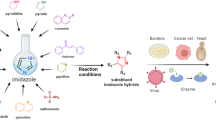
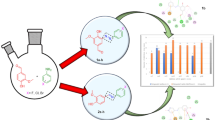
Abstract
Quinazoline derivatives, as an important category of heterocyclic compounds, have received much attention for the design and development of new drugs due to their various pharmacological properties. Besides, there is a great deal of evidence showing pyrimidine analogs as anticancer agents. Thus, in the present study, for the design of new target compounds with cytotoxic activity, we focused on various quinazolinone and pyrimidine hybrids. A new series of quinazoline-pyrimidine hybrid derivatives (6a-6n) have been designed and synthesized as novel antiproliferative agents. All the synthesized compounds characterized based on their IR, NMR and Mass spectroscopic data. Antiproliferative activities of the new compounds were evaluated against three human cancer cell lines (MCF-7, A549, SW-480). The compounds were found to have appropriate potential with IC50 values ranging from 2.3 ± 5.91 to 176.5 ± 0.7 μM against the tested cell lines. Compound 6n exerted the highest antiproliferative activity with IC50 values of 5.9 ± 1.69 μM, 2.3 ± 5.91 μM and 5.65 ± 2.33 μM against A549, SW-480 and MCF-7 respectively. The results indicated that 6n could induce apoptosis in A549 cell line in a dose dependent manner and arrest in the S phase of cell cycle. Docking studies were also done to investigate the detailed binding pattern of the synthesized compounds against EGFR. Furthermore, molecular dynamic simulation and binding free energy calculation have been done to rescore initial docking pose of the synthesized compounds using ensemble-based MMGB/PBSA free energy method. According to the results, free energy calculation confirmed biological activity of compounds and also, Arg 817 and Lys 721 residues had the pivotal role in the high potency of 6n. Finally, the drug likeness and in silico ADME study were also predicted.
Similar content being viewed by others
Introduction
Cancer, one of the principal causes of death, is a multifactorial disease that comprises numerous genetic defects and is characterized by abnormal growth of cells1, 2. According to the World Health Organization (WHO) nearly 10 million cancer deaths occurred in 2022 in worldwide. Chemotherapy is considered a basis in the management of many types of cancer, however, its effectiveness in the curing of cancer is partly troubled by drug resistances3. However for most types of disseminated cancers, no effective treatment is available, and development of new active chemotherapeutic agents is urgently needed4. More than 90% of the novel drugs bear heterocycle rings in their structure and among them, nitrogen containing heterocyclic compounds display more notable pharmaceutical effect than non-nitrogen compounds5. Quinazoline scaffolds represent an important class of biologically active nitrogen heterocyclic compounds and a variety of marketed drugs are based on these moieties6 with broad spectrum of pharmacological activities and minimum side effects7. Pharmacological activities of quinazoline and its related scaffolds include anti-cancer, antifungal, anti-tumor, anti-malaria, anticonvulsant, anti-microbial, anti-inflammatory and antihyperlipidemic activities8,9,10,11,12. Quinazoline compounds have been also shown to inhibit tyrosine kinase activities, and accordingly are useful to prevent tumor growth13. Quinazoline derivatives are one of the largest chemical groups reported as potent Epidermal Growth Factor Receptor (EGFR) inhibitors. EGFR, among different receptors and their associated signaling proteins, widely play a critical role in tumor genesis14, 15. Evidences for the role of EGFR in inhibition and pathogenesis of various cancers have led to the development of agents that target this receptor. Activation of the EGFR signaling pathway in cancer cells has been linked with increased cell proliferation, angiogenesis and metastasis16. In recent years, several quinazoline scaffolds such as Gefitinib (2003), Erlotinib (2004), Lapatinib (2010), Vandetanib and Icotinib (2011), Afatinib (2013) clinically approved as EGFR inhibitors (Fig. 1)13.
In addition, literature review has discovered that many effective EGFR tyrosine kinase inhibitors were derived from pyrimidine nuclei i.e. Osimertinib17, Rociletinib18 and Olmutinib (Fig. 2)19. Recently, efforts have been made to synthesize novel hybrid derivatives of quinazoline and pyrimidine20. Hybrid molecules with two or more pharmacophores usually have a better potential to reduce side effects and overcome drug resistance. There are many hybrid molecules in different phases of clinical trials for treatment of various diseases. This suggests that hybridization is a useful strategy for development of the novel drugs21. Notably, pyrimidine, quinazoline and their analogues have been reported to have beneficial effects in varieties of cancer22. In our previous study we reported a series of quinazolinone–pyrimidine hybrids as dipeptidyl peptidase-4 (DPP4) inhibitors23. Here, we have focused on design and synthesis of new series of quinazolinone–pyrimidine hybrids as EGFR inhibitors. In this regard 14 hybrids (6a-6n) were synthesized and characterized by different spectroscopic methods. Biological evaluations of the synthesized compounds were tested against three cancerous cell lines (A549, SW-480 and MCF-7). Molecular docking study was also performed to explore the possible mechanisms of the newly synthesized compounds. Finally, we used ensemble-based MMGB/PBSA free energy method to simulate molecular dynamic and calculate binding free energies to rescore initial docking pose of the synthesized compounds.
Results and discussion
Design approach
EGFR tyrosine kinase is considered as one of the most effective clinically targets for treatment of non-small-cell lung cancer (NSCLC)24, 25. Tyrosine kinase inhibitors (TKIs) can be classified into reversible and irreversible inhibitors. These inhibitors bind reversibly and competitively to the ATP-binding site of the tyrosine kinase domain of the intracellular EGFR and prevent its phosphorylation and downstream pathways26. Erlotinib and Gefitinib were approved as selective EGFR tyrosine kinase inhibitors by Food and Drug Administration (FDA) for treatment of NSCLC cancer patients27, 28. Pyrimidine derivatives such as Osimertinib show significant activity against EGFR with an IC50 value of 12.92 nM29. Osimertinib was approved by FDA in 2015 and can be used as first-line treatment for advanced NSCLCs according to the current European Society of Medical Oncology (ESMO) recommendation30. In this study, we used a combinatorial pharmacophore approach to design novel antiproliferative compounds as EGFR inhibitors. To achieve synergistic cytotoxic effects, the pyrimidine ring was attached to the 6-boromoquinazoline backbone having different benzyl substitution at N-3 position with different electronic profiles (Fig. 3).
Synthesis and characterization of 6-boromo-quinazolinone–pyrimidine hybrids (6a–6n)
The synthesis route of 6-boromo-quinazolinone–pyrimidine hybrids (6a-6n) is shown in Fig. 15. First, a mixture of anthranilic acid (1) and N-bromo succinimide in acetonitrile was used to prepare 5-boromoantranilic acid (2). Then 5-boromoantranilic acid was reacted with chloroacetyl chloride under the basic conditions (DIPEA) in DCM at room temperature for 2 h to synthesize 6-bromo benzoxazine-4-one (3). Then, various substituted anilines (4a-4n) were reacted with 3 via nucleophilic attack under acidic conditions in acetonitrile. Finally, 6-boromoquinazoline derivatives (5a-5n) were refluxed with 6-chloro-3-methyl-uracil at 80 °C in the presence of DIPEA to obtain the final compounds (6a-6n) in good yield. The final compounds were purified by plate chromatography using chloroform/n-hexane as eluents (4:1) (Table 1). Chemical structures of the final compounds were confirmed by IR, 1H-NMR, 13C-NMR, and Mass spectroscopies. In IR analysis, the stretching frequency of carbonyl bonds were observed at 1674–1719 cm−1. In 1H NMR spectra of compounds 6a-6n compounds a signal at 4.72–5.24 ppm associated with the CH2 protons which are between 6-chloro-3-methyl-uracil and quinazolinone. The singlet peak at 8.29–8.41 ppm indicated the presence of a proton at C5 position of the quinazoline moiety. A single signal at 5.98–6.14 ppm corresponds to the proton of uracil. In the 13C NMR spectra, the chemical shift at 157.82–164.31 ppm and 47.57–66.4 ppm related to the carbonyl groups and CH2 moieties, respectively.
