Abstract
Next-generation sequencing (NGS) is becoming more relevant for medical diagnostics, especially for using cell-free DNA to monitor response to therapy in cancer management, as high sensitivity of NGS enables detection of rare events. Sequencing Library preparation is a time-consuming and complex process, and large-scale liquid handlers are often used for automation. However, for smaller labs and low-to-medium throughput samples, these liquid handlers are expensive and need experts for handling. This work presents a proof-of-concept for library preparation on a commercially available and open lab-on-a-chip platform, which provides an alternative automation for low-to-medium throughput requirements. It covers common library preparation steps optimized to a microfluidic environment that include customizable PCR for target enrichment, end-repair, adapter ligation, nucleic acid purification via magnetic beads, and an integrated quantification step. The functionality of the cartridge is demonstrated with reference cfDNA containing different allelic frequencies of seven known mutations. Processing the samples in the cartridge reveals highly comparable results to manual processing (Pearson r = 0.94) based on amplicon sequencing. Summarized, the proposed automated lab-on-a-chip workflow for customizable library preparation could further pave the way for NGS to evolve from a technology used for research purposes to one that is applied in routine cancer management.
Similar content being viewed by others
Introduction
Next-generation sequencing (NGS) has become a widely used and developed method in research1. NGS plays a crucial role in diagnostics, especially for cancer, as the methodology allows sensitive detection of mutations, which can be critical for therapy decisions. Regular screenings could identify therapy resistances quickly by observing tumor progression, as opposed to testing once at the beginning of therapy. As biopsies require surgical intervention, they are unsuitable for regular sampling, while liquid biopsies, like blood samples, are non-invasive. The high sensitivity of the NGS method is needed to find rare events like mutations in cfDNA of tumorigenic identity.
Since NGS has also become important for clinical diagnostics in the context of precision medicine, automated solutions for library preparation have become increasingly relevant. Factors including protocol complexity, contamination, and costs are the three main challenges in preparing a sequencing library that can be overcome by automated systems, according to Hess et al.2. Automation enables higher throughput, leading to lower costs and higher reproducibility with less risk of contamination3.
Liquid handlers enable high throughput of samples, thus decreasing the cost per sample, but require high investments and experts for handling. As sequencers become smaller, including the iSeq (Illumina, San Diego) or the portable MinIon (Oxford Nanopore Technologies, Oxford), point-of-care (PoC) sequencing seems to be within reach. Microfluidic lab-on-a-chip (LoC) devices could drive the PoC-sequencing approach forward to enable small hospitals, ambulatory healthcare centers with small labs, or doctor’s offices to use sequencing for diagnostics and therapy monitoring. Here, the focus is not on high sample throughput but on small investment costs and quick results. Beyond this, the device should be able to be used by non-experts with a simple and flexible workflow, which allows for different library preparation methods to address several diagnostic approaches. For automated library preparation, different kinds of microfluidic systems were already demonstrated3,4,5,6,7,8,9,10. Systems produced by multilayer soft lithography, like the automated library preparation of Kim et al. based on the tagmentation chemistry, can be easily designed for small volumes and high sample throughput, but a lack of scalability results in high costs per single sequencing reaction6. The microfluidic device by Murphy et al. was manufactured using a computer numerical control (CNC) milling machine and enabled the preparation of sequencing libraries with a DNA input of only 10 pg7. Nevertheless, the system only offers a semi-automated solution so far, as the library amplification has to be finalized manually7. However, for a PoC diagnostic device, a low number of sample processing steps in a fully automated way and inexpensive production of the microfluidic unit would be more applicable3. One microfluidic system that addresses these challenges is the centrifugal microfluidics approach by Hess et al.3,8,11. They used disposable cartridges that can be produced cheaply and process samples with pre-stored reagents3,8,11. They implemented a two-step library preparation with multiplex PCR and a library preparation for a whole genome sequencing approach by tagmentation3,8,11. Using a one-step PCR that targets the region of interest and attaches adaptors necessary for sequencing makes the library preparation reasonably short for a microfluidic workflow. However, primers for a multiplex PCR with sequencing adaptors and barcodes, which are still specific enough to target the defined regions, require a high development effort for diverse diagnostic approaches. Microfluidic library preparation by tagmentation chemistry is often used in microfluidic systems, where a transposase enzyme adds adaptors on both ends of the DNA and fragments the DNA in one step. This allows a shorter library preparation compared to a classical procedure, where several consecutive steps such as fragmentation, end-repair, and adaptor ligation are necessary12. However, a defined transposase complex-to-DNA ratio is critical to get the required fragment size distribution for sequencing12. Since fragmentation is an essential part of adaptor ligation, the use of already small DNA fragments, like cfDNA, or small amplicons is not recommended. As the transposase cannot add adaptors at the end of a DNA fragment, it can lead to a drop in sequencing coverage at the distal ends13. For this reason, the classical ligation procedure is preferred for short DNA fragments like cfDNA and enables moreover higher flexibility using a microfluidic system for several diagnostic sequencing approaches, even though this involves more effort in transferring the individual experimental steps on the LoC system.
