Abstract
Fatty acid metabolism (FAM) is associated with prognosis and immune microenvironment remodeling in many tumors. It is currently unknown how FAM affects the immunological microenvironment and prognosis of Gastric cancer (GC). Therefore, the current work aims to categorize GC samples based on the expression status of genes involved in FAM and to identify populations that might benefit from immunotherapy. In total, 50 FAM genes associated with overall survival (OS) were determined through univariate Cox proportional hazard regression analysis by mining the public TCGA and GEO databases. The GSE84437 and TCGA-STAD cohort samples were divided into two clusters using the "NMF" R package. According to the survival curve, patients in Cluster-1 showed considerably longer OS than those in Cluster-2. Patients in Cluster-1 exhibited earlier T stages, more intestinal GCs, and were older. MSI molecular subtypes were mainly distributed in Cluster-1, while GS molecular subtypes were distributed primarily in Cluster-2. There were 227 upregulated and 22 down-regulated genes (logFC > 1 or logFC < − 1, FDR < 0.05) in Cluster-2 compared with Cluster-1. One hub module (edges = 64, nodes = 12) was identified with a module score of 11.636 through Cytoscape plug-in MCODE. KEGG and GO analysis showed that the hub genes were associated with the cell cycle and cell division. Different immune cell infiltrates profile, and immune pathway enrichment existed between the subtypes. In conclusion, the current findings showed that practically all immunological checkpoint and immunoregulatory genes were elevated in patients with Cluster-2 GC, indicating that FAM subtypes may be crucial in GC immunotherapy.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is one of the most common malignant tumors of the digestive tract. Globally, GC ranks fifth in incidence and fourth in mortality, according to Global Cancer Statistics 20201. In 2020, 770,000 deaths (65% men) were reported worldwide due to GC. GC is among the top primary causes of cancer deaths in 42 countries and the leading cause in 13 countries. Eastern Asia was home to 56.6% of all deaths worldwide, 48.6% of which were in China alone. Male mortality rates varied by area, from under 5 per 100,000 in New Zealand/Australia, Northern Europe, and Northern America to over 20 per 100,000 in Eastern Asia. Tajikistan (15.4) and Mongolia (36.5 per 100,000) had the highest male and female mortality rates, respectively1. The strategy for treating GC is a comprehensive treatment based on surgery. Although with the popularization of gastroscopy, the proportion of early GC increases year by year2, and with the promotion of precise medical concepts and the development of surgical technology and instruments, the prognosis of GC patients has been improved3, the overall prognosis is still not ideal. Classification of GC into molecular subtypes and metabolic subtypes suitable for precision therapy may further improve the prognosis of GC4.
A carbon, hydrogen, and oxygen compound forms fatty acids (FA). It is the primary constituent of glycolipid, phospholipid, and neutral fat. It is also the main substance for cell energy supply. Fatty acid metabolism (FAM) includes catabolism and anabolism. Under oxygen, FA can oxidize and break down, producing energy known as fatty acid oxidation (FAO). Acetyl-CoA, a byproduct of glycolysis, the tricarboxylic acid cycle, and amino acid breakdown is used in fatty acid synthesis (FAS) to create 16-carbon intermediates and transform them into different fatty acids. The importance of FAM in cancer progression, survival, and metastasis has received increasing attention in recent years5,6,7.
According to studies, lipid metabolism assists in helping cancer cells proliferate rapidly, survive, migrate, invade, and metastasize. Additionally, increased lipid production or uptake aids in the growth of cancer cells and accelerates the development of tumors8. Lipid metabolism changes are important metabolic phenotypes of cancer cells9. Therefore, blocking lipid supply in cancer cells has significant implications for cell bioenergetics, membrane biosynthesis, and intracellular signaling processes10. Studies have shown that the gene set related to FAM can distinguish the clinicopathological features of glioma, and there is a potential association between FAM and the immunophenotype of glioma11. However, studies on the characteristics and prognosis of GC subtypes based on genes related to FAM are still limited12.
The emergence of immunotherapy has brought a glimmer of hope to treating patients with advanced GC, but there are also some difficulties, such as unsatisfactory screening population13. Specific populations with GC benefit from PD-1 antibodies. Currently, patients with high microsatellite instability (MSI-H) and Epstein-Barr Virus (EBV) positive have unique characteristics, and the efficacy and benefits of immunotherapy are relatively clear14. However, MSI-H and EBV-positive patients account for only about 7% to 8% of the total population with advanced GC, and there is no clear and effective screening method for most of the rest of the population15,16.
