Abstract
Previous studies showed that Australian wheat cultivars Janz and Sunco carry leaf rust and stem rust resistance genes Lr24 and Sr24 derived from Thinopyrum ponticum chromosome arm 3AgL. However, the size of the alien segments carrying Lr24 and Sr24 in the lines were not determined. In this study, we used non-denaturing fluorescence in situ hybridization (ND-FISH), genomic in situ hybridization (GISH), and PCR-based landmark unique gene (PLUG) markers to visualize the alien segments in Janz and Sunco, and further compared them with the segments in US cultivars Agent and Amigo. The fraction length (FL) of the alien translocation in Agent was 0.70–1.00, whereas those in Janz, Sunco, and Amigo were smaller, at FL 0.85–1.00. It was deduced that the alien gene RAg encoding for red grain color and rust resistance genes Lr24 and Sr24 on chromosome arm 3AgL were in bins of FL 0.70–0.85 and 0.85–1.00, respectively. We retrieved and extracted nucleotide-binding site-leucine-rich repeat (NBS-LRR) receptor genes corresponding to the region of Lr24 and Sr24 on chromosomes 3E, and 3J, 3Js and 3St from the reference genome sequences of Th. elongatum and Th. intermedium, respectively. A set of molecular markers developed for Lr24 and Sr24 from those extracted NBS-LRR genes will provide valuable information for fine mapping and cloning of these genes.
Similar content being viewed by others
Introduction
The hard red winter wheat cultivar Agent, CI 13523, carrying a spontaneous wheat-Thinopyrum ponticum translocation T3DS.3DL-3AgL was released by Oklahoma Agricultural Experiment Station in 19671. At that time, Agent was resistant to all known races of Puccinia triticina (Pt), the leaf rust pathogen, and moderately resistant to “some races of stem rust” caused by P. graminis f. sp. tritici (Pgt)1. The leaf rust and stem rust resistances in Agent were derived from Th. ponticum, shown to be completely linked, and designated Lr24 and Sr24, respectively2. The translocation carrying resistance was incorporated into wheat cultivars, and the Agent source of Lr24 and Sr24 was widely exploited in the Americas and South Africa. However, attempts to use the resistance in Australia failed because all derivatives had red grain color.
Sears3 used induced homoeologous recombination to produce several 3D-3Ag translocation lines carrying Lr24 in Chinese Spring (CS) which has the red grain allele R1 on chromosome 3D. Thus, the source of the red grain allele (i.e. R1 or RAg) in these lines was unknown4. Following discussions with Sears, four of these lines were backcrossed with white seeded Australian genotypes and white seeded rust resistant lines were obtained when Sears’ lines CS 3D-3Ag#3 and CS 3D-3Ag#14 were used as donors5. Based on chromosome pairing studies Sears3 had predicted that these lines had the smallest 3Ag chromosome segments. The first white-seeded Australian cultivar carrying Lr24 and Sr24, Torres, was released in Queensland in 1983. Subsequently, more than 60 cultivars, including Janz and Sunco, carrying Lr24 and Sr24 were released and widely grown throughout Australia. This is one of the most successful examples of overcoming linkage drag in exploiting high value resistance genes4,6. Lr24 remained effective in Australia for almost 20 years until a virulent pathotype was detected in 20007. Sr24 continues to be effective and remains an important source of stem rust resistance despite its early failure against race Ug99 in Eastern Africa and earlier failure in South Africa in 19878.
Sr24 and Lr24 were identified in some white seeded, stem rust resistant Australian backcross lines with Amigo as donor. Amigo was believed to carry resistance from cereal rye; however, some derivatives also had leaf rust resistance that was later attributed to Lr24. There was no evidence of an association with red grain color. Jiang et al.9 and Friebe et al.10 reported that Amigo had a non-homoeologous translocation T1BL.1BS-3AgL with the 3Ag segment lacking the RAg allele. In this study, we used ND-FISH, GISH, PLUG markers, and comparative genomic analysis to: (1) visualize the alien segments carrying Lr24 and Sr24 in Agent, Janz, Sunco and Amigo; (2) further map the Lr24/Sr24 and RAg genes to smaller regions on chromosome arm 3AgL; and (3) develop a set of new chromosome 3AgL-specific NBS-LRR-related molecular markers within the same bin as the Lr24/Sr24 genes.