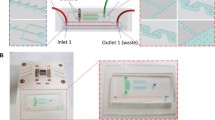
Within this work, we used the commercially available Vivalytic cartridge from Bosch Healthcare Solutions GmbH (Waiblingen, Germany) to prepare a targeted sequencing library (Fig. 1a), which is open for different kinds of downstream diagnostic approaches. We designed a multiplex PCR targeting multiple tumor-associated single nucleotide variants (SNVs) of a cell-free DNA (cfDNA) reference material. Due to variable allelic frequencies of 0%, 0.1%/0.13%, 1%/1.3%, and 5%/6.3%, this biologically relevant reference material allows determination of the performance of the automated library preparation (hereafter referred to as 'on-chip') compared to the manual workflow (hereafter referred to as 'off-chip'). All necessary steps have been integrated into two cartridges to implement a sample preparation for NGS sequencing (Fig. 1b). These include all required enzyme reactions for targeted PCR, end-repair, ligation, and enrichment PCR. Besides the enzyme steps, a short DNA purification with carboxylated magnetic beads using solid-phase reversible immobilization was applied. To process the sample into a ready-to-load sequencing library, quantification has also been integrated separately. Thus, the cartridge of the Vivalytic system allows automated sample preparation for cfDNA samples with enriched targets and is still open to different downstream sequencing approaches.
Schematic Vivalytic cartridge (a) and a workflow for a proof-of-concept for automated library preparation (b) a: Reagents can be pre-stored on the cartridge and transported via the microfluidic network to the different process units like the mixing chamber, PCR strands, or array chamber. The read-out of arrays and qPCRs can be analyzed with a camera system above the detection area. b: The library preparation requires 48 manual steps, which can be fully-automated on the Vivalytic cartridges. Pur.: Purification; iPCR: index PCR.
Methods
Automated library preparation on the commercially available Vivalytic lab-on-a-chip platform
The lab-on-a-chip (LoC) system "Vivalytic" from Bosch Healthcare Solutions GmbH (Waiblingen, Germany) was launched in 2020 and comprises the Vivalytic Analyser and the assay-specific Vivalytic cartridges. The analyzer contains a pneumatic unit to control the microfluidics, a thermal system consisting of 12 heaters, and an optical readout system for qPCRs and microarrays. The cartridge consists of four layers, combined by laser welding. The fluidic layer and a pneumatic layer are separated by a thermoplastic polyurethane (TPU) membrane, which enables the actuation. The principle is described by Rupp et al.14. At this point, it should be mentioned that polyurethanes have the property to show slight swelling in aliphatic alcohols like ethanol or propan-2-ol. For this reason, it should be avoided that fluids containing aliphatic alcohols come into contact with the microfluidic network of the Vivalytic cartridge. The fluids are transported with pump chambers and valves within the cartridge (Fig. 1 a). There are different pumping mechanisms to transport the fluids inside the microfluidic environment. In this work, we tested the chamber pumping against the peristaltic pump mechanism. Chamber pumping is defined as the transportation of fluids by opening and closing a unit operation consisting of a valve, a pump chamber, and another valve. The peristaltic pump mechanism works the same way; however, instead of a pump chamber, one or more valves are involved. This allows the transport of smaller volumes during one pump cycle. For a PCR reaction, the mix is shuttled between three pumping chambers with different temperature zones (PCR strands) within the Vivalytic cartridge (shuttling PCR). To avoid manual steps, reagents for PCR or purifications can be directly stored on-chip, in reagent reservoirs for buffers, and lyophilized beads inside the pump chambers for the enzymes. In addition, the cartridge consists of a sample input chamber, a mixing chamber, and a waste chamber.
The microfluidic workflow was developed by programming each step separately and was optimized by processing each workflow with fluorescein-dye labeled buffers, which were used for visual confirmation of filling and control procedures. Temperature-dependent processes were also controlled using thermal elements, which were integrated into the cartridge. Ultimately, each microfluidic module of the automated library preparation was tested individually and then sequentially combined. Modules included a multiplex PCR to enrich the relevant targets on reference cfDNA samples, three enzyme steps (end-repair, ligation, and adapter finalization), three magnetic bead nucleic acid purifications by solid phase reversible immobilization (SPRI), and an Index-PCR (iPCR). The final microfluidic workflow for the complete library preparation is explained in Supplementary File 1 in detail.
The automation of library preparation was realized by using the reagents of the NEBnext Ultra II Library Kit for Illumina (New England Biolabs, Ipswich). The microfluidic processed samples were obtained and quantified by Qubit 1X dsDNA HS Assay-Kit with the Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham) and analyzed with the HS NGS Fragment Analysis Kit and the Fragment Analyzer (Agilent, Santa Clara). The libraries were sequenced on the MiSeq system (Illumina, San Diego), using the Miseq Reagent Kit v2 and sequenced with 2 × 150 cycles.
Demultiplexing, FASTQ data generation, and alignment of the internal PhiX control were processed using Local Run Manager software (Illumina, San Diego) immediately after sequencing on the MiSeq system. Adapter and base quality trimming of the data was performed using BBDuk (algorithm by Brian Bushnell). To align the data to the human reference genome (Genome Reference Consortium Human Build 37), mapping was performed using the Burrows-Wheeler Algorithm (BWA-MEM), and the processed data were finally analyzed using the Integrative Genomic Viewer (Broad Institute and the Regents of the University of California).