Many studies have shown that FAM affects the main immune cells in tumor microenvironment17,18. Therefore, we hypothesized that classifying GC samples based on the expression status of FAM genes might further screen out the population that may benefit from immunotherapy.
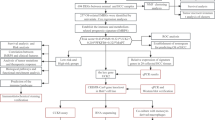
This study used the GSE84437 data set to screen out prognostic fatty acid metabolization-related gene sets. We also used this gene set to perform non-negative matrix factorization (NMF) consensus clustering in GC patients to identify fatty acid metabolization-related subtypes. TCGA-STAD data sets were used to verify the conservatism of subtypes. Then, we compared the differences in prognosis and immune microenvironment characteristics among different subtypes, analyzed the clinicopathological features associated with subtypes, and evaluated the FAM subtypes as independent prognostic factors of GC. We also examined the relationship between FAM subtypes19, the expression of immunological checkpoint and immune regulatory genes, and the molecular properties of FAM subtypes.
Results
Identification of FAM genes associated with prognosis in GC
The publicly available microarray dataset GSE84437, composed of 433 GC samples, was downloaded from GEO along with the complete clinicopathological and follow-up data. 309 out of 332 FAM genes had expression data in this dataset. 50 FAM genes related to OS were picked out by univariate Cox proportional hazard regression (Supplementary Table S2).
Identification of FAM-related subtypes in GC and analysis of the prognosis value of the subtypes
Based on 50 survival-related FAM gene expressions, we divided the 433 GC samples into two subtypes through NMF consensus clustering. For any desired rank k, the NMF algorithm groups the samples into k clusters. To find out the robust rank k, we performed the NMF with rank k from 2 to 10 (Fig. 1A, Supplementary Fig. S1). According to the cophenetic coefficient and silhouette score, the stable and strong k was 2 (Fig. 1B). Cluster-1 included 261 GC samples, and Cluster-2 comprised 172 GC samples. The heatmap showed that Cluster-1 and Cluster-2 exhibited distinct gene expression profiles (Fig. 1C). PCA analysis was used to confirm the distribution patterns of the clusters (Fig. 1D).
Identification of FAM-related subtypes in GC and analysis of the prognosis value of the subtypes. (A) The consensus map of NMF clustering results in the GSE84437 dataset with rank k = 2. (B) Heatmap of prognostic FAM gene expression. (C) The trend of the cophenetic, dispersion, evar, residuals, rss, silhouette, and sparseness coefficients at different ranks k. (D) The principal component analysis results for GSE84437 cohort samples. (E) Kaplan–Meier survival curves of overall survival in Cluster-1 and Cluster-2 GC patients. (F) Forest plot of GSE84437 cohort univariable Cox regression analysis. (G) Forest plot of GSE84437 cohort multivariable Cox regression analysis.
To explore the clinical significance of classification, we performed the survival analysis, and the result showed that the GC patients of Cluster-1 have a better prognosis than that of Cluster-2 (p = 5e−05) (Fig. 1E). The Univariate Cox regression analysis and multivariate Cox regression analysis showed that the FAM-related subtypes were independent prognostic factors (Fig. 1F, G). We used the TCGA STAD cohort as a validation cohort to verify the stability and robustness of our clustering. The findings demonstrated that the TCGA STAD cohort was also robustly categorized into two clusters (Supplementary Fig. S2, S3) and that the PCA analysis divided these two clusters (Supplementary Fig. S4). Additionally, the findings revealed that GC patients in Cluster-1 had a better prognosis than those in Cluster-2 (p = 0.00522) (Supplementary Fig. S5, S6, and S7).
Clinicopathological features of FAM-related subtypes
Since the TCGA-STAD cohort has relatively complete clinicopathological data, we used the data of this cohort to explore the differences in clinicopathological characteristics among GC FAM subtypes. As shown in Table 1 and Fig. 2A, compared with Cluster-2, Cluster-1 patients were older, and the proportion of intestinal-type GC and T1/T2 stage GC were higher. However, the two groups had no significant differences in gender, pathological stage, presence or absence of lymph node metastasis, and distant metastasis. At the same time, considering that gastric-esophageal junction carcinoma has unique characteristics, we further analyzed whether there are differences in the distribution of gastric-esophageal junction carcinoma and GC between Cluster-1 and Cluster-2. The results showed no statistically significant difference between the two groups in gastroesophageal junction carcinoma and GC distribution.