Results
Cytogenetic identification of four wheat lines carrying Lr24 and Sr24
ND-FISH and GISH images of mitotic metaphase cells of Agent, Janz, Sunco and Amigo are shown in Fig. 1. ND-FISH analysis using Oligo-pSc119.2-1 and Oligo-pTa535-1 allowed us to identify individual chromosomes. Agent (Fig. 1a), Janz (Fig. 1c), and Sunco (Fig. 1e) all had 42 chromosomes. Combined with GISH analysis using Pseudoroegneria stipifolia (St) genomic DNA as a probe (Fig. 1b,d,f), it was revealed that Agent, Janz and Sunco each had 40 wheat chromosomes and two wheat-Thinopyrum translocation chromosomes, T3DS.3DL-3AgL. The 3AgL chromosome segments in Janz and Sunco were similar in size, and smaller than that in Agent. As expected, Amigo had 38 wheat chromosomes, two T1BL.1BS-3AgL wheat-Thinopyrum translocation chromosomes and a pair of T1RS.1AL wheat-cereal rye translocation chromosomes (Fig. 1g,h). The 3AgL segment in Amigo was similar in size as those in Janz and Sunco.
FISH-GISH images of metaphase chromosomes of four wheat lines carrying Lr24 and Sr24, Agent (a,b), Janz (c,d), Sunco (e,f) and Amigo (g,h). Probes Oligo-pSc119.2-1 (green) and Oligo-pTa535-1 (red) were used in FISH (a,c,e,g) and Pseudoroegneria stipifolia (St) genomic DNA was used as the probe (yellow-green) for GISH (b,d,f,h). (a–f) Arrows point to the breakpoint in T3DS.3DL-3AgL. (g,h) Arrows point to the breakpoint in T1BL.1BS-3AgL, and arrowheads point to the breakpoint for T1RS.1AL. Bars, 10 μm.
Locating the Lr24/Sr24 and red grain color genes on chromosome arm 3AgL
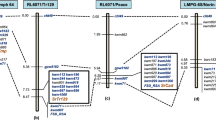
The chromosomal segment sizes and breakpoint positions of four translocation lines carrying Lr24 and Sr24 were determined using GISH. As shown in Table 1 and Fig. 2, the 3Ag segment in translocation chromosome T3DS.3DL-3AgL in Agent was in the long arm bin FL 0.70–1.00, and that in Janz, Sunco and Amigo were in bins FL 0.85–1.00. Because Agent, Janz, Sunco, and Amigo all carry Lr24 and Sr24, they were mapped to 3AgL bin FL 0.85–1.00 (Fig. 2). In addition, Agent had red grain color, whereas Janz, Sunco and Amigo derivatives had white grain color, so the gene encoding the red grain color in Agent (RAg) must be in 3AgL bin FL 0.70–0.85 (Fig. 2).
Chromosomal locations of the red grain color gene RAg and Lr24/Sr24 genes in chromosome arm 3AgL. Dashed lines separate chromosome 3AgL-specific molecular markers specific for Lr24/Sr24 and RAg, respectively. Green, blue, and red colors indicate Th. ponticum chromosome 3AgL segments, and wheat chromosomes 3D and 1B, respectively. +/+ (R) indicates that the line carries Lr24 and Sr24. Red and white, grain colors.
Screening of chromosome 3AgL-specific PLUG markers identifying R Ag, Lr24 and Sr24
Twenty-four PLUG primer pairs from the long arms of homoeologous group 3 chromosomes were used to identify chromosome 3AgL-specific markers. Five markers, TNAC1285, TNAC1286, TNAC1382, TNAC1383, and TNAC1707, generating specific fragments in Agent, Janz, Sunco and Amigo, but not in Aroona, CS, Avocet S (AvS) and Schomburgk were chromosome 3AgL-specific and were in 3AgL bin FL 0.85–1.00 (Table 2). Another two markers, TNAC1282 and TNAC1342, amplified specific bands in Agent but not in Janz, Sunco and Amigo, or Aroona, CS, AvS and Schomburgk, indicating their locations in 3AgL bin FL 0.70–0.85 along with RAg (Table 2). The images for two representative PLUG markers, TNAC1282 and TNAC1707, are shown in Fig. 3. The primer sequences of the chromosome 3AgL-specifc PLUG markers are listed in Table 2.
