Abstract
Microclimate ecology is attracting renewed attention because of its fundamental importance in understanding how organisms respond to climate change. Many hot issues can be investigated in desert ecosystems, including the relationship between species distribution and environmental gradients (e.g., elevation, slope, topographic convergence index, and solar insolation). Species Distribution Models (SDMs) can be used to understand these relationships. We used data acquired from the important desert plant Nitraria tangutorum Bobr. communities and desert topographic factors extracted from LiDAR (Light Detection and Ranging) data of one square kilometer in the inner Mongolia region of China to develop SDMs. We evaluated the performance of SDMs developed with a variety of both the parametric and nonparametric algorithms (Bioclimatic Modelling (BIOCLIM), Domain, Mahalanobi, Generalized Linear Model, Generalized Additive Model, Random Forest (RF), and Support Vector Machine). The area under the receiver operating characteristic curve was used to evaluate these algorithms. The SDMs developed with RF showed the best performance based on the area under curve (0.7733). We also produced the Nitraria tangutorum Bobr. distribution maps with the best SDM and suitable habitat area of the Domain model. Based on the suitability map, we conclude that Nitraria tangutorum Bobr. is more suited to southern part with 0–20 degree slopes at an elevation of approximately 1010 m. This is the first attempt of modelling the effects of topographic heterogeneity on the desert species distribution on a small scale. The presented SDMs can have important applications for predicting species distribution and will be useful for preparing conservation and management strategies for desert ecosystems on a small scale.
Similar content being viewed by others
Introduction
The Ulan Buh Desert is the 8th largest desert in China, and is located in the mid-latitudinal zone. It is characterized by a typical continental climate with little precipitation. The desert is located in a transitional zone with a temperate climate ranging from semiarid to arid conditions1. It is one of the driest regions at similar latitudes in the world and has a very sensitive ecological environment, which creates a fragile desert ecotone. The desert ecotone is the key ecoregion and is suitable for researching ecosystem degradation and recovery mechanisms. Desert ecotones may have high ecological significance, which quantitatively reveals the interactions between plants and environmental factors. The spatial distribution pattern of the desert plants is therefore the result of the interactions between the plant communities and the harsh environmental factors2.
Spatial distributions are fundamental to species ecology3 and help identify threats to the conservation of plant species, manage the impacts of biological invasion4, and plan conservation strategies. Climate, edaphic, and topographic factors are the main factors affecting species distributions5. These factors are affected by a variety of biophysical processes including competitive interactions between plant species and their dispersal history6,7. SDMs, like habitat suitability or ecological models, are developed with the observed species occurrence as a response variable and environmental factors as predictor variables8. Many modelling techniques are available to developed SDMs9,10,11. Although previous SDMs studies have compared different modelling techniques for their precision12,13,14,15,16,17, most of them describe species distributions over a large scale, and therefore have low predictive performance in small-localized condition6,18,19,20. Most SDMs are calibrated only with climate data at a low spatial resolution, such as the climate data acquired from WorldClim(https://www.worldclim), with a 1 km2 grid resolution. However, some studies have indicated that the predictive performance of SDMs can be substantially improved by incorporating other environmental factors in addition to climate factors21 or by considering finer-scale processes, such as topo-climatic processes22. Topographic heterogeneity is the most important factor affecting the spatial distribution of desert plants23. In the mountainous regions, topography may precisely describe local temperature, light, and humidity, Topography significantly affects light and water availability and soil development. Furthermore, topography affects species composition, structure, appearance, and dynamics of the plant communities24,25,26.
Building on the previous work done in this field, the current study intends to answer a few important questions including ‘What distribution patterns are followed by the desert plant species?’ and ‘Which topography factors play a major role in affecting the species distribution?’ Intense solar radiation, high evapotranspiration, extreme temperatures, degraded soil and vegetation cover, low atmospheric moisture, and harsh topographic features are important factors in the desert environment. The desert is extremely arid with < 60–100 mm mean annual precipitation, and therefore, its moisture regime is the most critical factor for regulating biological processes and species distribution. The rainfall spatially varies at different scales, not only at the regional scale ranging from 0.1 to 10 km, but also at a local scale of few meters. Rainfall is the main source of moisture for desert plants, which triggers species' survival and growth27. Many studies have explored the appropriate methods for understanding the relationships between vegetation dynamics, soil moisture, and temperature in the desert ecosystem28,29,30. However, under similar climate and desert environmental conditions, the influence of topography on species distribution has rarely been investigated in a small scale.