TCGA database divided GC patients into four molecular subtypes, namely chromosomal instability (CIN), EBV, genomically stable (GS), and MSI, each with unique clinical characteristics. We also explored the relationship between FAM-related subtypes and TCGA GC molecular subtypes. The results showed little difference in the distribution of CIN and EBV between the two groups, but MSI was mainly distributed in Cluster-1 GC patients. In contrast, GS was distributed primarily among Cluster-2 GC patients (Table 2, Fig. 2B).
Differential expression gene (DEG) analysis, PPI, and Functional enrichment analysis of FAM-related subtypes
DEG analysis, PPI analysis, and GO and KEGG functional enrichment analysis were performed to explore the characteristic molecular characteristics between subtypes. Differential expression analysis showed 227 upregulated genes and 22 down-regulated genes (logFC > 1 or logFC < −1, FDR < 0.05) in the Cluster-2 subtype compared with the Cluster-1 subtype (Fig. 3A, B and Supplementary Table S3). String database was used to analyze the PPI of differentially expressed genes, and Cytoscape plug-in MCODE was used to predict the hub module. The threshold value was set as MCODE score ≥ 4 and node ≥ 6. One hub module (edges = 64, nodes = 12) was identified with a module score of 11.636. The nodes included AURKB, TPX2, BUB1, UBE2C, CDCA3, CCNA2, AURKA, CCNB2, TOP2A, MAD2L1, CENPA, and CDC20 (Fig. 3C). GO and KEGG functional enrichment analysis were performed on the hub module genes (Fig. 3D).
Differential expression gene(DEG) analysis, PPI, and Functional enrichment analysis of FAM-related subtypes. (A) Heatmap of differentially expressed genes of FAM subtypes. (B) Volcano map of differentially expressed genes of FAM subtypes. (C) Cytoscape plug-in MCODE identified one hub module (edges = 64, nodes = 12) with a module score 11.636. (D) GO and KEGG functional enrichment analysis of the hub module genes.
The results show that the most significant enrichment analysis of biological processes is a nuclear division (GO:0000280), organelle fission (GO:0048285), and mitotic nuclear division (GO:0140014) (Table 3). The most significant results of cell component enrichment analysis were: Condensed nuclear idea, Centromeric region (GO:0000780), Condensed idea (GO:0000793), Condensed idea, Centromeric Region (GO:0000779) (Table 3). The most significant results of molecular function enrichment analysis are Histone kinase activity (GO:0035173), protein C-terminus binding (GO:0008022), histone kinase activity (GO:0035173), Protein C-terminus binding (GO:0008022) Protein serine/threonine/tyrosine kinase activity (GO: 0004712) (Table 3). The most significant KEGG enrichment analysis was Progesterone mediated oocyte maturation (HSA04914), Cell cycle (HSA04110), and oocyte meiosis (HSA04114) (Table 3).
Characteristic differences of immune microenvironments between FAM-related subtypes
Cibersoft and MPCcounter were used to evaluate the immune cell infiltration between different subtypes. Tumor purity, immune, and stromal scores differences between subtypes were assessed using an ESTIMATE algorithm; GSVA was used to determine the enrichment of immune-related pathways between subtypes. The results showed that the stroma score, immune score, and estimation score of Cluster-2 were significantly higher than those of Cluster-1 (Fig. 4A). CIBERSORT results showed that the infiltration of natural B cells, CD8+ T cells, M2-type macrophages, and mast cells in Cluster-2 group were higher than that in Cluster-1 group. In contrast, the infiltration of M0 macrophages, M1 macrophages, and resting dendritic cells in the Cluster-1 group was higher than in the Cluser2 group (Fig. 4B).
Characteristic differences of immune microenvironments between FAM-related subtypes. (A) Tumor purity, immune and stromal scores of FAM subtypes were assessed using an ESTIMATE algorithm. (B) Analysis of immune cell infiltration of FAM subtypes using the CIBERSORT algorithm. (C) Analysis of immune cell infiltration of FAM subtypes using the MPCcounter algorithm. (D) Enrichment analysis of immune-related pathways in FAM subtypes by GSVA algorithm.