PCR amplification patterns of two representative chromosome 3AgL-specific markers in common wheat and Lr24/Sr24-carrying lines. (a) TNAC1282 (Taq I); (b) TNAC1707 (Taq I). M, GeneRuler (Thermo Scientific, Waltham, MA) 1 kb DNA Ladder. Arrows point to chromosome arm 3AgL-specific bands. Uncropped gel images are provided in Supplementary Information (Supplementary Fig. 1).
Development of chromosome 3AgL-specific NBS-LRR-related markers
As showed in Table 2, the 3AgL bin FL 0.85–1.00 containing Lr24 and Sr24 contained five chromosome 3AgL-specific PLUG markers. Based on the syntenic relationships among Th. elongatum, Th. intermedium and Th. ponticum species, the 3AgL bin FL 0.85–1.00 carrying Lr24 and Sr24 were located into the physical intervals 597.72–675.27, 586.76–664.51, 464.48–526.13 and 435.61–493.34 Mb in chromosomes 3E, 3J, 3Js and 3St, respectively. We then retrieved and extracted 26, 30, 42 and 30 NBS-LRR genes (related to disease response) in the respective physical intervals (Table S1). Using Primer-BLAST software we designed and developed four pairs of chromosome 3AgL-specific primers from the CDS sequences of three NBS-LRR genes (Table 3). Three of these markers mapped to 3AgL bin FL 0.85–1.00, and one mapped to 3AgL bin FL 0.70–0.85. PCR amplification results for these markers are shown in Fig. 4. Combining all results, eight markers were in bin FL 0.85–1.00 containing Lr24 and Sr24, and three were in bin FL 0.70–0.85 containing RAg (Fig. 2).
PCR amplification patterns of the newly developed chromosome arm 3AgL-specific NBS-LRR-related markers. (a) TLS24-1; (b) TLS24-8; (c) TLS24-9; (d) TLS24-18. M, GeneRuler (Thermo Scientific, Waltham, MA) 1 kb DNA Ladder. Arrows point to chromosome arm 3AgL-specific bands. Uncropped gel images are provided in Supplementary Information (Supplementary Fig. 2).
Discussion
Although neither Lr24 nor Sr24 is a source of durable rust resistance, they are still useful for resistance breeding in many parts of the world, especially in combination with other genes. Several publications reported the development of molecular markers linked to Lr24 and/or Sr2411,12,13,14. These resistance genes originated from Th. ponticum mainly in the form of chromosome T3DS.3DL-3AgL translocations10,15. Janz and Sunco are white-seeded Australian wheat cultivars with CS 3D-3Ag#3 and CS 3D-3Ag#14 as donors of Lr24 and Sr24. Although Sears3 predicted that CS 3D-3Ag#3 and CS 3D-3Ag#14 might carry smaller alien segments than that in Agent based on chromosome pairing studies, their exact chromosomal structures were not determined. Zwart et al.16 constructed a genetic linkage map of chromosome 3D using 111 doubled haploid lines derived from a cross between synthetic hexaploid wheat accession CPI133872 and Janz. Their finding that Lr24 was flanked by P37M54e and Xgwm169b at 0.4 cM and 1.0 cM, respectively, inspired our curiosity to characterize the 3AgL chromosomal segment size in Janz and Sunco. In the present study, ND-FISH and GISH revealed that different translocation events in Janz (Fig. 1c,d) and Sunco (Fig. 1e,f) were of similar size and significantly smaller than the translocation in Agent (Fig. 1a,b), as predicted by Sears3. Based on the cytological results, RAg and Lr24 and Sr24 were in 3AgL bins FL 0.70–0.85 and 0.85–1.00, respectively (Fig. 2). Combined with the PLUG marker results, the 3AgL bin FL 0.85–1.00 containing Lr24 and Sr24 was estimated to be at least 60 Mb (Table S1). Due to the presence of the Ph1 (pairing homoeologous 1) gene in wheat, it is highly unlikely to have recombination between the Th. ponticum segment in the translocation chromosome T3DS.3DL-3AgL and homoeologous wheat chromosome regions in Janz and Sunco, as also mentioned by others2,11,12,14. All these results made us question the high recombination frequency between the translocated 3AgL chromosomal fragment carrying Lr24 in Janz and homoeologous wheat chromosome 3D reported by Zwart et al.16.