In vitro cytotoxicity screening
Antiproliferative activities of all the synthesized compounds (6a-6n) were investigated against three human cancerous cell lines, A549 (human lung adenocarcinoma), SW-480 (colorectal cancer), and MCF-7 (human breast cancer). The best cytotoxic effect was obtained for lung cancer (A549) followed by colorectal cancer (SW-480). Compound 6n with two electron withdrawing atoms (Cl and F) at positions 3 and 4 of the phenyl ring showed good activity with IC50 = 5.9 ± 1.7 µM, 2.3 ± 0.91 µM and 5.65 ± 2.33 µM compared to Cisplatin with IC50 = 15.37 µM, 16.1 µM and 3.2 against A549, SW-480 and MCF-7 cell lines respectively (Table 2).
The synthesized compounds could be classified in two categories as mono-substituted (6a-6k) and di-substituted (6l-6n). In the mono substituted category compounds 6h and 6k with bromine and phenoxy groups at para position of the phenyl ring displayed significant antiproliferative effects (Table 2). The presence of electron-withdrawing groups (halogen atoms) at para position of the phenyl ring in compounds 6f., 6g and 6h seems to cause these compounds more active than compounds 6i and 6j with electron-donating groups in order of Br > Cl > F. Changing the halogen substitutions from para to meta position resulted in 1.95–8.44 folds’ decrease in activity. In the di-substituted groups, all compounds had low cytotoxic activity in the range of > 80 µM except 6n, which is consistent with the mono-substituted compounds. The unsubstituted compound 6a was less active compared to all other substituted compounds. Structure activity relationship (SAR) studies showed that compounds containing one or two halogen substitutions on the phenyl ring, have significant antiproliferative activity (Fig. 4). On the other hand, the presence of OMe and Me groups reduced the efficacy. In addition, the presence of substitution at para position was more effective than in the meta position. Having bulky substitution (4-phenoxy) on the phenyl ring (6k) also increased the inhibitory activity. In addition, to evaluate the selectivity activity of compounds between normal and cancerous cell lines, the cytotoxic activity on the normal lung cell line (MRC-5) was applied for three potent compounds (6h, 6k and 6n). The results showed a desire selectivity (Table 2).
Apoptosis detection
Cell death mechanism of compound 6n as promising compound was investigated via Annexin V-Propodium Iodide (PI) double staining technique. As could be seen, using this technique, four staining patterns could be detected: viable cells Q4: (Avneg/PIneg), early apoptotic cells Q3: (Avpos/PIneg), late apoptotic cells Q2: (Avpos/PIpos) and necrotic cells Q1: (Avneg/PIpos). Annexin V was applied to detect the phosphatidyl serin, which moved to outer membrane during apoptosis, and PI was used to distinguish live and dead cells. The apoptotic activity of 6n on A549 cell line in three different concentrations is shown in Fig. 5. The results showed that 6n significantly induced apoptotic death in the dose dependent manner from (2.61% early apoptosis and 21.2% for late apoptosis in 5 µM concentration), to 33.26% and 65.08% apoptotic death in 10 µM and 15 µM concentration.
Cell cycle analysis
To investigate the effect of compound 6n on the cell cycle distribution of A549 cell line, a flow cytometric based method was used. For this purpose, A549 cells were treated with two concentrations of 6n (10 and 15 μM) for 72 h (Fig. 6). As could been seen, the A549 cells exposed to compound 6n showed accumulation in the S-phase with a percentage of 8.91% and 11.6% following treatment with 10 and 15 μM, respectively, compared to untreated cells (4.46%). A slight decrease in the G2/M population was also observed. These results collectivel suggest that compound 6n could induce arrest at S-phase.
Molecular docking study
Molecular docking simulations based on the 3D structure of EGFR (PDB code: 1M17) was performed using Auto Dock Vina software for the synthesized compounds (6a-6n) and Erlotinib as an internal ligand31. In Fig. 7, the superimposing of the internal ligand (Erlotinib) before and after the docking operation was displayed. The binding free energies and interaction details of Erlotinib and the synthesized compounds (6a-6n) against EGFR were summarized in Table 3.
All of the compounds exhibited better affinity to EGFR protein and also, had higher binding energy compared to Erlotinib. According to the proposed binding mode of Erlotinib (Fig. 8), the planar region of Erlotinib (quinazoline moiety) interacted via hydrogen bonds and pi interactions with Lys 721, Asp 831, and Val 702. Additional hydrogen bond interactions were observed between Cys773, Met769 and Gln 767 and aliphatic chain. Besides, hydrophobic region (phenyl ring) formed pi interactions with Phe 699 and Asp 831.
The binding mode of the most potent compound (6n) and the less active compound (6a) were showed in Fig. 9. In compound 6n, the planar region (quinazoline pharmacophore) interacted via two hydrogen bonds with Lys 721 and Gly 772 and also, formed pi interactions with PHE 699, Asp 831 and Val 702. The pyrimidine moiety involved in pi-alkyl interactions with Lys 721 and ALA 719 and halogen bond with Glu 738. The 3-chloro-4-fluoro phenyl substituents of compound 6n formed two interactions including hydrogen and pi-alkyl with Gly 772, Leu 820, Val 702 and Leu 694. Some hydrophobic interactions with Met 769, Met 742, Gln 767, Thr 766, Thr 830, Cys 751, Cys 773 were also, seen. These additional interactions may explain the reason of the highest cytotoxic activity of compound 6n among the other compounds. The proposed binding mode for compound 6a includes hydrogen bonding and pi interactions with Lys 721 and Asp 831, Phe 699 by the planar quinazoline ring. Additionally, it formed alkyl bond with Val 702 through Br atom. The pyrimidine moiety formed one pi-alkyl interaction with Pro 853 and also, some hydrophobic interactions with Arg 817, Asp 813, Leu 834, Glu 734 and Gly 833 were observed. The results of docking studies showed that most compounds have key amino acids in the active site of EGFR like Erlotinib, such as Cys 773, Lys 721, Val 702, Phe 699.
Molecular dynamic study
The MMPB/GBSA binding energies of all ligands (6a-6n) and the corresponding experimental binding energies obtained from the cytotoxicity data related to A549, SW-480 and MCF-7 cell lines are shown in Table 4. All binding free energy values are ordered based on experimental values of A549 cell line. Compound 6n has the maximum binding energy for both MMPBSA and MMGBSA and this is in line with cytotoxicity (IC50) values after converting them to binding energies using ΔG = − RT ln (IC50) equation. Pearson and Spearman's correlation coefficients were used to measure the correlation between values of MMPB/GBSA and experiment data (Fig. 10). The Pearson correlation coefficient (r) is known as the most usable correlation coefficient that summarizes the characteristics of a dataset. Also, it describes the strength and linear relationship direction between two quantitative variables. Spearman's rank correlation is a nonparametric statistical dependence coefficient between the rankings of two variables. Pearson coefficient works with a linear relationship between the two variables whereas the Spearman coefficient works with monotonic relationships as well. On the other hand, Pearson works with raw data values of the variables whereas Spearman works with rank-ordered variables. Conferring to Fig. 10, MMPBSA results correlate well with experimental binding energies according to its Pearson (rP = 0.724, 0.711 and 0.632 for A549, SW-480 and MCF-7, respectively) and Spearman correlation coefficient (rs = 0.533, 0.535 and 0.578 for A549, SW-480 and MCF-7, respectively). In addition, the correlation is stronger when we rank MMPBSA binding energy values with binding energies obtained from the cytotoxicity of A549 cell lines (Fig. 10a). Pearson correlation predicts the binding energy ranking of ligands 6n, 6 h, 6 k, 6 g, 6f. and 6a in the correct order. These ligands are chosen for further analysis.
Pearson and Spearman Rank correlation between MMPBSA and experimental binding energy obtained from anti-proliferative activities toward (a) A549, (c) SW-480, and (e) MCF-7, and between MMGBSA and experimental binding energy obtained from anti-proliferative activities toward (b) A549, (d) SW-480 and (f) MCF-7.