The results indicated that the Cluster-1 group had a stronger inflammatory response. In comparison, the Cluster-2 group had stronger specific and non-specific immune responses. In the MCPcouunter method, neutrophils in the Cluster-1 group were significantly increased (p < 0.001), indicating a stronger inflammatory response (Fig. 4C). However, T cells, B cells, Monocytic lineage, Myeloid dendritic cells, endothelial cells, and fibroblasts were significantly increased in Cluster-2 group, indicating stronger immune response and more stromal cell infiltration (Fig. 4C). GSVA analysis showed that there were significant differences in the enrichment of immune pathways among GC FAM-related subtypes. More immune pathways were activated in Cluster-2 (Fig. 4D).
Correlation between FAM subtypes and expression of immune checkpoint genes and immunomodulatory genes
To explore whether FAM subtypes of GC are associated with predicting immune checkpoint inhibitor therapy in GC, we analyzed the correlation between the expression of immune checkpoint genes and immunomodulatory genes among different subtypes. Results showed that almost all immune checkpoint genes were upregulated in Cluster-2 GC patients. The differences between CD276, CD40, CD80, CTLA4, and PDCD1 were statistically significant (Fig. 5B). Cluster analysis of immunoregulatory genes, including Chemokine, Immunoinhibitor, Immunostimulator, MHC, and Receptor showed that the expression of immunoregulatory genes was significantly upregulated in Cluster-2 compared with Cluster-1 (Fig. 5A).
Discussion
The therapeutic effect of GC has been enhanced to some extent with the introduction of minimally invasive techniques, concepts, related instruments, and immunotherapy drugs. Nevertheless, the overall therapeutic outcome remains unsatisfactory. Specific GC populations, such as M SI–H and EBV-positive individuals, can benefit from PD-1 antibodies. Nonetheless, this cohort represents approximately 7% to 8% of the entire population with advanced GC, and there is no clear and effective screening tool for most of the remaining population. Tumor microenvironment (TME) refers to the microenvironment around tumor cells, mainly composed of stromal cells, immune cells, blood vessels, and various signaling molecules. It is usually in a state of immune tolerance20.
Immune cells in TME can undergo metabolic reprogramming under the regulation of tumor cells and other signaling molecules, as well as nutrients, to obtain special metabolic characteristics and affect their survival and effector function, thus reducing the efficacy of various immunotherapies such as ICBand tumor vaccine21,22,23. FAM is a key metabolic pathway that regulates immune response, providing energy for immune cells and substrates and precursors for synthesizing cell components and signal molecules7,24. Consequently, employing prognostic FAM-related genes, we conducted comparable clustering categorization of GC samples using bioinformatics analysis. Two clusters of GC samples were identified, and it was discovered that each cluster had distinct immunological states, clinicopathological characteristics, and substantial predictive differences. This may, to a certain extent, predict the effectiveness of immunotherapy for GC.
More and more studies have shown that abnormal FAM is involved in the occurrence and development of various cancers, including lung cancer25, prostate cancer26, colorectal cancer27, bladder cancer28, etc. FAM-related genes signature is associated with the prognosis of various cancers29,30,31. We first downloaded gene sets related to FAM from the MSigDB database and Genecards database. GSE84437 data set was used to screen out prognostic FAM-related gene sets, including 50 FAM-related genes. GC patients can be divided into 2 groups by the non-negative matrix consensus clustering algorithm and consensus clustering matrix surface. The survival curve showed that Cluster-1 patients had significantly longer overall survival than Cluster-2 patients. Using this gene collection, PCA dimension reduction analysis may efficiently differentiate between the two subtypes of GC patients. Univariate and multivariate Cox regression analyses revealed that FAM-related subtypes were independent predictive variables for patients with GC. We used TCGA-STAD cohort data for validation to test the gene clustering results' stability. The results showed that the TCGA-STAD cohort was still clustered into two groups by non-negative matrix consensus, and the survival curve showed that Cluster-1 patients had significantly longer overall survival than Cluster-2 patients. Changes in energy metabolism in cancer cells compared with normal cells are a new hallmark of most cancers32. The results of our study show that variations in FAM may be correlated with the prognosis of GC patients, although the particular mechanism needs to be investigated further.