Library preparation reference workflow
The reference workflow was realized with the NEBnext Ultra II Library Kit for Illumina (New England Biolabs, Ipswich), according to the manufacturer’s specifications with the following modifications. The end-repair reaction was performed in half volume and incubated for 10 min at 20 °C and 10 min at 65 °C. Ligation and USER reactions were also performed using half the volume, and a working concentration of 1.5 μM of NEB adapters was applied. For the index PCR, the reaction mix was prepared with 7 μl adapter-ligated DNA, 10 μl NEBnext Ultra II Q5 Master Mix, and 5 μl each of i5 and i7 Primer (NEBnext Multiplex Oligos for Illumina, Dual Index Primers Sets). The quantity and quality control (QC) as well as sequencing were performed as described before.
Multiplex PCR
To show a targeted sequencing approach as a technical proof-of-principle, a multiplex PCR was designed to enrich clinically-relevant mutation regions of the Multiplex I cfDNA reference standard set (Horizon, Cambridge). The sequences contain the regions of the mutations EGFR-L858R (0.2 µM), EGFR-delE746-A750 (0.6 µM), EGFR-T790M (0.2 µM), KRAS-G12D (0.6 µM), NRAS-Q61K, NRAS-A59T (both NRAS mutations flanked by one primer pair with 0.4 µM), and PIK3CA-E545K (0.6 µM). The final concentration for the adapted approach is shown in brackets. The equimolar approach was performed with a final concentration of 0.2 µM. The primers were produced by Biomers (Ulm, Germany). Q5 High-Fidelity 2X Master Mix (New England Biolabs, Ipswich) was used for the PCR. The temperature program started with an initial denaturation of 30 s at 98 °C. The mix was then cycled 35 times with denaturation of 10 s at 98 °C, annealing/extension of 10 s at 65 °C, and a final extension of 5 min at 65 °C. To analyze the amplicon distribution, the products were prepared for sequencing as described before, and the amplicon reads were related to the totalized reads.
Microfluidic magnetic bead purification
For the implementation of a magnetic bead-based purification on the Vivalytic LoC cartridge, SPRIselect beads (Beckman Coulter, Indianapolis) were used according to the manufacturer’s information. The purification efficiencies were determined and compared to each other before and after purification by concentration measurement with the Qubit 1X dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham). Therefore, a 280 bp amplicon was used, corresponding to the library size. To make the protocol compatible with the material composition of the Vivalytic LoC cartridge, the elution was performed with an appropriate elution buffer (EB, Bosch Healthcare Solutions, Waiblingen) instead of water, and a wash buffer (WB, Bosch Healthcare Solutions, Waiblingen) instead of ethanol. To test the impact of dried pre-storage of magnetic beads on their purification performance, beads were separated, washed with 80% ethanol, and then dried and stored at room temperature. At different time points, up to 12 weeks, the beads were resuspended in binding buffer and the purification was done as described before. The dried beads were resuspended in polyethylene glycol 8000 (Sigma Aldrich, St. Louis), transferred to the cartridge, and dried for 5 h at room temperature. Because the Vivalytic Analyser currently does not consist of a magnet, a neodymium plate magnet (RS Components, Frankfurt/Main) was fixed with double-faced adhesive tape on the cartridge.
Quantification
All quantifications were performed with the Qubit 1X dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham) on the Qubit 4 Fluorometer (Thermo Fisher Scientific, Waltham). To analyze the fragment sizes of the samples, the quality controls were performed with the HS NGS Fragment Kit on the Fragment Analyzer (Agilent, Santa Clara). The Qubit 1X dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham) was also used for the on-chip quantification. A serial dilution was automatically performed with ratios of 1:2, 1:4, and so forth until a ratio of 1:64 was reached. Every step was documented and analyzed with Matlab (Mathworks Inc.) by gray-scale determination.
Results
Amplicon distribution of the multiplex PCR are comparable for on- and off-chip processed samples
NGS is mainly used as a targeted approach in clinical diagnostics. Targeted sequencing allows for higher sensitivity, better quality, lower turnaround time, and reduced costs15,16. However, the target sequences must already be known. For microbial meta-analysis, e.g. sepsis diagnostic17,18 or hospital contamination screenings19, whole genome sequencing (WGS) is more useful. Therefore, targeted enrichment should be feasible as an optional module in the microfluidic process, but it should not exclude a universal enrichment process. For the proof-of-concept, we developed a low-multiplex PCR with six primer pairs to enrich cancer treatment-relevant mutations. The multiplex PCR initially showed imbalanced amplicon distribution using equimolar primer concentrations of 0.2 µM, as only a few reads could be sequenced of the amplicons, targeting the regions of interest of PIK3CA-E545K, KRAS-G12D and EGFR delE746-A750 (Fig. 2—Off-chip equimolar). To detect low-abundant mutations, each amplicon must be sequenced often enough to statistically detect these mutations. One possible solution is to increase the sequencing throughput to generate more reads for every amplicon. As the distribution of the different amplicons is imbalanced, higher sequencing throughput would generate enough reads for the low-represented amplicons. However, too many reads of the over represented amplicons lower cost efficiency. Therefore, we adapted the primer amount to allow a more consistent amplicon distribution (Fig. 2—Off-chip adapted). In addition, we compared on- and off-chip processed samples to determine possible influences on the amplicon distribution due to different PCR cycling mechanisms (thermoblock versus microfluidic shuttle PCR). The adapted on-chip library processing showed a similar amplicon distribution (Fig. 2). Hence, the shuttling between temperature zones in the microfluidic platform has no influence on the target enrichment of a multiplex PCR.