Binding free energy, energy decomposition analysis, hydrogen bond and clustering analysis
To show the factors responsible for the inhibitor activities of 6n, 6 h, 6 k, 6 g, 6f. and 6a ligands, the MM/PBSA method was run. Figure 11 illustrated the changing amount of binding free energy for each replica in the ensemble as well as the associated uncertainties. The final binding free energy can be estimated by the average binding energy of each replica. The total binding free energies are found in a range of − 24.45 and − 17.08 kcal·mol−1 (Table 5). Table 4 shows the contribution terms in the binding free energies of these inhibitors against EGFR target. The more contributed of the net binding free energy of 6n is belong to VDW energy term with − 53.56 kcal·mol−1 energy. Another contribution is GNon-Polar term which is essentially always small and similar for all selected ligands (ranging from − 4.71 to − 3.90), the intermolecular electrostatic interactions is normally calculated using Coulomb’s law with atomic charges taken from the MM force field. Consequently, the results depend on the charges used for the receptor and the ligand. This energy term has a more positive effect on binding energy of 6n and 6 k (− 22.42 and − 19.51 kcal·mol−1).
The strength of intermolecular electrostatic interactions is depending on the number of hydrogen bonds, water bridges and other polar interactions between the protein and the ligand. To evaluate the type and strength of hydrogen bonds between selected ligands and the protein, MD simulation of ligand-enzyme was accomplished during the whole ensemble-based trajectories. We used the H-bond module of AmberTools 22 to check the hydrogen-bonding profiles between the chosen ligands and the enzymes. As default values, the angle and H-bonding distance threshold were set to 135’ and 3.0 Å, respectively. Figure 12 demonstrates the hydrogen bonding occupancy plots versus time for all aforementioned ligands. The number of hydrogen bonds for all compounds were varied between 0 and 1. Compounds 6n, 6 h and 6 k with the highest number of hydrogen bonds and the highest occupancies are expected to have stronger interactions. By exact inspection of hydrogen bonds from all selected ligands, we found Arg 817 and Lys 721 had strong hydrogen bonds with oxygen atom of quinazolinone motif which has pivotal role in binding of these ligands. It is supposed that the close contacts between oxygen in the pyrimidine ring (O12) of 6n with –OG atom of Thr 766 has the highest occupancies during the MD simulation (Fig. 12a). Likewise, the same contact between oxygen in pyrimidine ring (O12) of 6 h with –NH group of Arg 817 with moderate occupancies acknowledged its high potency toward EGFR (Fig. 12b). It is believed that another interaction which has a great contribution to the inhibition activities of these ligands is forming hydrogen bond between –NH2 group of Lys 721 with the oxygen atom of the quinazolinone group of these ligands. Similarly, compound 6 g forms the close contact between oxygen atom of its quinazolinone group and –NH2 group in Arg 721.
The MMGBSA formulation can provide supplementary information to evaluate the comparative contribution of every residue toward the binding free energy of the enzyme/ligand complex. Val 702 and Asp 831 are a common contribution in the binding energy of 6n, 6 h and 6 k in the active site of EFGR target. Among these residues, Val 702 plays a key role in the stablishing of these ligands through VDW interactions. However, Asp 831 disfavours the binding of these ligands through their high polar solvation energy term (Fig. 13).
Conformational clustering was conducted on the molecular dynamic trajectories of the previously described ligand–protein complexes to select intellectual representative conformations for advanced evaluation32,33,34. Clustering was carried out using k-means algorithms and the RMSD of residues with a sieving frame of 10. The representative structures of the more populated cluster were choosing as a model to illustrate the much often interactions between aforementioned ligands and the active site of EGFR. The results depicted in Fig. 14a,b,c,d,e,f, nevertheless, these models do not necessarily reflect all main interactions and can be refer to the other analysis such as H-bond analysis for more accurate data. According to Fig. 14, visual analysis of the protein active site discloses 2 primary interactions that influence the accommodation of aforementioned ligands in the active site: hydrophobic "clamp" that provides affinity; and hydrogen-bonding network that determines binding mode. Figure 14a shows the halogen bond manifestation between bromine atom of 6n and Met 769. According to Fig. 14a, residues such as Leu 694, Val 702, Lys 721, Leu 764, Thr 830 stabilize the 6n in the active site of enzyme by forming several alkyl-alkyl staking interaction and this is in line with what we observed in decomposition analysis of this ligand. Figure 14b shows the main electrostatic interactions of 6 h including H-bond between oxygen atom in quinazolinone moiety with Lys 721 (Fig. 14b). In addition, the π-π and alkyl-alkyl stacking interactions between pyrimidine moiety and -NH2 group of Lys 721 and Phe 699 is participated in ligand stabilization. Although hydrogen bond analysis proved forming of H-bond between oxygen atom in quinazolinone moiety of the 6 k with Lys 721, same as the 6 h, this is not appearing in its interaction map of representative configuration. Figure 14c illustrates the possible hydrophobic cavities involving Leu 694, Val 702, Lys 721, Asp 776 and Thr 830 with 6 k in the active site of the protein. Figure 14d and 12e show the representative configuration of 6 g and 6f., respectively. 6 g and 6f. had the same configuration in the active site of the protein. These two structures formed H-bond between pyrimidine oxygen atoms with Arg 817. Despite the fact that the main interaction, is forming H-bond between quinazoline oxygen atom with Lys 721, this interaction has not been considered as a stable one for 6f. justifying the underestimation of MMPBSA free energy for this ligand in our calculation (Fig. 14d,e).
In silico Physicochemical parameter (ADME) prediction
In this part of the study, all the synthesized compounds (6a-6n) and Erlotinib were conducted to determine their physicochemical properties using http://www.swissadme.ch/ online server (Table 6). According to the Lipinski’s rule, compound's absorption will be better if they perform at least three of these following rules: all of the compounds fulfilled the Lipinski rule except molecular weight (MW) of some compounds.
In silico study of all compounds and Erlotinib were evaluated for ADME properties by using preADMET online server (www.preadmet.bmdrc.org). According to ADME profiles and the obtained results (Table 7), we can suggest that the synthesized compounds have better intestinal absorption in humans (97.54–98.24) than Erlotinib (96.28). Preferred properties of these compounds enable them to facilitate the movement across various biological membranes (30). The Caco-2 permeability parameters indicate that all compounds have moderate ability to penetrate biological membranes. Also, the designed compounds demonstrate low to moderate crossing of the blood–brain barrier (0.58–1.92). All compounds similar to Erlotinib have relatively high binding to plasma protein. Furthermore, compounds 6k and 6l can inhibit cytochrome P3A4, which is the main enzyme involved in the metabolism of drugs, while Erlotinib doesn’t have this ability.
Conclusion
In summary, we designed and synthesized some new quinazoline-pyrimidine hybrid derivatives incorporating different phenyl moieties at position 3 of the quinazoline ring (N-3). All the synthesized compounds (6a-6n) were successfully characterized using IR, NMR (1H & 13C) and Mass spectroscopic techniques. The antiproliferative activities were examined against a panel of three human cancer cell lines (A549, SW-480, and MCF-7) using MTT assay. Among the tested compounds, 6n showed the highest antiproliferative activities against the tested cell lines. This compound could also induce apoptosis in A549 cell line in a dose dependent manner, and also could arrest the cells in the S phase of cell cycle. Molecular docking study and predicting the possible interactions between target compounds and EGFR showed that the binding site of the proposed compounds with EGFR active site and docking simulation were in good agreement with the results of biological screening. The ensemble-based MMPB/GB calculations was used to inhibitory ranking of all synthesized ligands against different cell lines. The MMPBSA calculation is well correlated with the cytotoxic activity in A549 cell line. We used the ensemble trajectories for H-bond, clustering and energy decomposition analysis to investigate the vital residues associated with the binding of ligand in the active site of the protein. Accordingly, we found the crucial role of Arg 817 and Lys 721 in binding affinities of potent ligands (6n and 6h). Furthermore, the ADME study was calculated and the designed compounds were shown to be compatible with the Lipinski's rule.
Experimental
Chemistry
All solvents and reagents were obtained from the Sigma, Aldrich and Merck. Reaction progress was monitored using thin-layer chromatography (TLC) on silica gel plates. Melting points were measured by Electrothermal 9200 apparatus (Electrothermal, UK). Infrared spectra (KBr) were recorded on a VERTEX70 spectrometer (Bruker, Germany). 1HNMR and 13CNMR spectra were obtained using VARIAN-INOVA 500 MHz Bruker in CDCl3 or DMSO‑d6 solution. Mass spectra were determined with Agilent Technologies (HP).