Furthermore, we explored the clinicopathological features of the two subtypes. Cluster-1 patients were older, with more intestinal GC and earlier T stages. However, there were no significant differences between the two groups in gender, pathological stage, presence or absence of lymph node metastasis, distant metastasis, and anatomical tumor site. In addition, the distribution of the four types of GC molecular subtypes19 reported by TCGA was also different between the two groups. MSI molecular subtypes were mainly distributed in Cluster-1 GC patients, while GS molecular subtypes were primarily distributed in Cluster-2 GC patients. These findings demonstrate that the expression status of genes involved in FAM is regulated by patient age, tumor T stage, and tumor Lauren type and is connected with microsatellite instability. Developing the schedule for combination treatments and identifying new cancer therapy targets may benefit from a better understanding the metabolic requirements during the cell cycle.
To explore the internal mechanism of the differences between the two clusters, we analyzed the gene differential expression between the two clusters. The threshold of LogFoldchange and adjusted P values were set to 1 and 0.05, and 276 differential genes were obtained, most of which were highly expressed in Cluster-2. Cytoscape software MCODE plug-in was used to screen out the hub modules (threshold set MCODE score ≥ 4, node ≥ 6). The hub module contains 12 genes, which are AURKA, AURKB, CCNA2, CCNB2, CDC20, CDCA3, TOP2A, MAD2L1, CENPA, TPX2, BUB1 and UBE2C. These genes have been reported to promote proliferation and metastasis in multiple tumors33,34,35,36,37,38,39. Hub genes' functional enrichment study also indicated that these genes were associated with the cell cycle and mitosis. This outcome is also in agreement with the poor prognosis for Cluster-2 patients.
Fatty acids are energy storage and signaling molecules that control defensive mechanisms and developmental processes. They also supply crucial building blocks for the development of membranes40. Previous research suggested cell cycle arrest might occur if fatty acid synthesis (FAS) was inhibited. The acetyl-CoA carboxylase (cut6) and fatty acid synthetase (lsd1) mutants have reduced intracellular fatty acid levels, affecting the nucleus and cell division41.
Cibersoft and MPCcounter were used to evaluate the immune cell infiltration between different subtypes to explore the effect of FAM on the immune function of GC. Tumor purity, immune, and stromal scores differences between subtypes were assessed using an ESTIMATE algorithm; GSVA was used to evaluate the enrichment of immune-related pathways between subtypes. Our study showed that the tumor immune microenvironment significantly differed between the two subtypes. The infiltration degree of specific and non-specific immune response cells in Cluster-2 was increased considerably, and the immune pathway was greatly enriched, indicating stronger specific and non-specific immune responses in Cluster-2.
Such results demonstrate that FAM-related genomes may be associated with tumor immune microenvironment. FAM has also been found to influence the function of several immune cells in the tumor microenvironment, including T cells and macrophages. In addition, we explored the expression of immune checkpoint (IC) related genes and immune regulation between the two subtypes. In Cluster-2, we found relatively high expression of IC-related genes and immunoregulatory genes. FAM-related genes are associated with biomarkers of the immune checkpoint, which may play an important role in GC immunotherapy. Previous studies have also shown that since PD-1 inhibitor does not affect the metabolic phenotype of tumor-infiltrating lymphocytes42, the combination of metabolic reprogramming drugs and PD-1 inhibitor can achieve additive effects. The membrane receptor CD36 targeting fatty acid uptake can inhibit fatty acid uptake by Treg cells, down-regulate fatty acid oxidation of Treg cells, and reduce the number and function of Treg cells 43,44 . Targeted CD36 has been shown to act synergistically with PD-1 inhibitors in mouse models45. According to Cui et al., aberrant FAM impacted GC growth. Chemotherapy drug resistance and recurrence were linked to the abnormal expression of FAM-related genes46. Sterol O-acyltransferase (SOAT)1 was highly expressed in GC tissues and negatively correlated with GC prognosis via regulation of SREBP-1 and SREBP-2 expression. SREBP-1 and SREBP-2 increased the expression of vascular endothelial growth factor C (VEGF-C), which promoted lymphangiogenesis47.
Additionally, FAM is crucial for the malignant proliferation of tumors through invasion and metastasis. For instance, stimulation of the TGFβ signaling pathway in lung cancer cells decreases fatty acid production by inhibiting ChREBP, and knockdown of FASN decreases E-cadherin expression, which increases lung cancer cell invasion and metastasis48. Since only a few investigations have determined the role of FAM in GC, possible novel FAM pathways in GC require further exploration.