Additionally, several studies have been conducted on the topographic factors affecting animal communities in the desert. In recent decades, some studies have been conducted to model the environmental variables influencing kit fox distribution. For example, Maxent fixed-effects and generalized linear mixed-effects models have been developed to describe kit fox distribution30. These modelling studies show the large effects of elevation on kit fox survival and growth, followed by that of the slope and canopy height of the vegetation at the landscape scale. The Maxent algorithm was used to predict the habitat in the Mojave desert and parts of the Sonoran desert considering the geographical and topographical factors at the spatial grid of one square kilometer31. The field-validated site—and landscape-level SDMs were developed to identify the potentially rare and endemic plant habitats in the Great Basin of Western North America32, in which the potential habitat combined with the elevation, slope, aspect, rock type, and geologic processes are mapped with the resolution of 100 square meters. Similar to climatic factors, non-climate predictors (e.g., topography and habitat) are also important at a finer scale33. Thus, the analysis of these factors on a finer scale could be the most appropriate for developing SDMs and increasing their predictive accuracy34. As mentioned above, topography is one of the most important predictors of SDMs. It controls the habitat structure, biota, and all growing conditions locally35, especially in the desert, where topographic features are more unfavorable than other land features. Thus, a high-resolution topographic map is necessary to develop SDMs.
For dominant species, such as Nitraria tangutorum Bobr. in the desert, a small geographic range and narrow habitat may lead to acute habitat limitation, which is strongly correlated with other environmental factors. The microclimate, which influences ecosystem dynamics and processes, is often disregarded in ecology and evolution36. It is meaningful to investigate the geographical variations of Nitraria tangutorum Bobr. species in the desert on a small scale, such as within the scale of 100 m, where topographic factors significantly affect species distribution. Existing studies have shown that geographic and topographic predictors can substantially improve the fitting performance of SDMs37.
High-resolution terrain data are rarely available. Remote sensing technology, which offers alternative solutions to various data sets, can supply accurate high-resolution terrain data using the light detection and ranging (LiDAR) technology. Remote sensing data is important for determining species distribution38,39. In recent years, LiDAR datasets have become useful for modelling biodiversity and habitat analyses. For example, LiDAR data could improve the prediction of composition and changes in the plant communities40,41, improve the descriptions of animal behaviors and their spatial distributions42, and enhance the species richness prediction17.
In this study, we developed SDMs for revealing the relationships between desert plants and topographic factors affecting species distributions. Topographic variables used in the SDMs were derived from LiDAR data (resolution: 0.15 m). The SDMs could be used to predict the precise distribution of species covering an area of one square kilometer with a resolution of 10 m. Different modelling algorithms proposed in this study were evaluated based on the dataset obtained from 67 Nitraria tanguotrum species in the Ulan Buh Desert, an arid land in northern China. The objectives of this study were to: (1) compare various modelling algorithms including parametric and non-parametric modeling based on statistical indexes, and determine the best to develop SDMs; (2) evaluate the contribution of topographic factors to the precise prediction of distribution of the plant species of interest in the desert; (3) determine the effects of topographic heterogeneity on the distribution of Nitraria tangutorum Bobr; and (4) present the habitat suitability map for Nitraria tangutorum Bobr. and show the corresponding management implications of the presented SDMs.
Results
Model performances
Random sampling and each of the seven models were run ten times, and the Area Under Curve (AUC), true positive rate (TPR), and false positive rate (TNR) were calculated. With the presence of 67 and the absence of 100, the results indicated the AUC varied from low (0.59) to high (0.81), indicating different qualities of the prediction SDMs of Nitraria tangutorum Bobr. (Fig. 1). The RF produced the highest AUC value, and the Domain had the lowest AUC value (Table 1). All the model performances were fair and satisfactory, except for BIOCLIM and Domain. RF produced the largest AUC, but not with maximum TPR and TNR; TPR and TNR of RF were the third lowest of seven models. Mahalanobis produced the highest TPR + TNR value.