Amplicon distribution of the multiplex PCR after sequencing. The multiplex PCR was performed with an equimolar concentration (0.2 µM of each primer, n = 4) and with adapted primer concentrations off-chip (n = 12) and on-chip (n = 12). The used adapted primer concentration for each amplicon is from top down as follows: 0.6 µM, 0.4 µM, 0.6 µM, 0.2 µM, 0.6 µM, 0.2 µM.
Magnetic bead purification as a new purification module of the microfluidic Vivalytic platform
Solid phase reversible immobilization (SPRI) beads are the gold standard for the purification and size selection of library preparations for sequencing. Different ratios of sample to bead-buffer-mixture allow scaling of the purification and can also be rapidly adapted to the desired fragment length for size selection16,17. We used a microfluidic compatible buffer for elution and washing, which showed the same or slightly better performance during off-chip analysis than the commercial reference buffer (Fig. 3a). Moreover, the purification efficacy of dried magnetic beads was tested for up to 12 weeks (Fig. 3a). To implement a magnetic bead purification protocol on the Vivalytic LoC cartridge, we tested the beads on-chip as supplied by the manufacturer in binding buffer (liquid) and in dried form. In addition, the fluid transport for washing and eluting was performed by two different pump strategies (chamber versus peristaltic) and compared (Fig. 3c). Chamber pumping, defined as straight actuation of the chamber in which the beads are immobilized with a magnet, led to the loss of beads. Therefore, for liquid and dried beads, a purification efficiency of only 25% and 5%, respectively, was observed. To minimize bead-loss, the microfluidic procedure was adapted. Instead, the buffer could be transported bead-free by using peristaltic pumping out of the chamber. Thus, a purification efficiency of around 50% and 40% for liquid and dried beads could be reached, respectively. The dried beads show slightly poorer outcomes, although off-chip tested dried beads did not show any performance loss within a storage time of 12 weeks. Based on these results, the purification with beads suspended in binding buffer in combination with the peristaltic microfluidic procedure was further optimized and used for the automated library preparation (the final detailed microfluidic purification protocol is described in Supplementary File 1).
Implementation of a magnetic bead based DNA purification process on the Vivalytic LoC cartridge. A 280 bp amplicon was purified with SPRIselect beads in different ways, quantified with the Qubit 1X dsDNA HS kit, and set to an off-chip reference (ref.). The reference (ref.) was processed according to the manufacturer's manual. (a) Off-chip testing (ref. n = 18) of microfluidic compatible elution buffer (EB, n = 5) and wash buffer (WB, n = 5). (b) Purification efficiency of dried magnetic beads after different storage times (n = 5, ref. n = 15). (c) Purification efficiency of liquid and dried magnetic beads resuspended either with chamber or peristaltic pumping on the Vivalytic cartridge. The beads were either applied in binding buffer (liquid: chamber pumping n = 5, peristaltic pump n = 4) or dried in the PCR chamber (dried: chamber pump n = 3, peristaltic pump n = 3, ref. n = 14). Error bars display standard deviation; x = median.
Translation of the complex process chain for library preparation into the microfluidic environment
We used the commercially available kit (NEBNext Ultra II Library Prep Kit, New England Biolabs, Ipswich) as a proof-of-concept for the core modules that are required for generating a sequencing library in the microfluidic LoC cartridge (e.g. the Vivalytic system): DNA end-repair (containing simultaneous dA-tailing and 5’ phosphorylation), adapter ligation, and index PCR. Typically, sequencing library preparations begin with a fragmentation step. However, as the amplicons used in this study are designed for cfDNA analysis, the DNA is already short enough, and a fragmentation step is not required. If a fragmentation step would be required, a module that combines the fragmentation with the end-repair could be easily used instead (e.g. NEBnext Ultra II FS Library Prep Kit, New England Biolabs, Ipswich). The end-repair reaction prepares the DNA for adapter ligation, which is followed by purification. The subsequent index PCR (iPCR) finalizes the adapters and enriches the library. Before sequencing, a final purification step is required to remove abundant PCR reagents.
Implementation challenges of these enzymatic reactions were the application in small volumes, the dependence of buffers between end-repair/dA-tailing and ligation, and the minimum process temperature of the Vivalytic Analyser of 40 °C, even though end-repair and ligation should be performed optimally at 20 °C according to the manufacturer manual. Additionally, the end-repair step requires an inactivation temperature of 65 °C to avoid inhibition of the following on-chip ligation. This is also critical for residual amounts of reaction mix in the microfluidic channels. Therefore, we adapted and optimized all the fluidic unit operations according to the requirements of the reaction and the boundary conditions of a microfluidic environment (details summarized in Supplementary File 1). For example, to avoid functional enzyme contamination (Supplementary Fig. 1d), we developed fluidic workflows where the entering path is not the exit path. The ligation enzyme steps (ligation and USER, to cut the NEB hairpin adapters) were integrated into the mixing chamber to allow short distances to the reagent reservoirs and to be able to mix different volumes (Supplementary Fig. 1d,e). We prefilled a channel with adapters of a defined length to add a few defined microliters to the mixture (Supplementary Fig. 1d2). After implementing a functional microfluidic workflow for each enzymatic and purification step, the modules were combined and tested with gDNA. During the first stage, the multiplex PCR was combined with the purification, end-repair and the ligation step (Fig. 4a). For the second stage, a defined amount of manually processed ligated samples were purified, amplified (index PCR), and again purified on-chip (Fig. 4b). The complete workflow, stage I and II combined, is shown in (Fig. 4c).