General procedure for the synthesis of 2-amino-5-bromobenzoic acid (2)
Anthranilic acid (1) (1 mmol) was treated with N-bromo succinimide (1.2 mmol) in acetonitrile at room temperature for 2 h, then the remained solvent was removed by vacuum evaporation and the reaction mixture washed with acetonitrile to give the pure white solid.
General procedure for the synthesis of 6-bromo-2-(chloromethyl)-4H-benzo[d][1,3]oxazin-4-one (3)
2-Amino-5-bromobenzoic acid (2) (1 mmol), diisopropylethylamine (DIPEA) (1.5 mmol) and chloroacetyl chloride (1.2 mmol) were stirred in dichloromethane (10 mL) at room temperature for 2 h. After completion the reaction, 20 mL H2O was added to the reaction mixture and extracted with ethyl acetate (3 × 20 mL). The organic layers were dried by sodium sulfate and concentrated under vacuum to give the white solid compound.
General procedure for the synthesis of 6-bromo-2-(chloromethyl)-3-substituted quinazoline-4(3H)-one (5a-5n)
Intermediate 3 was reacted with various substituted anilines (4a-4n) (1 mmol) under acidic condition in the presence of 1.5 mmol PCl3 through refluxing in acetonitrile (CH3CN) at 80 °C for 24 h. Then the reaction was poured into a saturated NaHCO3 solution and extracted with ethyl acetate (3 × 20 mL). The organic layers were dried by sodium sulfate and concentrated under vacuum to give the white solid compounds.
General procedure for the synthesis of 1-((6-bromo-4-oxo-3- substituted -3,4-dihydroquinazolin-2-yl) methyl)-6-chloro-3-methylpyrimidine-2,4(1H,3H)-dione (6a–6n)
In the final step, intermediates 5a–5n (2 mmol) and 6-chloro-3-methyl uracil (2 mmol) were refluxed in acetonitrile in the presence of 1.5 mmol DIPEA at 80 °C for 24 h. After completion the reaction, the crude products extracted with ethyl acetate (3 × 20 mL). The organic layers were dried by sodium sulfate and concentrated under vacuum to give the final target compounds (Fig. 15).
1-((6-Bromo-4-Oxo-3-Phenyl-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6a).IR (KBr, cm−1): 2961 (CH), 2928 (CH), 1715 (C = O), 1684 (C = O), 1670 (C = O), 1611 (C = N), 1519 (C = C), 1476 (C–N), 1441 (C–O), 847 (C–Br). 1H NMR (500 MHz, CDCl3) δH (ppm): 8.35 (s, 1H, H-5-quinazoline), 7.81 (d, 1H, J = 10 Hz, H-7-quinazoline), 7.467 (d, 1H, J = 10 Hz, H-8-quinazoline), 7.36–7.39 (m, 3H, phenyl), 7.31 (t, 2H, J = 10 Hz, phenyl, 6.01 (s, 1H, uracil), 4.81 (s, 2H, quinazoline-CH2-uracil), 3.33 (s, 3H, CH3). 13C NMR (125 MHz, CDCl3) δC (ppm): 164.31, 162.30, 160.71, 151.12, 149.87, 145.62, 145.43, 137.95, 131.05, 130.01, 129.94, 129.65, 129.53, 122.33, 121.12, 117.84, 117.66, 102.38, 47.68, 28.47.
3-(6-Bromo-2-((6-Chloro-3-Methyl-2,4-Dioxo-3,4-Dihydropyrimidin-1(2H)-Yl)Methyl)-4-Oxoquinazolin-3(4H)-Yl) Benzonitrile (6b). IR (KBr, cm−1): 3065 (CH), 2957 (CH), 2239 (C≡N), 1719 (C = O), 1674 (C = O), 1666 (C = O), 1611 (C = N), 1580 (C = C), 1464 (C–N), 1437 (C–O), 847 (C–Br). 1H NMR (500 MHz, CDCl3) δH (ppm): 8.33 (s, 1H, H-5-quinazoline), 7.89 (d, 1H, J = 10 Hz, H-7-quinazoline), 7.83(d, 1H, J = 10 Hz, H-8-quinazoline), 7.76–7.79 (m, 1H, phenyl), 7.72 (s, 1H, phenyl), 7.67 (d, 1H, J = 5 Hz, phenyl), 7.49 (d, 1H, J = 5 Hz, phenyl), 6.02 (s, 1H, uracil), 4.72–4.81 (m, 2H, quinazoline-CH2-uracil), 3.32 (s, 3H, CH3). 13C NMR (125 MHz, CDCl3) δC (ppm): 160.60, 160.40, 151.04, 149.03, 145.45, 145.26, 138.27, 136.25, 133.90, 132.94, 131.88, 131.60, 129.73, 129.51, 122.08, 121.47, 117.02, 114.99, 102.46, 47.60, 28.47. MS (m/z, %): 499.0 (M + 1, 9.89)+, 464.0 (100), 406.0 (30.44), 377.0 (11.16), 339.0 (62.34), 259.1 (39.07), 221.0 (20.85), 170.0 (15.87).
1-((6-Bromo-3-(3-Fluorophenyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6c). 1H NMR (500 MHz, CDCl3) δH (ppm): 8.36 (s, 1H, H-5-quinazoline), 7.82 (d, 1H, J = 5 Hz, H-7-quinazoline), 7.63–7.59 (m, 1H, phenyl) , 7.48 (d, 1H, J = 5 Hz , H-8-quinazoline), 7.31 (t, 1H, J = 10 Hz, phenyl), 7.20 (d, 1H, J = 10 Hz, phenyl), 7.14 (d, 1H, phenyl), 6.01 (s, 1H, uracil), 4.79–4.88 (m, 2H, quinazoline-CH2-uracil), 3.33 (s, 3H, CH3). 13C NMR (125 MHz, CDCl3) δC (ppm): 164.41, 162.40, 160.69, 160.42, 151.10, 149.50, 145.57, 138.01, 136.49, 131.92, 129.66, 123.96, 122.30, 121.19, 117.76, 116.02, 115.84, 102.38, 47.57, 28.46. MS (m/z, %): 492.1 (M + 1, 13.60)+, 457.1 (100), 399.1 (33.63), 332.0 (31.72), 252.1 (28.50), 223.0 (26.51), 170.0 (2.53), 75.1 (9.23).
1-((6-Bromo-3-(3-Chlorophenyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6d). IR (KBr, cm−1): 2961 (CH), 2928 (CH), 1709 (C = O), 1680 (C = O), 1665 (C = O), 1609 (C = N), 1587 (C = C), 1468 (C–N), 1437 (C–O), 837 (C–Br). 1H NMR (500 MHz, CDCl3) δH (ppm): 8.36 (s, 1H, H-5-quinazoline), 7.82 (d, 1H, J = 10 Hz, H-7-quinazoline), 7.57 (s, 2H, phenyl), 7.48 (d, 1H, J = 10 Hz, H-8-quinazoline), 7.41 (s, 1H, phenyl), 7.30 (s, 1H, phenyl), 6.01 (s, 1H, uracil), 4.83 (s, 2H, quinazoline-CH2-uracil), 3.33 (s, 3H, CH3). 13C NMR (125 MHz, CDCl3) δC (ppm): 160.70, 160.44, 151.10, 149.47, 145.57, 145.41, 138.04, 136.27, 136.26, 131.54, 130.71, 129.67, 129.53, 128.44, 126.37, 122.28, 121.22, 102.39, 47.66, 28.48. MS (m/z, %): 508.1 (M+, 0.85), 473.1(100), 416.1 (24.38), 348 (50.29), 233.1 (40.02), 111.1 (23.59).