The boxplots of different models (BIOCLIM: classic climate envelope model; Domain computes the Gower distance between environmental variables and locations of occurrence; Mahalanobis model based on the mahalanobis distance; GLM: Generalized Linear Model; GAM: Generalized Additive Model; RF: Random Forest; SVM: Support Vector Machine).
For the profile models, Mahalanobis produced the highest AUC, while AUC for Domain, which was the lowest, and AUC for BIOCLIM were more stable (Table 1). For GLM and GAM, the AUC values were almost identical. For the machine learning models, AUC for RF was higher than that for SVM, and the same was true for max TPR + TNR. RF proved to be better than SVM in modelling the distribution of Nitraria tangutorum Bobr. in the desert.
Habitat suitability and prediction maps for Nitraria tangutorum Bobr. distribution
We produced the maps (Sup. Figures 1–7) using R 3.4.3 and ARCGIS 10.5, which show the habitat suitability for Nitraria tangutorum Bobr. The potentially suitable habitat distribution maps of Nitraria tangutorum Bobr. in the study area were obtained from different algorithms. Furthermore, the corresponding species' presence/absence with each algorithm was mapped. The Nitraria tangutorum Bobr. species distributed mainly in the eastern part of the study area. The spatial distributions of Nitraria tangutorum Bobr. simulated by the different models were significantly different. Concerning the most suitable area for Nitraria tangutorum Bobr, the most suitable habitat area simulated by the Domain model was the largest, and the most suitable habitat area simulated by BIOCLIM model was the smallest. The suitable habitat areas simulated by GLM and GAM models were identical; the suitable habitat simulated by GLM and GAM was the largest, while that simulated by BIOCLIM was the smallest.
The driven factors of topography on the Nitraria tangutorum Bobr. distribution
The responses of the driven factor- topography with different models were compared for Nitraria tangutorum Bobr. Different models delineated the topography characteristics of the potential distribution areas. With BIOCLIM, the existence probability followed a trend of increase at first and then decreasing trend, and aspect of the south part was on the top. For DEM, the existence probability trend of the species was steeper, reaching a peak at an altitude of approximately 1010 m. For the profile factor, the species Nitraria tangutorum Bobr. appeared more suitable at the predicted value of zero, and was also suitable for slope of 0°–20°. With the Domain model, the existence probability trend of the species was similar to that of the BIOCLIM model, but it seemed steeper. In the Malanhanobis model, at the aspect of the north by east 10°, the species Nitraria tangutorum Bobr. exhibited minimum fitness, and at the altitude range of 1007–1013 m, exhibited maximum fitness. Regarding the topography factor of the profile, there were two crests at the predicted value near zero. With the GLM and GAM models, the existence probability trend of the species was nearly the same as the linear trend line, but in the RF, the trend was nonlinear. Regarding the factor of aspect, predicted value fluctuated between 0.4 and 0.8, but for DEM factor, predicted value was the highest at an altitude of approximately 1010 m. With the factors of profile and slope, at the degree of zero, predicted value reached the top, at a slope of 20°–60°, exhibiting a rising tendency.
Analysis of the maps showed the distribution of the species Nitraria tangutorum Bobr. was more suitable in the south, with an altitude of approximately 1010 m, from a slope between 0° and 20°, and general trend was the same with different models. The GLM or GAM was found inadequate for the distribution modelling of Nitraria tangutorum Bobr.
Discussion
Importance of LiDAR data and its ecological implications with selected predictor variables
How does climate change affect organisms? In the past, our limited ability to map and monitor the microclimatic variations using modelling through the incorporation of the microclimate. The microclimate sensors set in the networks provided point-based approaches and weather stations provided macroclimate data. However, means of interpolating and downscaling data were lacking43. LiDAR is valuable for mapping microclimates, as it provides spatially continuous and sub-meter-scale information about ground topography44. Aerial photography provides an approach for assessing topography. However, these are less accurate than LiDAR at deriving terrain elevation45. An important advantage of UAVs is that they are flexible, enabling the collection of time series of aerial imagery over a period at a reasonable cost. In our study, LiDAR data were used to obtain DEM for SDMs, thereby supporting more effective conservation monitoring, management, and policy decision for a sustainable future46.
Our results showed that Nitraria tangutorum Bobr. species are specialists in the range of their habitat resources. In other words, Nitraria tangutorum Bobr. has a relatively narrow ecological niche.