Quantity and quality control of on-chip prepared libraries to manually (off-chip) processed ones. (a) Stage I prepared libraries from targeted enrichment PCR until fully processed ligation (n = 4). Libraries were finished off-chip with 10 cycles in the index PCR. (b) Ligated DNA was normalized to one concentration and on-chip purified, amplified via index PCR (15 cycles), and again purified on cartridge (off-chip n = 4, on-chip n = 6). (c) Whole workflow (Stage I + II) with 10 ng gDNA input and 15 cycles in the index PCR (n = 4). Dashed lines indicate the minimum DNA library amount for sequencing of 4 nM for 25 μl of an average library size of 270 bp. Quantity measurements were performed with the Qubit 1X dsDNA HS kit. Quality controls were performed with the HS NGS Fragment Kit (digital blot images are cropped). Error bars display standard deviation.
A library preparation was successful when generating an amount of > 4 nM (shown as a dashed line) without side products, as indicated by the quality control. In both cases (on- and off-chip), yields well above the minimum library amount were achieved, and did not show any interfering side products. Due to the different PCR processes of the shuttling PCR on-chip and classical thermal block PCR off-chip, the cycles were adapted. Taking the same increased number of cycles for the off-chip control as optimized for on-chip processing, some side products between 100 and 200 bp occurred in the off-chip samples. Therefore, the off-chip controls for further sequencing analysis were performed with a lower cycling amount, which eliminated side products. In summary, the implementation of the complex enzymatic and purification processes to prepare a sequencing library on-chip could be shown to be functional and successful.
Microfluidic workflow performs equally to the manual processed samples
For analyzing the performance of the automated library preparation on a sequencing level, a reference cfDNA with different mutational frequencies (0%, 0.1%/0.13%, 1%/1.3%, and 5%/6.3%) was prepared and compared with the off-chip processed samples (read counts are summarized in Supplementary File 2). The mutation frequency shows the ratio of mutated to non-mutated target genes. For example, a mutation frequency of 0.1% means that the mutation is present in 1 of 1000 sequences. The frequencies vary within a sample, depending on the mutation. The mutation frequencies are 0.1%, 1%, and 5% for the EGFR gene. NRAS, KRAS, and PIK3CA mutations have frequencies of 0.13%, 1.3%, and 6.3%. Therefore, the samples are labeled with two mutation frequencies (e.g., 1%/1,3%). Overall, the sequencing results show a great Pearson correlation coefficient of 0.94 between all on- and off-chip libraries (Fig. 5).
For the samples with 5%/6.3% mutational frequency, the mutations could be detected independently of the processing with similar accuracy (Fig. 6). The average mutation frequency of the EGFR mutations is off-chip 4.7% ± 0.9%, and on-chip 4.2% ± 1.4%. For the KRAS, NRAS, and PIK3CA mutations, the average mutation frequency off-chip was 6.2% ± 0.4% and on-chip was 6.4% ± 0.5%. In general, the EGFR-delE746-A750 variant shows a lower frequency, as indicated by the supplier. This is the case for off- and on-chip prepared samples and is therefore process-independent. Comparing the samples of 1%/1.3% shows an average mutational frequency for EGFR mutations of 0.9% ± 0.3% off-chip and 0.7% ± 0.2% on-chip. For the KRAS, NRAS, and PIK3CA mutations, the average is 1.4% ± 0.2% off-chip and 1.0% ± 0.2% on-chip. The sample shows three outliers, which can only be determined for individual mutations and not for the entire sample. In addition, the outliers occur on- and off-chip, so a process difference cannot be assumed. High mutation frequencies are easier to detect with small sample inputs, as PCR and sequencing errors are less important. Detecting mutational frequencies of 0.1%/0.13% is difficult without using unique molecular identifiers (UMIs), which bioinformatically help correct PCR and sequencing errors. This is demonstrated by the fact that mutation events could rarely be detected for the wild type, which has a mutational frequency of 0% ± 0.02% for off- and on-chip processed samples (Supplementary File 3). Nevertheless, EGFR mutations with a 0.1% mutational frequency could be detected on average with 0.1% ± 0.08% for on-chip samples and 0.08% ± 0.01% for off-chip samples. The KRAS, NRAS, and PIK3CA mutations are specified by the manufacturer with a frequency of 0.13%. On average, the mutation frequencies are off-chip 0.17% ± 0.09% and on-chip 0.17% ± 0.04%. In general, it can be assumed, that due to the slight deviation, it is irrelevant whether the samples were processed manually or automatically on the cartridge.
Comparison of sequencing performance of off- and on-chip processed samples. Reference cfDNA with different mutational frequencies were used with 10 ng DNA input for targeted enrichment PCR. The 5%/6.3% sample was also processed with only 1 ng DNA input. Dotted lines divide mutations with different frequencies.