PLUG markers17 based on orthologous gene conservation between rice and wheat have proven to be highly efficient and reliable molecular markers for confirming homoeologous relationships between wheat and alien chromosomes18,19,20. In this study, we used 24 PLUG primer pairs identified at the distal ends of the long arms of homoeologous group 3 chromosomes to genotype Agent, Janz, Sunco and Amigo, and found that five markers amplified Thinopyrum-specific bands. Two additional PLUG markers amplified fragments specific to Agent (Fig. 3, Table 2), indicating that: (1) the translocated Thinopyrum chromosomal segments in Agent, Janz, Sunco and Amigo belonged to homeologous group 3 (3AgL); and (2) the 3AgL chromosomal segments in Janz, Sunco and Amigo were similar in size, but smaller than that in Agent.
With the rapid development of sequencing technologies and availability of whole genome sequences of T. aestivum, Hordeum vulgare L. and Secale cereale, comparative genomic analysis is becoming increasingly achievable in developing molecular markers for use in non-sequenced species or in fine mapping of target genes21,22,23,24. In this study, based on the syntenic relationships among Th. elongatum chromosome 3EL, Th. intermedium chromosomes 3JL, 3JsL and 3StL, and the current Th. ponticum chromosome 3AgL, we developed four pairs of chromosome 3AgL-specific NBS-LRR-related primers (Fig. 4, Table S1) from the coding sequences (CDS) of three NBS-LRR genes in the Lr24/Sr24 region. Moreover, the eight Th. ponticum chromosome 3AgL-specifc molecular (five PLUG and three NBS-LRR) markers developed in this study will be useful in future fine mapping and cloning Lr24 and/or Sr24. No study thus far has separated the two genes indicating relatively close linkage, but we have no reason to believe they are the same gene.
Grain color is an important trait affecting flour yield and quality in wheat. Previous studies indicated that red-seeded wheats are more tolerant to preharvest sprouting than white-seeded wheats25. Hence Agent or another of Sears’ translocation lines may be preferred as donors of Sr24 and Lr24 in locations where red seeded wheat is preferred. In countries such as Australia and India where white seededness is considered a key aspect of quality or marketing advantage the use of these resistance genes was made possible only by their separation from RAg. In hindsight, mutation of RAg in an appropriate derivative lacking R2 and R3 from Agent or in other Sears’ derivatives other than CS 3D-3Ag#3 and CS 3D-3Ag#14 could have achieved the same result.
Methods
Plant materials
The materials used in this study included four common wheat (Triticum aestivum L., AABBDD, 2n = 6x = 42) cultivars Aroona, Chinese Spring, Avocet S and Schomburgk, and four cultivars carrying Lr24 and Sr24, namely Agent, Janz, Sunco and Amigo. Agent is a hard red winter wheat cultivar carrying a spontaneous wheat-Th. ponticum translocation T3DS.3DL-3AgL1. Janz and Sunco are white-seeded Australian wheat cultivars with CS 3D-3Ag#3 and CS 3D-3Ag#14 translocations, respectively, as donors of Sr24 and Lr24. The red-seeded wheat cultivar Amigo with a T1AL.1RS Robertsonian translocation also carried a 3Ag chromosome segment, but as a T1BL.1BS-3AgL translocation9. The above materials are maintained at the Plant Breeding Institute, The University of Sydney.
Cytological analysis
Chromosome preparation
Slides for examination of mitotic metaphase chromosomes of Agent, Janz, Sunco and Amigo were prepared according to the procedure in Lang et al.26 with minor modifications.
Non-denaturing fluorescence in situ hybridization (ND-FISH)
ND-FISH with oligonucleotide probes Oligo-pSc119.2-1 and Oligo-pTa535-1 was used to identify individual wheat chromosomes27. Oligo-pSc119.2-1 and Oligo-pTa535-1 were labelled with 6-carboxyfluorescein (6-FAM) generating green signals and 6-carboxytetramethylrhodamine (Tamra) generating red signals, respectively. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen Life Science, Carlsbad, CA, USA) in Vectashield (Vector Laboratories, Burlingame, CA) and pseudo-colored blue. Slides were analyzed with a Zeiss Axio Imager epifluorescence microscope. Images were captured with a Retiga EXi CCD (charge-coupled device) camera (QImaging, Surrey, BC, Canada) operated with Image-Pro Plus version 7.0 software (Media Cybernetics Inc., Bethesda, MD, USA) and processed with Photoshop version CC 2022 software (Adobe Systems, San Jose, CA).