1-((6-Bromo-3-(3-Bromophenyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6e). IR (KBr, cm−1): 2965 (CH), 2922(CH), 1709 (C = O), 1680 (C = O), 1661 (C = O), 1605 (C = N), 1574 (C = C), 1472(C–N), 1431 (C–O), 835 (C–Br). 1H NMR (500 MHz, CDCl3) δH (ppm): 8.36 (s, 1H, H-5-quinazoline), 7.83 (d, 1H, J = 10 Hz, H-7-quinazoline), 7.73 (d, 1H, J = 10 Hz, H-8-quinazoline), 7.56 (s, 1H, phenyl), 7.48–7.52 (m, 2H, phenyl), 7.34 (d, 1H, J = 5, phenyl), 6.02 (s, 1H, uracil), 4.83 (s, 2H, quinazoline-CH2-uracil), 3.34 (s, 3H, CH3). 13C NMR (125 MHz, CDCl3) δC (ppm): 160.71, 160.44, 151.09, 149.45, 145.55, 145.40, 138.04, 136.37, 133.61, 131.75, 131.22, 129.67, 129.54, 126.83, 123.94, 122.27, 121.23, 102.39, 47.67, 28.48. MS (m/z, %): 551.9 (M+, 10.18), 516.9 (100), 459.9 (24.65), 391.9 (56.95), 312.0 (23.77), 233.1 (30.9), 157.0 (14.68).
1-((6-Bromo-3-(4-Fluorophenyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6f.). IR (KBr, cm−1): 2964 (CH), 2922(CH), 1715(C = O), 1686 (C = O), 1670 (C = O), 1607 (C = N), 1510 (C = C), 1473 (C–N), 1441 (C–O), 847(C–Br). 1H NMR (500 MHz, CDCl3) δH (ppm): 8.35 (s, 1H, H-5-quinazoline), 7.79 (d, 1H, J = 10 Hz, quinazoline), 7.44 (d, 2H, J = 15 Hz, phenyl), 7.24 (s, 2H, quinazoline), 7.08 (d, 2H, J = 10 Hz, phenyl), 5.98 (s, 1H, uracil), 4.81 (s, 2H, quinazoline-CH2-uracil), 3.86 (s, 3H, CH3). 13C NMR (126 MHz, DMSO-d6) δC (ppm): 161.05, 160.92, 160.06, 155.15, 155.11, 151.05, 145.58, 137.83, 131.60, 130.96, 130.87, 129.83, 128.49, 122.68, 120.16, 116.56, 116.33, 105.36, 66.88, 27.91; MS (m/z, %): 490.1 (M+, 10.72), 457.2 (100), 399.1 (81.43), 331.1 (29.89), 223.1 (31.89), 170.0 (16.87).
1-((6-Bromo-3-(4-Chlorophenyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6g). IR (KBr, cm−1) : 2959 (CH), 2922 (CH), 1713 (C = O), 1682 (C = O), 1674 (C = O), 1609 (C = N), 1493 (C = C), 1470 (C–N), 1435 (C–O), 847 (C–Br). 1H NMR 500 MHz, CDCl3) δH (ppm): 8.37 (s, 1H, H-5-quinazoline), 7.83 (d, 1H, J = 5 Hz, H-7-quinazoline), 7.61 (d, 2H, J = 5 Hz, phenyl), 7.49 (d, 1H, J = 5 Hz, H-8-quinazoline), 7.34 (d, 2H, J = 5 Hz, phenyl), 6.03 (s, 1H, uracil), 4.82 (s, 2H, quinazoline-CH2-uracil), 3.34 (s, 3H, CH3). 13C NMR (125 MHz, CDCl3) δC (ppm):160.70, 160.56, 151.10, 149.63, 145.60, 145.40, 137.99, 136.56, 133.64, 130.89, 129.66, 129.54, 129.42, 122.29, 121.17, 102.40, 47.67, 28.47. MS (m/z, %):508.1 (M+, 13.47), 473.1 (100), 415.1 (26.23), 394.1 (0.63), 233.2 (32.79).
1-((6-Bromo-3-(4-Bromophenyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6h).IR (KBr, cm−1): 2959 (CH), 2932 (CH), 1715 (C = O), 1674 (C = O), 1659 (C = O), 1607 (C = N), 1578 (C = C), 1470 (C–N), 1437 (C–O), 833 (C–Br). 1H NMR (500 MHz, CDCl3) δH (ppm): 8.35 (s, 1H, H-5-quinazoline), 7.82 (d, 1H, J = 10.Hz, H-7-quinazoline), 7.76 (d, 2H, J = 10 Hz, phenyl), 7.48 (d, 1H, J = 10 Hz, H-8-quinazoline), 7.27 (d, 2H, J = 10 Hz, phenyl), 6.01 (s, 1H, uracil), 4.81 (s, 2H, quinazoline-CH2-uracil), 3.33 (s, 3H, CH3). 13C NMR (125 MHz, CDCl3) δC (ppm): 160.68, 160.48, 151.08, 149.56, 145.58, 145.38, 137.98, 134.17, 133.87, 129.69, 129.65, 129.52, 124.63, 122.26, 121.16, 102.38, 47.66, 28.46. MS (m/z, %): 551.9 (M+, 13.14), 517.0 (100), 457.0 (42.69), 391.9 (16.34), 314.0 (22.34), 233.1 (23.81), 170.0 (13.71).
1-((6-Bromo-4-Oxo-3-(P-Tolyl)-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6i). IR (KBr, cm−1): 3065–3046 (C–H), 2963–2924 (C–H), 1709 (C = O), 1682 (C = O), 1688 (C = O), 1605 (C = N), 1510 (C = C), 1333 (C–N), 1395 (C–O), 762 (C–Br).1H NMR (500 MHz, CDCl3) δH (ppm): 8.38 (s, 1H, H-5-quinazoline), 7.81 (d, 1H, J = 5 Hz, H-7-quinazoline), 7.47 (d,1H, J = 5 Hz,H-8-quinazoline), 7.41 (d, 2H, J = 5 Hz, phenyl), 7.24 (d, 2H, J = 5 Hz, phenyl), 6.01 (s, 1H, uracil), 4.83 (s, 2H, quinazoline-CH2-uracil), 3.34 (s, 3H, CH3), 2.47 (s, 3H, CH3 phenyl). 13C NMR (125 MHz, CDCl3) δC (ppm): 160.78, 160.77, 151.14, 150.19, 145.73, 145.56, 140.56, 137.74, 132.49, 131.23, 129.54, 127.57, 122.49, 120.88, 102.29, 100.76, 47.74, 28.45, 21.32. MS (m/z, %): 488.1 (M + 1+, 15.04), 453.1 (100), 394.1 (22.57), 327.0 (33.98), 233.1 (15.31), 146.1 (26.51), 170.0 (2.53), 75.1 (4.37).
1-((6-Bromo-3-(4-Methoxyphenyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6j). IR (KBr, cm−1): 3011 (CH), 2957(CH), 1704 (C = O), 1682 (C = O), 1668 (C = O), 1611 (C = N), 1437 (C = C), 1431(C–N), 1422 (C–O), 837 (C–Br) .1H NMR (500 MHz, CDCl3) δH (ppm): 8.35 (s, 1H, H-5-quinazoline), 7.78 (dd, 1H, J = 10 Hz, J = 5Hz, H-7-quinazoline), 7.44 (d, 1H, J = 15 Hz, H-8-quinazoline), 7.47 (d, 1H, J = 5 Hz, phenyl), 7.08 (d, 1H, J = 10 Hz, phenyl), 5.98 (s, 1H, uracil), 4.81 (s, 2H, quinazoline-CH2-uracil), 3.86 (s, 3H, CH3), 3.31 (s, 3H, OCH3). 13C NMR (125 MHz, CDCl3) δC (ppm): 160.99, 160.83, 160.73, 151.19, 150.46, 145.75, 145.59, 137.78, 129.64, 129.58, 128.98, 127.47, 122.50, 120.91, 115.82, 102.34, 55.68, 47.78, 28.49. MS (m/z, %): 504.0 (M + 1+, 19.33), 469.0 (100), 411.0 (26.62), 343.0 (32.78), 249.1 (12.35), 221.0 (9.19).