For the sample with a 5% mutational frequency, we also tested a DNA input of 1 ng, which is close to the given minimal input amount of the library kit (0.5 ng). The comparison of the average standard deviations of off- and on-chip with ± 1.4% and ± 2.4%, respectively, shows a more variable processing performance than with 10 ng as starting material (off-chip ± 0.4%, on-chip ± 0.5%). Thus, decreasing input material down to 1 ng affects the microfluidic performance between on- and off-chip, but the variation of off-chip prepared samples also increases from 0.4% with 10 ng to 1.4% with 1 ng. These observations show that the workflow, off-chip as well as on-chip, is getting less robust towards lower starting material, while on-chip processing is getting towards its limits when starting with 1 ng DNA. It is likely that, as the input complexity of the samples is tenfold lower than before, the increased loss of material, especially during the purification steps, is too high to detect the mutations in the same way as off-chip. However, a more complex sample, with a low input amount of 10 ng allowed for robust automatic library preparation. Taken together, the sequencing results show that library preparation on the Vivalytic LoC cartridge can be performed successfully.
Automated serial dilution allows microfluidic DNA quantification
Not included in the whole workflow of library preparation, but an essential part before sequencing is the quantification of the library. Quantification at the end of the library preparation can reduce further manual steps before sequencing. Consequently, we also integrated an on-chip quantification method with an intercalating dye and corresponding fluorometric readout separately, using the Qubit 1X dsDNA high sensitivity working solution. If the concentration is higher than the upper limit of detection (LOD) during the wet lab procedure, the sample is diluted and measured again. An automated dilution to find the optimal dilution step for quantification could overcome the challenge of excessive concentrations on the Vivalytic LoC system.
On the cartridge, one chamber filled with sample is diluted with one chamber filled with Qubit Working Solution (QWS), mixed, and analyzed by a picture, taken with the Vivalytic Analyser. After that, one chamber is discarded, refilled with QWS, and again diluted. That way, a serial dilution of the library of 1:2, 1:4, 1:8, etc. can be generated. We tested different sample concentrations on the cartridge and fitted measurement points with a polynomial equation (Fig. 7a). The manufacturer specifies the upper limit concentration of 0.5 ng/μl, but as shown for a sample with 1 ng/μl (Fig. 7a), the measured point displays severe deviation from linearity. As the 1:2 ratio is not designated for the QWS, sufficient fluorescent molecules are likely not available. In numbers, the manual workflow has a lower limit of 180 µl QWS mixed with 20 µl of the sample. On-chip, we generated a mixture of 10 µl QWS with 10 µl sample for a 1:2 dilution in one chamber. If the number of fluorescent molecules is too low due to a low QWS ratio, 0.5 ng/µl is too high for on-chip quantification. Thus, only measurements up to a lower limit of the 1:4 dilution were used for quantification, as the linearity here is given (see Fig. 7, 1 ng/µl). Measured concentrations above the maximum were out of range. Using the first dilution step left-sided to the maximum, the lowest deviation for the concentration determination could be reached (blue labels in Fig. 7b). For example, after fitting the dilution series of a 5 ng/µl sample the maximum could be determined. The third dilution step is within linearity and should therefore be used (1:8; first dilution step to the left of the maximum) for the concentration calculation. This provides a fairly accurate concentration determination, which is only 11% lower than the reference with the Qubit (Fig. 7b). Hence, quantification of DNA by fluorescent intercalating dye is feasible on the cartridge. Combined with an amplicon-based automated library preparation, it would generate higher value by reducing further manual steps and the necessary quantification devices.
DNA quantification on the Vivalytic LoC cartridge with an automated dilution system for highly concentrated samples. (a) Samples were diluted automatically with Qubit 1X dsDNA high sensitivity working solution by mixing filled chambers on-chip (n = 3). After mixing, a picture was taken (excitation/emission 469 nm/ 525 nm) and diluted again. (b) Concentration determination on-chip during different dilution steps of four samples in comparison with the Qubit 4 Fluorometer as a reference. The relative deviations from the reference are given numerically. ○ (blue labels): optimal dilution steps for quantification corresponding to the actual input concentration. Error bars display standard deviation.
Discussion
NGS has gained a crucial role in personalized medicine in combination with tumor diagnostic approaches. High throughput of samples can be prepared for sequencing by liquid handlers in sequencing centers and large laboratories. Using sequencing at the point-of-care, for example, to monitor tumor progression through liquid biopsies, requires smaller and more flexible systems for library preparation. The microfluidic automated library preparation presented in this work can overcome these challenges. The described proof-of-concept on a microfluidic cartridge showed successful preparation of targeted libraries with the possibility of also preparing a universal WGS library by skipping the multiplex-PCR step. These types of library preparations are widely distributed and commonly used for different types of NGS applications starting with DNA as input material22. In addition, the automated library preparation has to be validated with clinical samples, as shown by other microfluidic devices. For example, the centrifugal-based system by Hess et al. successfully prepared sequencing libraries from gDNA samples of leukemia patients7,11. Another clinical study by Shi et al. could successfully demonstrate the library preparations from gDNA samples of breast cancer and ovarian cancer patients9. For the Vivalytic LoC application shown in this manuscript, the next logical step is clinical testing of cfDNA from lung cancer patients. Moreover, for easy handling in a routine workflow at the point-of-care, further developments, which are discussed below, should be evaluated and implemented.