Genomic in situ hybridization (GISH)
Sequential GISH was performed after stripping off the oligo probes in 50% FA/1 × SSC and 50% FA/0.5 × SSC for 10 min each at 42 °C. GISH followed the procedure of Zhang et al.28. Total genomic DNA of Ps. stipifolia (StSt, 2n = 2x = 14, PI 314058, The National Small Grains Collection, USDA-ARS; kindly provided by Dr. L. Qi, USDA-ARS, ND) was used as probe, which was labelled with biotin-16-dUTP (Roche Diagnostics Australia, Castle Hill, NSW, Australia) using nick translation. Unlabeled total genomic DNA of CS wheat was used as blocker with a probe:blocker ratio of 1:120. Signals were detected with fluorescein–avidin DN (Vector Laboratories, Burlingame, CA). Chromosomes were counterstained with DAPI and pseudo-colored red.
Alien segment size and breakpoints in translocation lines
To estimate the size and breakpoints of the Th. ponticum 3AgL chromosomal segments in lines carrying Lr24 and Sr24, 10 cells at mitotic metaphase from each translocation line were photographed. Arm lengths were measured using ImageJ v2.0.0 software29, and the ratio between the length of 3AgL chromosomal segments and the long arm of translocation chromosome T3DS.3DL-3AgL in Agent was calculated as described by Endo and Gill30. The position of the centromere was considered zero, and the terminal end of long arm of T3DS.3DL-3AgL in Agent was considered one. Standard error calculations for fraction length (FL) measurements were performed with the software Excel 2021 (IBM Corporation, New York).
PLUG marker analysis
Genomic DNA was extracted from young leaves of Aroona, CS, AvS, Schomburgk, Agent, Janz, Sunco, and Amigo. Twenty-four PCR-based landmark unique gene (PLUG) markers from the long arm of wheat homoeologous group 317 were selected and synthesized (Sigma-Aldrich, Inc., St. Louis, MO, USA). PCR were performed according to the procedure in Li et al.31. PCR products were digested with TaqI (at 65 °C) or HaeIII (at 37 °C) (Genesearch Pty Ltd, Arundel, QLD, Australia) for improving levels of polymorphism, and separated in 1.5% agarose gels. PLUG markers present in Agent, Janz, Sunco and Amigo but absent in Aroona, CS, AvS and Schomburgk were identified as chromosome 3AgL-specific. The physical locations of PLUG markers were searched in the reference genome sequence of CS (IWGSC RefSeq v.2.1; https://urgi.versailles.infra.fr/blast/).
Development of NBS-LRR-related molecular markers identifying the smallest alien segment
Firstly, the chromosome 3AgL bin FL 0.85–1.00 containing Lr24 and Sr24 were mapped to chromosomes 3E, 3J, 3Js and 3St to determine their physical positions in the reference genome sequences of Th. elongatum32 (EE, 2n = 2x = 14) and Th. intermedium (JJJsJsStSt, 2n = 6x = 42; https://phytozome-next.jgi.doe.gov/), respectively. Depending on the physical positions, we retrieved and extracted all annotated genes and protein sequences in the targeted intervals in chromosomes 3E, 3J, 3Js and 3St from the WheatOmics 1.0 (Th. elongatum v1.0; http://202.194.139.32/jbrowse.html) and Phytozome 13 (Th. intermedium v3.1; https://phytozome-next.jgi.doe.gov/jbrowse/index.html) databases in combination with the TBtools-II (Toolbox for Biologists) v1.120 software33. The Hidden Markov Model (HMM) (https://www.ebi.ac.uk/Tools/hmmer/) profile of the NB-ARC domain (Pfam accession number: PF00931; http://pfam-legacy.xfam.org/) was used as query to perform HMMsearch using the HMMER-3.0 package against the protein sequences of Th. elongatum and Th. intermedium with an expected value (e-value) threshold of < 1e−30 using a local server MobaXterm. The results were further confirmed using the NCBI Conserved Domain Database (CDD) tool (e-value = 1e−2) (https://www.ncbi.nlm.nih.gov/Structure/cdd/). NBS-LRR candidate genes without a conserved NBS domain were manually removed. Finally, we used an online primer design website Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) to design and develop chromosome 3AgL-specific primers from the CDS sequence of the candidate genes.