1-((6-Bromo-4-Oxo-3-(4-Phenoxyphenyl)-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6k). IR (KBr, cm−1): 2963 (CH), 2826 (CH), 1719 (C = O), 1684 (C = O), 1676 (C = O), 1609 (C = N), 1504 (C = C), 1487 (C–N), 1437 (C–O), 847 (C–Br). 1H NMR (500 MHz, CDCl3) δH (ppm): 8.38 (s, 1H, H-5-quinazoline), 7.82 (d, 1H, J = 10 Hz, H-7-quinazoline), 7.48 (d, 1H, J = 10 Hz, H-8-quinazoline), 7.40–7.43 (m, 2H, phenyl), 7.30 (d, 2H, J = 10 Hz, phenyl), 7.20–7.23 (m, 1H, J = 5 Hz, phenyl), 7.17 (d, 2H, J = 5 Hz, phenyl), 7.12 (d, 2H, J = 5 Hz, phenyl), 6.02 (s, 1H, uracil), 4.86 (s, 2H, quinazoline-CH2-uracil), 3.34 (s, 3H, CH3). 13C NMR (125 MHz, CDCl3) δC (ppm): 160.85, 160.77, 159.34, 155.53, 150.22, 145.70, 145.53, 137.84, 130.14, 129.63, 129.56, 129.30, 129.21, 124.71, 122.45, 120.99, 120.20, 119.36, 102.35, 47.77, 28.47. MS (m/z, %): 566.2 (M+, 14.06), 530.2 (100), 473.2 (98.51), 392.2 (1264), 312.1 (18.20).
1-((6-Bromo-3-(2,4-Dimethoxyphenyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6l). IR (KBr, cm−1): 2940 (CH), 2841 (CH), 1705 (C = O), 1695 (C = O), 1624 (C = O), 1605 (C = N), 1587 (C = C), 1327(C–N), 1418 (C–O), 837 (C–Br). 1H NMR (500 MHz, CDCl3) δH (ppm): 8.41 (s, 1H, H-5-quinazoline), 7.84 (d, 1H, J = 10 Hz, H-7-quinazoline), 7.52 (d, 1H, J = 10 H-8-quinazoline), 7.16 (d, 1H, J = 5 Hz, phenyl), 6.61 (s, 1H, phenyl), 6.58 (d, 1H, J = 5 Hz, phenyl), 6.14 (s, 1H, uracil), 5.24 (d, 1H, J = 10 Hz, quinazoline-CH2-uracil), 5.08 (d, 1H, J = 15 Hz, quinazoline-CH2-uracil2), 3.85 (s, 3H, OCH3), 3.81 (s, 3H, OCH3), 3.45 (s, 3H, CH3). 13C NMR (126 MHz, CDCl3) δC (ppm): 162.23, 161.93, 160.58, 156.40, 155.47, 155.35, 151.27, 145.82, 137.71, 129.95, 129.77, 129.37, 122.83, 121.02, 116.00, 106.36, 105.30, 99.94, 66.40, 55.93, 55.67, 28.16. MS (m/z, %): 534.0 (M + 1+, 14.08), 348 (100), 374.0 (39.64), 359.0 (44.98), 343.0 (100), 315.0 (11.25), 220.1 (5.85), 153.1 (19.18).
1-((6-Bromo-3-(3,4-Dimethylphenyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6m). IR (KBr, cm−1): 2965–2918 (C–H), 2858 (C–H), 1713 (C = O), 1686 (C = O), 1672 (C = O), 1602 (C = N), 1503 (C = C), 1275 (C–N), 1395 (C–O), 762 (C–Br, stretch). 1H NMR (500 MHz, CDCl3) δH (ppm): 8.38 (s, 1H, H-5-quinazoline), 7.80 (d,1H, J = 5 Hz, H-7-quinazoline), 7.47 (d, 1H, J = 10 Hz, phenyl), 7.36 (d, 1H, J = 10 Hz, H-8-quinazoline), 7.12 (s, 1H, phenyl), 7.08 (d, 1H, J = 10 Hz , phenyl), 6.00 (s, 1H, uracil), 4.85(s, 2H, quinazoline-CH2-uracil), 3.34 (s, 3H, CH3), 2.34 (s, 3H, CH3-phenyl), 2.35 (s, 3H, CH3-phenyl). 13C NMR (125 MHz, CDCl3) δC (ppm): 160.81, 151.17, 150.26, 145.76, 145.60, 139.42, 139.26, 137.70, 132.67, 131.61, 129.59, 129.55, 128.50, 124.89, 122.52, 120.83, 102.29, 47.79, 29.69, 28.47, 20.00, 19.65. MS (m/z, %):502.1 (M + 1+, 14.43), 467.1 (100), 409.1 (20.61), 366.0 (9.48), 341.1 (40.57), 247.1 (16.68), 221.0 (4.81), 116.1 (17.35).
1-((6-Bromo-3-(3-Chloro-4-Fluorophenyl)-4-Oxo-3,4-Dihydroquinazolin-2-Yl)Methyl)-6-Chloro-3-Methylpyrimidine-2,4(1H,3H)-Dione (6n). IR (KBr, cm−1): 3071–3038 (C–H), 2959 (C–H), 1713 (C = O), 1678 (C = O), 1682 (C = O), 1611 (C = N), 1503 (C = C), 1267 (C–N), 1395 (C–O), 835 (C–Br, stretch). 1H NMR (500 MHz, CDCl3) δH (ppm): 8.29 (d, 1H, J = 5, H-5-quinazoline), 7.76 (dd, 1H, J = 10 Hz, J = 5 Hz, H-7-quinazoline), 7.41–743 (m, 2H, phenyl), 7.34 (t, 1H, J = 10 Hz, H-8-quinazoline) 7.22–7.26 (m, 1H, phenyl), 5.96 (s, 1H, uracil), 4.77 (s, 2H, quinazoline-CH2-uracil), 3.27 (s, 3H, CH3). 13C NMR (125 MHz, CDCl3) δC (ppm): 160.70, 157.82, 151.12, 149.42, 145.51, 145.35, 138.20, 131.54, 130.67, 129.74, 128.26, 123.56, 123.37, 122.18, 121.38, 118.63, 118.41, 102.48, 47.64, 28.51. MS (m/z, %): 526.0 (M+, 14.08), 491.0 (100), 434.0 (26.32), 366.0 (27.78), 251.1 (28.00), 223.0 (20.49), 221.0 (4.81), 116.1 (10.24).
Cell culture assay
Three human cancer cell lines with origin of breast (MCF-7), lung (A549), and colorectal cancer (SW-480) were obtained from the National Cell Bank, Pasteur Institute, Tehran, Iran. The cells were cultured in 1640 RPMI medium (Bio Idea, Iran), 10% fetal bovine serum (FBS; Gibco, USA) and 1% penicillin–streptomycin (Biosera, France) and were maintained at 37 °C in a 5% CO2 incubator. The cell lines were separately seeded in 96-well plates at a density of 8000 cells/well and were kept for 24 h to reattach. The cells were then treated with 6 different concentrations of the synthesized compounds and further incubated for 72 h. Then, the media were removed and replaced with 100 μL of MTT solution (0.5 mg/mL) was added and incubated for another 4 h. To dissolve the purple formazan crystals, 100 μL dimethyl sulfoxide (DMSO) (Sigma-Aldrich, Germany) was added to each well and left for 30 min. Finally, the absorbance of each well was obtained at 570 nm using an ELISA plate reader (Biotek, Winooski, VT, USA). To obtain inhibitory index of each compound, first the cell viability was normalized to the untreated control and then, a plot of inhibition index versus concentration was depicted. Curve Expert software version 1.40 was used to calculate the inhibitory concentration 50 (IC50) values.
Apoptosis analysis
The AnnexinV/PI detection kit was applied to assess the ability of compound 6n with the lowest IC50 in the induction of apoptosis in A549 cell line. To do this, 1 × 105 cells in 500 μL complete culture media were cultured for overnight followed by exposure to compound 6n for 72 h. After the incubation time, AnnexinV/PI staining using eBioscience™ Annexin V apoptosis detection kit (Invitrogen). The cells were gently harvested and washed once with 1X phosphate buffered saline (PBS), and once with 1000 μL 1X binding buffer. In the next step, the cells were suspended in 100 μL binding buffer containing 5 μL fluorescein isothiocyanate conjugated (FITC) Annexin V for 15 min and 5 μL Propodium Iodide (PI) solution. The cells were then acquired on BD FACS Calibur™ flow cytometry (BD Biosciences, San Jose, CA, USA) and apoptosis rates were calculated as the sum of early apoptosis and late apoptosis using Flowjo software.