It is limited to making the distribution by considering only topographic variables, the role of habitat heterogeneity in the desert ecosystem may be underestimated. If other environmental factors, such as light, soil, and water are considered, niche theory may be able to better explain the existence mechanism of species. Therefore, future studies need to consider more bio-variables to simulate the distribution of the species, and a permanent plot should be designed to contain the entire spatial gradient of the microclimate conditions in the study system. Long-term microclimate data series are required to complement the dynamics of the microclimate around the species. Gathering georeferenced microclimate data from different habitat types across the globe would significantly promote progress in microclimate ecology47. UAVs equipped with multispectral or hyperspectral sensors and laser scanner systems are very promising, providing high-resolution data in the context of microhabitats, vegetation structures, and topography48.
Suggestion for improvement of SDMs
In our study, not only AUC of the models but also the responses of the variables with the models were different. The parametric regression-based models (GLM and GAM) had lower mean performance, whereas nonparametric or machine learning-based models (RF and SVM) were more accurate. GLMs are widely used in SDM studies49, but have poor nonlinear response performance. At the same time, the results confirmed the effectiveness and robustness of the machine-learning techniques. Similarly, Marmion et al. found a single algorithm with lower accuracy than RF in predicting the plant distributions50. The parameters of the model affect its accuracy; for example, the performance of RF depends on two important parameters, ntree and mty. As for the ntree, the optimal value using only the default may not be the best; therefore, future research should test and adjust the model parameters to the optimum state. In this study, various parametric and nonparametric algorithms were evaluated for SDMs. In the next step, semi-parametric models51 shall be used for better results.
A stable and reliable mathematical algorithm is required to develop more accurate SDMs. In previous studies of community-level predictions of the species distributions, the choice of how species and site-level occurrence probabilities are combined into species assemblage predictions is more important than the choice of the model type used52. Following research shall use ensemble models to predict species distribution. As many researchers have expressed, no evidence has proven that one model is steadily accurate, but it is proven to be better than other models.
The number of environmental factors that accurately represent the habitat characteristics of a species was sufficient SDM has some limitations, for example, over-prediction of species richness per site and sensitivity to methodological biases. In addition, sample size, sample prevalence, sample design, model techniques, imperfect detection of species, or the choice of environmental variables could affect the uncertainty of the predictions53.
The relationships between species occurrence and sets of predictor variables explored by the models produce two kinds of useful outputs: estimates of the probability that species might occur at a given unrecorded location50, and estimates of an area’s suitability for species 16. Climate determines the distribution of desert species54. Previous research has shown that the roots of desert plant species develop and depend on the underwater55. As the study area was only one square kilometer, the variable of climate change could not be obvious, and species distribution could have been disturbed by the level of underground water. However, further studies are necessary in this regard. It is well known that biotic interactions play an important role in creating accurate SDMs for many species, particularly at small geographic scales, such as host requirements and competition6,56. In this study, the model did not include the interaction between species and the influence of predators or human activities. More efforts should be dedicated to include fine-scale environmental measures, such as micro-temperature, underwater, moisture, and rainfall, for fine-scale species mapping and management.
Management implication of SDMs
Species conservation and management are key to maintaining regional ecological balance, especially in ecologically fragile areas such as the Ulan Buh Desert. This study could distinguish the habitat requirements of Nitraria tangutorum Bobr., especially for the topography, to provide a reference for protection. Nitraria tangutorum Bobr. was found to be distributed mainly in the eastern part of the study area. This finding is consistent with the results of the actual survey. The suitable habitat area is larger than the real area, which indicates that the area of Nitraria tangutorum Bobr. is more widely distributed under current environmental conditions. Some of the suitable habitat characteristics shown in our study were consistent with those of other studies57. For example, the population size of Nitraria tangutorum Bobr. increases with sand depth, making it suitable for dunes. To expand the area of Nitraria tangutorum Bobr., the land should be dune, and will be a good choice at an altitude of approximately 1010 m, better to face the aspect of the south part, within the slope of 20°. Because of this characteristic, Nitraria tangutorum Bobr. can prevent quicksand from moving forward and land desertification. Nitraria tangutorum Bobr. is an excellent species for sand fixation.
Materials and methods
Study workflow
The study process shown in the flowchart (Fig. 2) consists of four parts: (1) preparing data, including species data and factors of topography; (2) fitting different models; (3) evaluating models by Area Under the Curve (AUC) and comparing the driving factors of different models; and (4) estimating areas suitable for habitat using the developed SDMs.