As the demonstrated library preparation consisted of many process steps and fluidic operations, the workflow for the technical proof-of-concept was divided into two cartridges. Currently, the number of reaction steps is limited due to the number of reagent reservoirs. A complete workflow on a single cartridge that can be stored at room temperature, thus PoC-compatible, would require lyophilized enzyme beads stored inside the pumping chambers instead of the reservoirs used here. Lyophilized PCR beads are already commercially available and easily transferable into microfluidic platforms23. For end-repair and ligation, lyophilized enzymes, which are not yet commercially available, would be necessary. This way, the number of reagent reservoirs would be sufficient to perform the library preparation workflow on one cartridge.
It would also be useful to have a variable system to switch between universal WGS and targeted approaches. A flexible choice of the library preparation approach could be realized by using PCR beads that can be inserted by the user. Diverse application-specific multiplex PCR beads would allow the cartridge to be used for various diagnostic approaches. Panels are often used to detect therapy-relevant mutations, especially for cancer diagnostics. Library preparation for a whole genome sequencing approach can be realized by skipping the target enrichment and using an end-repair module with simultaneous fragmentation. The iPCR bead should also be adjustable, as for a sequencing run, samples are pooled and distinguished bioinformatically employing indexes. By changing the iPCR-bead with different barcodes, the samples could be flexibly pooled after library preparation as well.
Currently, DNA nucleic acid extraction and purification from a sample are provided via the silica filter on the Vivalytic system. However, since one purification is not sufficient for library preparation, an alternative method was established. Magnetic particles instead of the integrated silica filter were used, which allows separate bead batches for each purification step and avoids cross-contamination of reagents for the different preparation steps. Furthermore, the magnetic particles can easily be adapted for specified size selection. The purification on-chip showed an efficiency of around 50% of the DNA compared to off-chip. In this work, the purification efficiencies were doubled by using a peristaltic pump mechanism. As only a simple disc magnet was used, which covered the chamber, we expect further optimization in future work. As no magnet has been installed in the Vivalytic system so far, a future integrated magnet could further minimize bead loss by calculating the optimal magnetic field strength. For the next generation of Vivalytic devices, this could be an attractive option to extend the number of applications. A magnet-free variant for immobilizing magnetic beads was implemented by Guo et al.10. The developed centrifugal microfluidic system uses the centrifugal force to sediment the magnetic particles for immobilization10. In this way, it has been possible to minimize the loss of material during library purification10. Another alternative solution to the magnet could be to use the already integrated filter matrices for size selection to separate the beads from the solution described by Kim and colleagues6.
A workflow from a sample to a ready-to-load library would also need a DNA extraction module. As the library preparation utilizes the capacity of the existing cartridge, separate sample type-specific extraction cartridges would be reasonable. A separate cartridge would also have the advantage of remaining flexible with the sample input material. Distinct cartridges could be used for processing liquid biopsies or extracting DNA from tissues. Moreover, the solution for DNA quantification shown here can complete the workflow for a fully automated process. The quantification option with an intercalating dye allows the determination of the library concentration. To optimize read generation, it is essential to use precise amounts of adaptor-ligated molecules when loading onto a DNA sequencer. The concentration and average fragment size of the library are used to calculate the molarity. If no external device is to be used for fragment analysis, this method is limited to amplicon-based approaches. As we used amplicons that have defined fragment size, the quantification module reduces further manual quantity control steps. Furthermore, the calculation of the proper dilution combined with an automated dilution of the library could allow a direct load of the microfluidic library on the sequencer.
In conclusion, the proof-of-concept of the automated library preparation compared to the manual workflow revealed that the mutation frequencies from 5%/6.3% down to 0.1%/0.13% could be detected for all mutations in a similar range in the microfluidic system. Decreasing input material to 1 ng slightly affected the microfluidic performance, reaching the current limit of sensitivity. For use as a part of a diagnostic PoC sequencing approach for small hospitals or ambulatory healthcare centers, real patient samples should be tested in future studies. Combined with further optimizations like the reduction to one cartridge, the microfluidic library preparation can bring point-of-care sequencing forward.
Data availability
The datasets supporting the conclusions of this article are available in the NCBI BioProject repository ID: PRJNA1047135 (https://www.ncbi.nlm.nih.gov/bioproject/1047135).
References
Goodwin, S., McPherson, J. D. & McCombie, W. R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17(6), 333–351. https://doi.org/10.1038/nrg.2016.49 (2016).
Hess, J. F. et al. Library preparation for next generation sequencing: A review of automation strategies. Biotechnol. Adv. 41, 107537. https://doi.org/10.1016/j.biotechadv.2020.107537 (2020).
Hess, J. F. et al. Automation of amplicon-based library preparation for next-generation sequencing by centrifugal microfluidics. Anal. Chem. 92(19), 12833–12841. https://doi.org/10.1021/acs.analchem.0c01202 (2020).
Tan, S. J. et al. A microfluidic device for preparing next generation DNA sequencing libraries and for automating other laboratory protocols that require one or more column chromatography steps. PLoS ONE 8(7), e64084. https://doi.org/10.1371/journal.pone.0064084 (2013).