Cell cycle analysis
The effect of 6n was also assessed according to our previous protocol23. Briefly, 1 × 105 A549 cells were seeded in a 24-well plate in 500 mL complete cell culture media and after 24 h, were treated with two concentrations of 6n (10 and 15 μM). After 72 h, the cells were harvested and washed twice with cold 1X pbs and stabilized with cold ethanol (70%) in drop-wised manner while vortexing. After keeping one day in the freezer, the fixed cells were washed with 1X pbs and treated with a mixture of ribonuclease (10 µg/ml) (Sigma-Aldrich, Germany) to remove RNA, and Propidium Iodide (PI, 20 µg/ml) (Sigma-Aldrich, Germany) to stain DNA. The cells were acquired on BD FACS Calibur™ flow cytometry (BD Biosciences, San Jose, CA, USA). The data were analyzed with FlowJo software packages version 10.
Statistical analysis
For each analysis, the data were presented as the mean SD. One-way ANOVA statistical analyses were carried out using GraphPad Prism 8.0 software (GraphPad Software Inc.).
Docking procedure
Molecular docking procedure was carried out using AutoDock 4.0 software. The 3D EGFR complex structure was obtained from Protein Data Bank (PDB code: 1M17). ACD chem BioDraw Uitra13.0 was used to draw the structures, then the molecules were minimized by molecular mechanics (MM+) and semi-empirical (AM1) method utilizing hyperchem software35. In order to prepare the protein, protein's unwanted water and the cognate ligand were removed and the missing hydrogen atoms were added then merged nonpolar hydrogens based on their carbon atoms. AutoDock Vina in conjunction with a batch script (DOCKFACE) was used to dock a grid box with a size of 70*70*70. Then the centroid of x = 20.143, y = 0.376, z = 52.210 and docking parameters were set with the default settings. Discovery studio 2021 software was used to analyze binding interactions between docked compounds and receptors. Lastly, Discovery studio 2021 client software was used to generate the pictures.
Molecular dynamics simulations
To generate full Amber starting files including the AMBERff14SB force field, the Ambertools package was used 36 for the protein, and the General Amber Force Field (GAFF)37 employing the AM1-BCC charge model was used for ligands38. The protonation states of all the titratable residues were set at their default values at pH 7.0 and the N- and C-termini of the receptor was acetylated and aminated, respectively. The TIP3P water model was presented to solvate the compounds and ions added to the box to accomplish a neutral system39. An electrolyte concentration of 150 mM KCl was added to the system to mimic physiological salt concentrations. The total size for each system was approximately 63,000 atoms.
Simulation setup protocol
All MD simulations were performed using the GPU version of the PMEMD in the Amber22 software package40. The periodic boundary condition (PBC) was used in three dimensions. The sharpest descent algorithm was utilized to minimalize their energy before the MD simulation of protein − ligand complexes, and the leap-frog algorithm was accounted for to assimilate their motions. Similar to previous research, the Particle Mesh Ewald (PME) method was used within this procedure to perceive the influence of long-range molecules' electrostatic interfaces41. In addition, the LINCS algorithm was surveyed in both equilibration and production runs to mimic the constraining of the H-bonds in this procedure42. The nonbonded interaction cutoff was set at 12.0 Å. We used ensemble-based ESMACS (Enhanced sampling of molecular dynamics with the approximation of continuum solvent) protocol43,44,45. This procedure is based on the setting of independent MD simulations to achieve a good estimation for binding free energy and associated uncertainties46,47,48. In this regard, a set of 25 replicas for ESMACS calculation, with 4 ns production runs, were established in this study. For each replica, the energy system was minimized using a steepest descent algorithm for 200 steps followed by 7000 steps of conjugated gradient. This was followed by 10 ns of equilibration simulations in which the protein was restrained using a harmonic potential on all heavy protein atoms. These restraints were gradually lowered over 5 consecutive 2 ns simulations, employing force constants of 1000, 500, 200, 100, and 50 kJ mol−1 nm−2 and hearing each replica throughout 2 ns to 300 K during only the first two steps of equilibration procedure. The same initial coordinates were used for a given ligand-receptor complex, with different initial velocities randomly assigned to the atoms according to a Maxwell–Boltzmann distribution.
Free energy calculations
Free energy calculation was obtained by the ESMACS approach and the binding free energy (ΔG) of every trivial molecule as an inhibitor was achieved with Eq. 1.
Com, Rec, and Lig subscripts are associated with the complex, receptor, and ligand, ESMACS approach is based on this equation, in which Gi is calculated from a set of structures from MD simulations. The molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) and generalized Born Surface Area (MM/GBSA) as continuum solvation was applied for the binding free energy calculations of selected compounds to EGFR receptor. For ESMACS, the trajectories for each replica containing 200 snapshots were further analyzed by MMPBSA.py.MPI to extract the energetic information for each snapshot. A script was then run to aggregate these results from the ensemble of simulations and values of ΔGESMACS computed along with bootstrap statistics (Figure S1–S44).
Hydrogen bonding, energy decomposition and clustering analysis
We used the H-bond module of AmberTools22 to consider a hydrogen bond formed between the chosen ligands and the protein (distance of 3.0 Å and an angle cutoff of 135). Therefore, all frames from all replicas has been used for this purpose. The residue energy decomposition reveals a congenial and displeasing data interface that might be used to improve lead quality. We evaluated the Water Swap residue-wise binding energy decompositions for the residues that made substantial influences on inhibitor binding during the MD simulation49. The most conventional and well-known similarity measure is the root mean square deviation (RMSD) values applied for partitioning MD trajectories that are gained by pairwise or matrix error distances50.
Data availability
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request. We have presented all data in the form of Tables and Figure. The PDB code (1M17) was retrieved from protein data bank (www.rcsb.org). https://www.rcsb.org/structure/1M17.
References
Almansour, A. I. et al. Design, synthesis and antiproliferative activity of decarbonyl luotonin analogues. Eur. J. Med. Chem. 138, 932–941 (2017).
Bieberich, A. A. et al. Optimization of the 4-anilinoquin (az) oline scaffold as epidermal growth factor receptor (EGFR) inhibitors for chordoma utilizing a toxicology profiling assay platform. Sci. Rep. 12, 12820 (2022).
Binkhathlan, Z. & Lavasanifar, A. P-glycoprotein inhibition as a therapeutic approach for overcoming multidrug resistance in cancer: Current status and future perspectives. Curr. Cancer Drug Targets 13, 326–346 (2013).
Kostova, I. Ruthenium complexes as anticancer agents. Curr. Med. Chem. 13, 1085–1107 (2006).
Hosseinzadeh, Z., Ramazani, A. & Razzaghi-Asl, N. Anti-cancer nitrogen-containing heterocyclic compounds. Curr. Org. Chem. 22, 2256–2279 (2018).
Hameed, A. et al. Quinazoline and quinazolinone as important medicinal scaffolds: a comparative patent review (2011–2016). Exp. Opin. Ther. Pat. 28, 281–297 (2018).
Karan, R., Agarwal, P., Sinha, M. & Mahato, N. Recent advances on quinazoline derivatives: A potential bioactive scaffold in medicinal chemistry. Chem. Eng. 5, 73 (2021).
He, D. et al. Pharmaceutical prospects of naturally occurring quinazolinone and its derivatives. Fitoterapia 119, 136–149 (2017).
Auti, P. S., George, G. & Paul, A. T. Recent advances in the pharmacological diversification of quinazoline/quinazolinone hybrids. RSC Adv. 10, 41353–41392 (2020).
Emami, L., Sabet, R., Khabnadideh, S., Faghih, Z. & Thayori, P. Quinazoline analogues as cytotoxic agents; QSAR, docking, and in silico studies. Res. Pharm. Sci. 16, 528 (2021).
Divar, M., Zomorodian, K., Bastan, S., Yazdanpanah, S. & Khabnadideh, S. Synthesis of some quinazolinone derivatives using magnetic nanoparticles-supported tungstic acid as antimicrobial agents. J. Iran. Chem. Soc. 15, 1457–1466 (2018).
Faghih, Z. et al. Synthesis of some novel dibromo-2-arylquinazolinone derivatives as cytotoxic agents. Res. Pharm. Sci. 14, 115 (2019).