Ethical approval
We comply with relevant guidelines and legislation regarding the sample collection and use in the present study. All materials in the present study are not endangered.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
References
Smith, E. L., Schlehuber, A. M., Young, H. C. & Edwards, L. H. Registration of agent wheat. Crop Sci. 8, 511–512. https://doi.org/10.2135/cropsci1968.0011183X000800040039x (1968).
McIntosh, R. A., Dyck, P. L. & Green, G. J. Inheritance of leaf rust and stem rust resistances in wheat cultivars ‘Agent’ and ‘Agatha’. Aust. J. Agric. Res. 28, 37–45. https://doi.org/10.1071/AR9770037 (1977).
Sears, E. R. Chromosome engineering in wheat. In Proc. 4th Wheat Genet. Symp. (eds. Sears, E. R. & Sears, L. M. S.) 23–38 (University of Missouri, 1972).
McIntosh, R. A. From Farrer to the Australian Cereal Rust Control Program. Aust. J. Agric. Res. 58, 550–557. https://doi.org/10.1071/AR07148 (2007).
McIntosh, R. A., Wellings, C. R. & Park, R. F. Wheat Rusts: An Atlas of Resistance Genes (CSIRO Publishing, 1995).
Park, R. F., Chhetri, M. & Singh, D. Rust resistance genotypes and expected rust responses of Australian cereal varieties. Cereal Rust Rep. 20(3), 1 (2023).
Park, R. F., Bariana, H. S., Wellings, C. R. & Wallwork, H. Detection and occurrence of a new pathotype of Puccinia triticina with virulence for Lr24 in Australia. Aust. J. Agric. Res. 53, 1069–1076. https://doi.org/10.1071/AR02018 (2002).
The, T. T. et al. Characterization of stem rust resistant derivatives of wheat cultivar Amigo. Euphytica 58, 245–252. https://doi.org/10.1007/BF00025256 (1992).
Jiang, J., Friebe, B. & Gill, B. S. Chromosome painting of Amigo wheat. Theor. Appl. Genet. 89, 811–813. https://doi.org/10.1007/BF00224501 (1994).
Friebe, B., Jiang, J., Raupp, W. J., McIntosh, R. A. & Gill, B. S. Characterization of wheat-alien translocations conferring resistance to diseases and pests: Current status. Euphytica 91, 59–87. https://doi.org/10.1007/BF00035277 (1996).
Schachermayr, G. M., Messmer, M. M., Feuillet, C., Winzeler, H. & Keller, B. Identification of molecular markers linked to the Agropyron elongatum-derived leaf rust resistance gene Lr24 in wheat. Theor. Appl. Genet. 90, 982–990. https://doi.org/10.1007/BF00222911 (1995).
Dedryver, F., Jubier, M.-F., Thouvenin, J. & Goyeau, H. Molecular markers linked to the leaf rust resistance gene Lr24 in different wheat varieties. Genome 39, 830–835. https://doi.org/10.1139/g96-105 (1996).
Mago, R. et al. Development of PCR markers for the selection of wheat stem rust resistance genes Sr24 and Sr26 in diverse wheat germplasm. Theor. Appl. Genet. 111, 496–504. https://doi.org/10.1007/s00122-005-2039-z (2005).
Gupta, S. K. et al. Development and validation of SCAR markers co-segregating with an Agropyron elongatum derived leaf rust resistance gene Lr24 in wheat. Euphytica 150, 233–240. https://doi.org/10.1007/s10681-006-9113-8 (2006).
Jiang, J., Friebe, B. & Gill, B. S. Recent advances in alien gene transfer in wheat. Euphytica 73, 199–212. https://doi.org/10.1007/BF00036700 (1994).
Zwart, R. S. et al. QTL mapping of multiple foliar disease and root-lesion nematode resistances in wheat. Mol. Breed. 26, 107–124. https://doi.org/10.1007/s11032-009-9381-9 (2010).
Ishikawa, G. et al. Localization of anchor loci representing five hundred annotated rice genes to wheat chromosomes using PLUG markers. Theor. Appl. Genet. 118, 499–514. https://doi.org/10.1007/s00122-008-0916-y (2009).
