Wang, M. et al. Paclitaxel-resistant gastric cancer MGC-803 cells promote epithelial-to-mesenchymal transition and chemoresistance in paclitaxel-sensitive cells via exosomal delivery of miR-155-5p. Int. J. Oncol. 54, 326–338. https://doi.org/10.3892/ijo.2018.4601 (2019).
Li, Y. et al. miR-424-3p contributes to the malignant progression and chemoresistance of gastric cancer. Onco Targets Ther 13, 12201–12211. https://doi.org/10.2147/OTT.S280717 (2020).
Lu, C. et al. Baicalein inhibits cell growth and increases cisplatin sensitivity of A549 and H460 cells via miR-424-3p and targeting PTEN/PI3K/Akt pathway. J. Cell Mol. Med. 22, 2478–2487. https://doi.org/10.1111/jcmm.13556 (2018).
Xu, J. et al. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene 32, 976–987. https://doi.org/10.1038/onc.2012.121 (2013).
Scholtz, B., Horváth, J., Tar, I., Kiss, C. & Márton, I. J. Salivary miR-31–5p, miR-345–3p, and miR-424–3p are reliable biomarkers in patients with oral squamous cell carcinoma. Pathogens https://doi.org/10.3390/pathogens11020229 (2022).
Chen, Y. et al. JAG1, Regulated by microRNA-424-3p, involved in tumorigenesis and epithelial-mesenchymal transition of high proliferative potential-pituitary adenomas. Front. Oncol. 10, 567021. https://doi.org/10.3389/fonc.2020.567021 (2020).
Shibata-Minoshima, F. et al. Identification of RHOXF2 (PEPP2) as a cancer-promoting gene by expression cloning. Int. J. Oncol. 40, 93–98. https://doi.org/10.3892/ijo.2011.1173 (2012).
Zhao, L., Zhang, X., Guo, H., Liu, M. & Wang, L. LOXL1-AS1 contributes to non-small cell lung cancer progression by regulating miR-3128/RHOXF2 Axis. Onco Targets Ther 13, 6063–6071. https://doi.org/10.2147/OTT.S247900 (2020).
Acknowledgements
We acknowledge and appreciate our colleagues for their valuable efforts and comments on this paper.
Funding
The costs of publication will be funded by grants from the Joint Funds of the National Natural Science Foundation of China (U2004132, N.L.), Leading Talents in Health Science and Technology Innovation for Young and Middle-aged People in Henan Province (YXKC2021008, N.L.), the National Natural Science Fund Youth Fund of China (82003041, C.W.), and Key Scientific and Technological Projects in Henan Province (222102310324, C.W.).
Author information
Authors and Affiliations
Contributions
Suxia Luo was the principal investigator who obtained the financial supports. Suxia Luo and Wei Shen conceived and designed the study. Wei Shen and Chen Wei drafted the manuscript. Chen Wei and Ning Li helped to performed the mRNA and miRNA expression profile analysis. Yan Li prepared the tables and designed figures. Wei Shen, Wenyue Yu and Xinyi Yang performed the experimental studies. All the authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shen, W., Wei, C., Li, N. et al. Oxaliplatin-induced upregulation of exosomal miR-424-3p derived from human bone marrow mesenchymal stem cells attenuates progression of gastric cancer cells. Sci Rep 14, 17812 (2024). https://doi.org/10.1038/s41598-024-68922-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-68922-6
- Springer Nature Limited