Workflow of the study. ARCGIS 10.5 is the software of geographic information system, R3.4.3 is the data processing platform, AUC is the area under the receiver operator curve, which evaluates the fitted models. Species distribution models include Bioclimatic Modelling (BIOCLIM), Domain, Mahalanobis, Generalized Linear Model (GLM), Generalized Additive Model (GAM), Random Forest (RF), Support Vector Machine (SVM). SHI denotes suitable area index.
Study area and data analysis
Study area
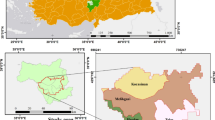
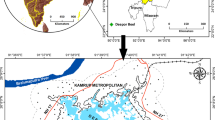
The Ulan Buh Desert is located in the northern part of Inner Mongolia Province, 106°38′42″E—106°57′00″ E, 40°17′24″ N–40°28′36″ N (Fig. 3), covering an area of 14,905.13 km2. The elevation ranges from 1009 to 1016 m, and the topographical profile on the surface varies from 1211.58 to 1339.63 m58. The climate is characterized by semi-arid to arid conditions, with a mean annual precipitation of 147.4 mm, mean annual evaporation of 2458.4 mm, mean annual relative humidity of 47.3%, and mean annual temperature of 6.8 °C in the desert59. The eastern edge of the desert is important for dividing the desert and grassland in Central Asia. Our study was based on control plots, such as desertification processes and control treatments60.
Location of study site in the Ulan Buh desert (the top of the figure is the northwest arid area in China; the bottom left is the study area located in Ulan Buh desert; and the bottom right is the plot of Ulan Buh desert. The number of rows are 0,1,2,3,…and the columns are A,B,C,…in the proper sequence).
Species data processing
Nitraria tangutorum Bobr. is a shrub species. It is 1–2 m tall, multi-branched, crooked, prone, and spreads with infertile needle-like apex branches and white tender branches. The leaves are clustered in 2–4 pieces on the young shoots, broadly lanceolate, 18–30 mm long, 6–8 mm wide, blunt apex, base narrowed into a wedge, and entire rare apex lobed. The flowers are densely arranged. The drupe is oval and dark red when ripe, juice rose-colored, 8–12 mm long, and 6–9 mm in diameter. (http://www.iplant.cn/) (State Key Laboratory of Systems and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences). Supplementary material Fig. 8 for photo of plant Nitraria tangutorum Bobr in the field investigation.
(1) Species data collection
The ground surveys were conducted between August and September 2019. 100 sample plots of Nitraria tangutorum Bobr. plant community was established in one square kilometer at the eastern edge of the study area. Each 100 × 100 m sample plot was divided into 100 small quadrats with an area of 10 × 10 m (Fig. 3). The center points GPS location is southeast corner of the sample F5, and the accuracy of the point is within a half meter. The grids that were created in the same direction as we sampled in field, all are facing north. 335 occurrence records for Nitraria tangutorum Bobr. were collected in the area (Fig. 5). The formal identification of the plant material used in my study has complied with the Chinese Virtual Herbarium (CVH): (https://www.cvh.ac.cn/spms/detail.php?id=f76aceff), and a specimen of this material has been deposited in a publicly available herbarium. And we were employed by the Chinese Academy of Forestry, which is also the State Forestry Administration Dengkou Desert ecosystem positioning observation station. To ensure that we have permission to collect Nitraria tangutorum Bobr. We confirm that all methods were performed following current guidelines and regulations in China, Huoyan zhou, Xiao Zhou, and Xiaodi Zhao undertook the formal plant identification, and field investigation were carried out.
(2) Processing species data
The data were cleaned for use in our modelling. The longitudes and latitudes of the sample plot center were recorded as coordinates, and if the record was out of the boundary of the study area, it was checked with the UAV-RGB (Unmanned Aerial Vehicle—Red Green Blue) data and overlaid on the map. If the canopy was outside the plot boundary, the root inside was preserved.
The coordinates were cross-checked by a visual or ‘overlay’ function using ArcGIS 10.5. This study applied the coordinate function from the ‘SP’ (spatial point) package to create a Spatial Point Data Frame, and then the over function from ‘SP’ to do a point-in-polygon query with the study boundary. Moreover, the occurrence of each grid would be only once, without duplicate records; the function ’duplicate’ could be used to remove duplicates in the software R3.4.3.