Zhang, H. et al. Molecular and cytogenetic characterization of new wheat-Dasypyrum breviaristatum derivatives with post-harvest re-growth habit. Genes 6, 1242–1255. https://doi.org/10.3390/genes6041242 (2015).
Li, G. et al. Characterization of wheat-Secale africanum chromosome 5Ra derivatives carrying Secale specific genes for grain hardness. Planta 243, 1203–1212. https://doi.org/10.1007/s00425-016-2472-z (2016).
Yu, Z. et al. Characterization of chromosomal rearrangement in new wheat-Thinopyrum intermedium addition lines carrying Thinopyrum-specific grain hardness genes. Agronomy 9, 18. https://doi.org/10.3390/agronomy9010018 (2019).
Zhang, J. et al. A strategy for identifying markers linked with stem rust resistance in wheat harbouring an alien chromosome introgression from a non-sequenced genome. Theor. Appl. Genet. 132, 125–135. https://doi.org/10.1007/s00122-018-3201-8 (2019).
Li, G. et al. Molecular and cytogenetic dissection of stripe rust resistance gene Yr83 from rye 6R and generation of resistant germplasm in wheat breeding. Front. Plant Sci. 13, 1035784. https://doi.org/10.3389/fpls.2022.1035784 (2022).
Zhu, K. et al. Fine mapping of powdery mildew resistance gene MlWE74 derived from wild emmer wheat (Triticum turgidum ssp. dicoccoides) in an NBS-LRR gene cluster. Theor. Appl. Genet. 135, 1235–1245. https://doi.org/10.1007/s00122-021-04027-2 (2022).
Gao, S. et al. Fine mapping of a Fusarium crown rot resistant locus on chromosome arm 6HL in barley by exploiting near isogenic lines, transcriptome profiling, and a large near isogenic line-derived population. Theor. Appl. Genet. 136, 137. https://doi.org/10.1007/s00122-023-04387-x (2023).
Bassoi, M. C. & Flintham, J. Relationship between grain color and preharvest sprouting-resistance in wheat. Pesq. Agropec. Bras. 40, 981–988. https://doi.org/10.1590/S0100-204X2005001000006 (2005).
Lang, T. et al. Precise identification of wheat-Thinopyrum intermedium translocation chromosomes carrying resistance to wheat stripe rust in line Z4 and its derived progenies. Genome 61, 177–185 (2018).
Tang, Z., Yang, Z. & Fu, S. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. https://doi.org/10.1007/s13353-014-0215-z (2014).
Zhang, P., Friebe, B., Lukaszewski, A. J. & Gill, B. S. The centromere structure in Robertsonian wheat-rye translocation chromosomes indicates that centric breakage-fusion can occur at different positions within the primary constriction. Chromosoma 110, 335–344. https://doi.org/10.1007/s004120100159 (2001).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. https://doi.org/10.1038/nmeth.2089 (2012).
Endo, T. R. & Gill, B. S. The deletion stocks of common wheat. J. Hered. 87, 295–307. https://doi.org/10.1093/oxfordjournals.jhered.a023003 (1996).
Li, J. et al. Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 133, 1095–1107. https://doi.org/10.1007/s00122-020-03534-y (2020).
Wang, H. et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 368, 5435. https://doi.org/10.1126/science.aba5435 (2020).
Chen, C. et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. https://doi.org/10.1016/j.molp.2020.06.009 (2020).
Acknowledgements
This work was financially supported by the Grains Research and Development Corporation (GRDC), Australia.
Author information
Authors and Affiliations
Contributions
P.Z. and R.A.Mc. conceived and designed the study. R.T. provided resources. J.L. and P.Z. performed the FISH and GISH experiments. J.L., H.G., and C.D. designed and developed chromosome 3AgL-specific NBS-LRR-related markers. J.L. and Y.W. conducted the molecular marker analysis. J.L., P.Z., and R.A.Mc. wrote the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, J., Guan, H., Wang, Y. et al. Cytological and molecular characterization of wheat lines carrying leaf rust and stem rust resistance genes Lr24 and Sr24. Sci Rep 14, 12816 (2024). https://doi.org/10.1038/s41598-024-63835-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-63835-w
- Springer Nature Limited