Conclusions
In conclusion, this study classified GC samples into two clusters by NMF consensus clustering using the FAM genes associated with prognosis, and there were significant prognostic differences between those two clusters. FAM-related subtypes are independent prognostic risk factors in GC patients. FAM-related subtypes have potential correlations with the immune microenvironment of GC. They are significantly correlated with biomarkers of immune checkpoints, which may play an important role in GC immunotherapy. In the next step, we will conduct cell biology experiments and clinical verification of our findings.
Methods and materials
Data sets and FAM genes
From Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE84437) to download GSE84437 datasets, which was a microarray data of mRNA expression in GC, and corresponding clinicopathological and prognostic information, as a training dataset. From The Cancer Genome Atlas (TCGA) data portal (https://gdcportal.nci.nih.gov/) to download the RNA sequencing data and corresponding clinicopathological and prognostic information of TCGA-STAD cohort as a validation dataset. From the Molecular Signature Database v.7.4 (MSigDB; https://www.gsea-msigdb.org/gsea/msigdb), three FAM gene sets (REACTOME_FATTY_ACID_METABOLISM, KEGG_FATTY_ACID_METABOLISM, HALLMARK_FATTY_ACID_METABOLISM) were downloaded. The keyword "fatty acid metabolism" was searched in the Genecards database, and genes with a score greater than 50 were regarded as Genecards FAM gene sets. A total of 332 genes related to FAM were obtained after removing duplicate genes (Supplementary Table S1).
Identification of prognostic FAM genes
Survival R package and Cox univariate regression analysis were used to analyze the training set, and the predictive FAM genes were screened out. A false discovery rate (FDR) < 0.05 was set as the cutoff criterion.
Identification and predictive analysis of FAM gene subtypes in GC
The "NMF" R package49 was used for unsupervised non-negative matrix consensus clustering of the normalized expression data of the prognostic FAM gene set. For each cluster number (k in 2:10), the NMF was run 50 times to evaluate cluster stability. The optimal cluster number k = 2 was selected for the magnitude of the cophenetic coefficient began to decrease. The "PCA" R package was used for dimension reduction analysis, the Survival "R package for subtype predictive analysis, and P < 0.05 was used as the significant difference threshold.
Differential expression gene analysis, protein–protein interaction analysis, and functional enrichment analysis of subtypes
Differential gene expression between subtypes was analyzed using the "Limma" R package50. The adjusted P values for multiple tests were calculated using Benjamini-Hochberg. The absolute value of logFC greater than 1 and FDR < 0.05 were regarded as the threshold of significantly differentially expressed genes. PPI analysis was performed on the differential gene set by the String database, and the MCODE plug-in of Cytoscape software screened the hub module. The threshold was set as MCODE score ≥ 4 and node ≥ 6. The "clusterProfiler" R package51 was used for GO and KEGG52 enrichment analysis of differentially expressed genes, with an adjusted p value less than 0.05 as the threshold of significant enrichment. The "ggplot2" R package53 was used to visualize the results.
Immune infiltration analysis
The CIBERSORT (https://cibersort.stanford.edu/) and MPCcounter platforms were utilized to evaluate the immune cell infiltration of GC patients. Only the samples with p < 0.05 were included in subsequent immune infiltration analysis in CIBERSORT. The ESTIMATE package was used to calculate the immune score, stromal score, and tumor purity of GC.
Statistical analysis
R software (https://www.r-project.org/) was used for all calculations and statistical analysis. Unpaired Student's T test or Mann Whitney U test were used to compare two groups of variables with normal or non-normal distribution. The chi-square test or Fisher exact test was used for categorical data. Survival analysis was performed using the "survival" R package. Log-rank test was used to determine whether there was a significant difference in survival curves.
Data availability
The datasets ANALYZED for this study can be found in The Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) and Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE84437).
References
Sung, H.A.-O., Ferlay, J., Siegel, R.A.-O., Laversanne, M., Soerjomataram, I., Jemal, A. & Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries.
Zhang, X. et al. Endoscopic screening in Asian countries is associated with reduced GC mortality: A meta-analysis and systematic review. Gastroenterology 155, 347–54.e9 (2018).
Ilson, D. H. Advances in the treatment of GC: 2019. Curr. Opin. Gastroenterol. 35, 551–554 (2019).