Ismail, R. S., Ismail, N. S., Abuserii, S. & Abou El Ella, D. A. Recent advances in 4-aminoquinazoline based scaffold derivatives targeting EGFR kinases as anticancer agents. Fut. J. Pharm. Sci. 2, 9–19 (2016).
Eldehna, W. M. et al. Design, synthesis, in vitro biological assessment and molecular modeling insights for novel 3-(naphthalen-1-yl)-4, 5-dihydropyrazoles as anticancer agents with potential EGFR inhibitory activity. Sci. Rep. 12, 12821 (2022).
Mass, R. D. The HER receptor family: A rich target for therapeutic development. Int. J. Radiat. Oncol. Biol. Phys. 58, 932–940 (2004).
Baselga, J. Why the epidermal growth factor receptor? The rationale for cancer therapy. Oncologist 7, 2–8 (2002).
Greig, S. L. Osimertinib: First global approval. Drugs 76, 263–273 (2016).
Piotrowska, Z. et al. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third-generation EGFR inhibitor. Cancer Discov. 5, 713–722 (2015).
Kim, D.-W. et al. Safety, tolerability, and anti-tumor activity of olmutinib in non-small cell lung cancer with T790M mutation: A single arm, open label, phase 1/2 trial. Lung Cancer 135, 66–72 (2019).
Zu, G., Chen, J., Song, B. & Hu, D. Synthesis, anti-tomato spotted wilt virus activities, and interaction mechanisms of novel dithioacetal derivatives containing a 4 (3 H)-Quinazolinone pyrimidine ring. J. Agric. Food Chem. 69, 14459–14466 (2021).
Xu, Z., Zhao, S.-J. & Liu, Y. 1, 2, 3-Triazole-containing hybrids as potential anticancer agents: Current developments, action mechanisms and structure-activity relationships. Eur. J. Med. Chem. 183, 111700 (2019).
Misra, A. et al. Synthesis, biological evaluation and molecular docking of pyrimidine and quinazoline derivatives of 1, 5-benzodiazepine as potential anticancer agents. J. King Saud Univ. Sci. 32, 1486–1495 (2020).
Emami, L. et al. Design, synthesis, molecular simulation, and biological activities of novel quinazolinone-pyrimidine hybrid derivatives as dipeptidyl peptidase-4 inhibitors and anticancer agents. New J. Chem. 44, 19515–19531 (2020).
Moasser, M. M. Targeting the function of the HER2 oncogene in human cancer therapeutics. Oncogene 26, 6577–6592 (2007).
Geyer, C. E. et al. Lapatinib plus capecitabine for HER2-positive advanced breast cancer. N. Engl. J. Med. 355, 2733–2743 (2006).
Ayati, A. et al. A review on progression of epidermal growth factor receptor (EGFR) inhibitors as an efficient approach in cancer targeted therapy. Bioorg. Chem. 99, 103811 (2020).
Barker, A. J. et al. Studies leading to the identification of ZD1839 (Iressa™): An orally active, selective epidermal growth factor receptor tyrosine kinase inhibitor targeted to the treatment of cancer. Bioorg. Med. Chem. Lett. 11, 1911–1914 (2001).
Rosell, R. et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 13, 239–246 (2012).
Wedge, S. R. et al. ZD6474 inhibits vascular endothelial growth factor signaling, angiogenesis, and tumor growth following oral administration. Can. Res. 62, 4645–4655 (2002).
Planchard, D. et al. Metastatic non-small cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 29, 192–237 (2018).
Zare, S., Ramezani, Z., Ghadiri, A. A. & Fereidoonnezhad, M. Synthesis of N-(2-(tert-Butylamino)-2-oxoethyl)-2, 2-dichloro-N-aryl (alkyl) acetamides as anticancer agents: molecular modeling and biological evaluations. Chem. Select 8, e202203931 (2023).
Khanal, P. et al. Integration of system biology tools to investigate huperzine A as an anti-Alzheimer agent. Front. Pharmacol. 56, 3362 (2021).
Sargazi, S. et al. 8-Alkylmercaptocaffeine derivatives: Antioxidant, molecular docking, and in-vitro cytotoxicity studies. Bioorg. Chem. 111, 104900 (2021).
Zargari, F. et al. Study of tyramine-binding mechanism and insecticidal activity of oil extracted from Eucalyptus against Sitophilus oryzae. Front. Chem. 10, 15–123 (2022).
Zare, S., Fereidoonnezhad, M., Afshar, D. & Ramezani, Z. A comparative QSAR analysis and molecular docking studies of phenyl piperidine derivatives as potent dual NK1R antagonists/serotonin transporter (SERT) inhibitors. Comput. Biol. Chem. 67, 22–37 (2017).
Alessandrini, S., Gauss, J. R. & Puzzarini, C. Accuracy of rotational parameters predicted by high-level quantum-chemical calculations: Case study of sulfur-containing molecules of astrochemical interest. J. Chem. Theory Comput. 14, 5360–5371 (2018).
Wang, W., Wolf, R. J., Caldwell, J. W. & Kollman, P. A. Case. DA J. Comput. Chem. 25, 92 (2004).
Jakalian, A., Bush, B. L., Jack, D. B. & Bayly, C. I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: I method. J. Comput. Chem. 21, 132–146 (2000).
Mark, P. & Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 105, 9954–9960 (2001).
Case, D. A. et al. AMBER 22 Reference Manual. (2022).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Hess, B., Bekker, H., Berendsen, H. J. & Fraaije, J. G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Coveney, P. V. & Wan, S. On the calculation of equilibrium thermodynamic properties from molecular dynamics. Phys. Chem. Chem. Phys. 18, 30236–30240 (2016).
Knapp, B., Ospina, L. & Deane, C. M. Avoiding false positive conclusions in molecular simulation: The importance of replicas. J. Chem. Theory Comput. 14, 6127–6138 (2018).
Wan, S., Sinclair, R. C. & Coveney, P. V. Uncertainty quantification in classical molecular dynamics. Phil. Trans. R. Soc. A 379, 20200082 (2021).
Wan, S., Knapp, B., Wright, D. W., Deane, C. M. & Coveney, P. V. Rapid, precise, and reproducible prediction of peptide–MHC binding affinities from molecular dynamics that correlate well with experiment. J. Chem. Theory Comput. 11, 3346–3356 (2015).
Bhati, A. P., Wan, S., Wright, D. W. & Coveney, P. V. Rapid, accurate, precise, and reliable relative free energy prediction using ensemble based thermodynamic integration. J. Chem. Theory Comput. 13, 210–222 (2017).
Wan, S. et al. Rapid and reliable binding affinity prediction of bromodomain inhibitors: A computational study. J. Chem. Theory Comput. 13, 784–795 (2017).
Kiani, Y. S., Ranaghan, K. E., Jabeen, I. & Mulholland, A. J. Molecular dynamics simulation framework to probe the binding hypothesis of CYP3A4 inhibitors. Int. J. Mol. Sci. 20(18), 4468 (2019).
Paris, R. D., Quevedo, C. V., Ruiz, D. D., Souza, O. N. D. & Barros, R. C. Clustering molecular dynamics trajectories for optimizing docking experiments. Comput. Intell. Neurosci. 2015, 32–32 (2015).
Acknowledgements
Financial assistance from the Shiraz University of Medical Sciences through grant number 98-
01-05-21702 is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
S.Z. synthesized all the compounds, performed the biological assays, ran computational section and prepared the manuscript. L.E. supervised the synthesis process and edit the manuscript. Z.F. supervised the biological assays. F.Z. performed and wrote the Molecular dynamic simulation section. Z.F. designed compounds. S.K. prepared the manuscript and supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zare, S., Emami, L., Faghih, Z. et al. Design, synthesis, computational study and cytotoxic evaluation of some new quinazoline derivatives containing pyrimidine moiety. Sci Rep 13, 14461 (2023). https://doi.org/10.1038/s41598-023-41530-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-41530-6
- Springer Nature Limited
This article is cited by
-
Synthesis, design, biological evaluation, and computational analysis of some novel uracil-azole derivatives as cytotoxic agents
BMC Chemistry (2024)
-
Design and investigation of interactions of novel peptide conjugates of purine and pyrimidine derivatives with EGFR and its mutant T790M/L858R: an in silico and laboratory study
Molecular Diversity (2024)