Kim, H. et al. A microfluidic DNA library preparation platform for next-generation sequencing. PLoS ONE 8(7), e68988. https://doi.org/10.1371/journal.pone.0068988 (2013).
Kim, S. et al. High-throughput automated microfluidic sample preparation for accurate microbial genomics. Nat. Commun. 8(1), 13919. https://doi.org/10.1038/ncomms13919 (2017).
Murphy, T. W., Hsieh, Y.-P., Zhu, B., Naler, L. B. & Lu, C. Microfluidic platform for next-generation sequencing library preparation with low-input samples. Anal. Chem. 92(3), 2519–2526. https://doi.org/10.1021/acs.analchem.9b04086 (2020).
Hess, J. F. et al. Automated library preparation for whole genome sequencing by centrifugal microfluidics. Anal. Chim. Acta 1182, 338954. https://doi.org/10.1016/j.aca.2021.338954 (2021).
Shi, C. et al. Development and clinical applications of an enclosed automated targeted NGS library preparation system. Clin. Chim. Acta 540, 117224. https://doi.org/10.1016/j.cca.2023.117224 (2023).
Guo, J. et al. Automated centrifugal microfluidic system for the preparation of adaptor-ligated sequencing libraries. Lab. Chip 24(2), 182–196. https://doi.org/10.1039/D3LC00781B (2024).
Hess, J. F. et al. Clinical pilot study on microfluidic automation of IGH-VJ library preparation for next generation sequencing. Clin. Chem. Lab. Med. CCLM 62(7), e164–e167. https://doi.org/10.1515/cclm-2023-1346 (2024).
Head, S. R. et al. Library construction for next-generation sequencing: Overviews and challenges. BioTechniques 56(2), 1. https://doi.org/10.2144/000114133 (2014).
Illumina. Nextera XT DNA Library Prep - Reference Guide. May 2019 (Online). Available: https://emea.support.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/samplepreps_nextera/nextera-xt/nextera-xt-library-prep-reference-guide-15031942-05.pdf
Rupp, J. et al. Rapid microarray processing using a disposable hybridization chamber with an integrated micropump. Lab. Chip 12(7), 1384. https://doi.org/10.1039/c2lc21110f (2012).
Zhong, Y., Xu, F., Wu, J., Schubert, J. & Li, M. M. Application of next generation sequencing in laboratory medicine. Ann. Lab. Med. 41(1), 25–43. https://doi.org/10.3343/alm.2021.41.1.25 (2021).
Di Resta, C. & Ferrari, M. Next generation sequencing: From research area to clinical practice. EJIFCC 29(3), 215–220 (2018).
Grumaz, S. et al. Next-generation sequencing diagnostics of bacteremia in septic patients. Genome Med. 8(1), 73. https://doi.org/10.1186/s13073-016-0326-8 (2016).
Grumaz, C. et al. Rapid next-generation sequencing-based diagnostics of bacteremia in septic patients. J. Mol. Diagn. 22(3), 405–418. https://doi.org/10.1016/j.jmoldx.2019.12.006 (2020).
Next Gen Diagnostics. Next Gen Diagnostics. Next Gen Diagnostics. Accessed March 05, 2022 (Online). https://www.nextgen-dx.com
Berensmeier, S. Magnetic particles for the separation and purification of nucleic acids. Appl. Microbiol. Biotechnol. 73(3), 495–504. https://doi.org/10.1007/s00253-006-0675-0 (2006).
Lis, J. T. & Schleif, R. Size fractionation of double-stranded DNA by precipitation with polyethylene glycol. Nucleic Acids Res. 2(3), 383–390. https://doi.org/10.1093/nar/2.3.383 (1975).
Mauger, F. et al. Comparison of commercially available whole-genome sequencing kits for variant detection in circulating cell-free DNA. Sci. Rep. 10(1), 6190. https://doi.org/10.1038/s41598-020-63102-8 (2020).
Xu, J. et al. Transferable, easy-to-use and room-temperature-storable PCR mixes for microfluidic molecular diagnostics. Talanta 235, 122797. https://doi.org/10.1016/j.talanta.2021.122797 (2021).
Acknowledgements
Special thanks go to Alexander Kempf, Michelle Hinrichs and Julia Schon for support in the experiments. Also, a special thanks goes to Hannah Bott for assistance during the polynomial fitting and Karin Lemuth as well as Franz Lärmer for their great supervision of the work.
Author information
Authors and Affiliations
Contributions
Conceptualization, A.H. and C.G.; bioinformatic evaluation, A.H. and C.G.; conducted the experiments, A.H.; images, A.H.; project administration, A.H., C.G., A.T., C.J. and S.R.; supervision, C.G., A.T., C.J., and S.R.; visualization, A.H., C.G.; writing original draft, A.H.; writing review & editing, C.G., A.T., C.J., and S.R.. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The PhD thesis of Anne Hoffmann was funded by Robert Bosch GmbH and she was an employee of Robert Bosch GmbH. Robert Bosch GmbH submitted a patent application regarding the process that leads to the DNA quantification using microfluidic in-range controls.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hoffmann, A., Timm, A., Johnson, C. et al. Automation of customizable library preparation for next-generation sequencing into an open microfluidic platform. Sci Rep 14, 17150 (2024). https://doi.org/10.1038/s41598-024-67950-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-67950-6
- Springer Nature Limited