Yuan, Q. et al. Integration of transcriptomics, proteomics, and metabolomics data to reveal HER2-associated metabolic heterogeneity in gastric cancer with response to immunotherapy and neoadjuvant chemotherapy. Front. Immunol. 4(13), 951137. https://doi.org/10.3389/fimmu.2022.951137 (2022).
Hoy, A. J., Nagarajan, S. R. & Butler, L. M. Tumour fatty acid metabolism in the context of therapy resistance and obesity. Nat. Rev. Cancer 21, 753–766 (2021).
Viens, K. et al. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature 566, 403–406 (2019).
Li, Z. & Zhang, H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. 73, 377–392 (2016).
Bian, X. et al. Lipid metabolism and cancer. J. Exp. Med. 218, 13 (2021).
Snaebjornsson, M. T., Janaki-Raman, S. & Schulze, A. Greasing the wheels of the cancer machine: The role of lipid metabolism in cancer. Cell Metab. 31, 62–76 (2020).
Broadfield, L. A., Pane, A. A., Talebi, A., Swinnen, J. V. & Fendt, S. M. Lipid metabolism in cancer: New perspectives and emerging mechanisms. Dev. Cell 56, 1363–1393 (2021).
Qi, Y., Chen, D., Lu, Q., Yao, Y. & Ji, C. Bioinformatic profiling identifies a fatty acid metabolism-related gene risk signature for malignancy, prognosis, and immune phenotype of glioma. Dis. Mark. 2019, 3917040 (2019).
Yang, S. et al. Fatty acid metabolism is related to the immune microenvironment changes of gastric cancer and RGS2 is a new tumor biomarker. Front. Immunol. 14(13), 1065927. https://doi.org/10.3389/fimmu.2022.1065927IF:7.3Q1 (2022).
Zhao, Q. et al. Immunotherapy for GC: Dilemmas and prospects. Brief Funct. Genomics 18, 107–112 (2019).
Chen, C. et al. Efficacy and safety of immune checkpoint inhibitors in advanced gastric or gastroesophageal junction cancer: A systematic review and meta-analysis. Oncoimmunology 8, e1581547 (2019).
De Meulder, S. et al. Prevalence of microsatellite instable and Epstein-Barr Virus-driven gastroesophageal cancer in a large Belgian cohort. Acta Gastroenterol. Belg. 85, 1–5 (2022).
Yoshida, T. et al. Clinicopathological features of PD-L1 protein expression, EBV positivity, and MSI status in patients with advanced gastric and esophagogastric junction adenocarcinoma in Japan. Cancer Biol. Ther. 23, 191–200 (2022).
DeBerardinis, R. J. Tumor microenvironment, metabolism, and immunotherapy. N. Engl. J. Med. 382, 869–871 (2020).
Wu, H. et al. Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. EMBO Mol. Med. 11, e10698 (2019).
Cristescu, R. et al. Molecular analysis of GC identifies subtypes associated with distinct clinical outcomes. Nat. Med. 21, 449–456 (2015).
Pitt, J. M. et al. Targeting the tumor microenvironment: Removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 27, 1482–1492 (2016).
Pingili, A. K. et al. Immune checkpoint blockade reprograms systemic immune landscape and tumor microenvironment in obesity-associated breast cancer. Cell Rep. 35, 109285 (2021).
Somarribas Patterson, L. F. & Vardhana, S. A. Metabolic regulation of the cancer-immunity cycle. Trends Immunol. 42, 975–993 (2021).
Bleve, A., Durante, B., Sica, A. & Consonni, F. M. Lipid metabolism and cancer immunotherapy: Immunosuppressive myeloid cells at the crossroad. Int. J. Mol. Sci. 21, 133 (2020).
Raud, B., McGuire, P. J., Jones, R. G., Sparwasser, T. & Berod, L. Fatty acid metabolism in CD8(+) T cell memory: Challenging current concepts. Immunol. Rev. 283, 213–231 (2018).
Ali, A. et al. Fatty acid synthase mediates EGFR palmitoylation in EGFR mutated non-small cell lung cancer. EMBO Mol. Med. 10, 7 (2018).
Xu, H. et al. Fatty acid metabolism reprogramming in advanced prostate cancer. Metabolites 11, 6 (2021).
Wang, Y. N. et al. CPT1A-mediated fatty acid oxidation promotes colorectal cancer cell metastasis by inhibiting anoikis. Oncogene 37, 6025–6040 (2018).
Ntaku, V. et al. Multi-omics integration analysis robustly predicts high-grade patient survival and identifies CPT1B effect on fatty acid metabolism in bladder cancer. Clin. Cancer Res. 25, 3689–3701 (2019).
Hu, B., Yang, X. B. & Sang, X. T. Construction of a lipid metabolism-related and immune-associated prognostic signature for hepatocellular carcinoma. Cancer Med. 9, 7646–7662 (2020).
Li, J. et al. Lipid metabolism gene-wide profile and survival signature of lung adenocarcinoma. Lipids Health Dis. 19, 222 (2020).
Li, X. et al. Identification of a novel metabolism-related gene signature associated with the survival of bladder cancer. BMC Cancer 21, 1267 (2021).
Pavlova, N. N. & Thompson, C. B. The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 (2016).
Xiao, J. & Zhang, Y. AURKB as a promising prognostic biomarker in hepatocellular carcinoma. Evol. Bioinform. Online 17, 11769343211057588 (2021).
Li, X. et al. Downregulation of CCNA2 disturbs trophoblast migration, proliferation, and apoptosis during the pathogenesis of recurrent miscarriage. Am. J. Reprod. Immunol. 82, e13144 (2019).
Shi, M. et al. APC(CDC20)-mediated degradation of PHD3 stabilizes HIF-1a and promotes tumorigenesis in hepatocellular carcinoma. Cancer Lett. 496, 144–155 (2021).
Dong, Y. et al. Type IIA topoisomerase (TOP2A) triggers epithelial-mesenchymal transition and facilitates HCC progression by regulating Snail expression. Bioengineered 12, 12967–12979 (2021).
Wang, Y. et al. miR-30a-3p targets MAD2L1 and regulates proliferation of GC cells. Onco Targets Ther. 12, 11313–11324 (2019).
Wang, Q. et al. CENPA promotes clear cell renal cell carcinoma progression and metastasis via the Wnt/β-catenin signaling pathway. J. Transl. Med. 19, 417 (2021).
Zhang, B. et al. TPX2 mediates prostate cancer epithelial-mesenchymal transition through CDK1 regulated phosphorylation of ERK/GSK3β/SNAIL pathway. Biochem. Biophys. Res. Commun. 546, 1–6 (2021).
Kwok, A. C. M. & Wong, J. T. Y. Lipid biosynthesis and its coordination with cell cycle progression. Plant Cell Physiol. 2, 1973–1986 (2005).
Al-Feel, W., DeMar, J. C. & Wakil, S. J. A Saccharomyces cerevisiae mutant strain defective in acetyl-CoA carboxylase arrests at the G2/M phase of the cell cycle. Proc. Natl. Acad. Sci. 54, 3095–3100 (2003).
Leone, R. D. et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 366, 1013–1021 (2019).
Hao, J. W. et al. CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nat. Commun. 11, 4765 (2020).
Rueda, C. M. et al. High-density lipoproteins selectively promote the survival of human regulatory T cells. J. Lipid Res. 58, 1514–1523 (2017).
Ma, X. et al. CD36-mediated ferroptosis dampens intratumoral CD8(+) T cell effector function and impairs their antitumor ability. Cell Metab. 33, 1001–12.e5 (2021).
Cui, M.-Y. et al. The role of lipid metabolism in gastric cancer. Front. Oncol. 41, 916661 (2022).
Zhu, T. et al. SOAT1 promotes gastric cancer lymph node metastasis through lipid synthesis. Front. Pharmacol. 4, 769647 (2021).
Li, C. et al. Key molecules of fatty acid metabolism in gastric cancer. Biomolecules 41, 706 (2022).
Gaujoux, R. & Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Bioinform. 11, 367 (2010).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 (2015).
Guangchuang, Y.L.-G.W., Han, Y. & He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 16, 284–287 (2012).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51(1), 587–592 (2023).
Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 3, 180–185 (2011).
Author information
Authors and Affiliations
Contributions
H.R. and Z.R. conceived the project and wrote the manuscript. L.T. participated in data analysis and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, R., Lu, TL. & Zhou, R. Identification and immune landscape analysis of fatty acid metabolism genes related subtypes of gastric cancer. Sci Rep 13, 20443 (2023). https://doi.org/10.1038/s41598-023-47631-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-47631-6
- Springer Nature Limited