The species data included the presence and absence data, the resolution of the grid was 10 m, and each grid retained only presence or absence data, reducing the bias of the model predictions by space sampling61.
Environment data processing
LiDAR data
The UAV technique with the SPAN_IGM_STIM300 IMU (Fig. 4a) was used to gather airborne LiDAR data during the study conducted on October 24, 2019. The LiDAR parameters were a pulse rate of 125 kHz, a scan angle (FOV(field of view)) of ± 22.5°, and a laser beam divergence (IFOV) of 0.5 mrad. An average point density of 41.62 points per m2 and a footprint diameter of 25 cm was obtained.
Topographic variables
The digital elevation model (DEM) was derived from the UAV LiDAR point cloud (Fig. 4b) with a resolution of 15 cm (Fig. 5a) and then resampled to 10 m (Fig. 5b) with image analysis tools. Other topographic variables, including slope, aspect, and profile (Fig. 6), were obtained using the spatial analysis model in ArcGIS 10.5. Predictor variables were organized as raster (grid) files for species distribution modelling. After generating the set of predictor variables (rasters) and the occurrence points, our next step was to extract the values of the predictors at the locations of the points using the raster tools of ‘extract’ in ArcGIS 10.5. To select the significant predictor variables, the variance inflation factor (VIF) was calculated. A VIF of 10 was acceptable as it showed no significant collinearity among the predictor variables62. All variables were resampled using ArcGIS 10.5 to unify the space dimension of 10 m, which was the same value as the resolution of the species samples.
Maps of the predictor variables. The horizontal and vertical coordinates represent latitude and longitude, respectively. The (a) indicates different aspect of the study area, (b) indicates different elevation of the study area, (c) indicates different profile of the study area, and (d) indicates different slope of the study area
Forms for SDMs
To predict species distribution, a number of statistical and machine-learning approaches have been used63,64. In conjunction with geographic information systems and remote sensing, the methods used in species distribution modelling can be classified as profile, regression, and machine learning methods. The profile method only considers the presence (occurrence) data, whereas the regression and machine learning methods use both the presence and absence data. The profile method includes BIOCLIM65, Domain66, and Mahalanobis67. In addition to these methods, we also analyzed and compared the predictive performance of SDMs developed using regression methods, such as the GLM68 and GAM69,70, and some machine learning methods, such as RF71 and SVM.
Sampling bias may have been present in the occurrence data used72. Random sampling was performed to record the presence or absence data with the random points function in the ‘dismo’ package. An ‘extent’ tool was used to further restrict the area from where random locations were drawn8,73. The ‘dismo’ package containing the k-fold function facilitates the data partitioning and creates a vector that assigns each row in the data matrix to a group (between 1 and k with k = 5).
We used seven modelling algorithms, including both nonparametric and parametric algorithms, to develop the SDMs, seven modelling algorithms are briefly described below.
Bioclimatic modelling (BIOCLIM)
BIOCLIM is a classic climate envelope model65. Although it generally does not perform as well as some other modelling methods74, particularly in the context of climate change72, it is still being used, because the algorithm is easy to understand.
Domain
The Domain algorithm66 has been extensively used for species distribution modelling. The Domain algorithm computes Gower’s metric75 between the environmental variables at any location and those at any of the known locations of occurrence, which were defined as training sites in this study.
A suitable means of quantifying the similarity between the two sites is provided by Gower’s metric. In Euclidean Q-dimensional space, the distance d between two points A and B is defined as
To equalize the contribution from each climatic attribute, Gower’s metric uses range standardization. In this application, the standardization method is preferred over variance standardization because it is less susceptible to bias arising from dense clusters of sample points. The complementary similarity measure (\({R}_{AB}\)) is defined as
\({R}_{AB}\) is constrained between 0 and 1 for points within the ranges used in Eq. 1 but may yield negative values for points outside this range. \({S}_{A}\) is defined as the maximum similarity between candidate point A and the combination of known record site Tm:
\({S}_{A}\) is evaluated for all grid points in a target area, where m denotes the number of known records, and a matrix of continuously varying similarity values is generated, which can be displayed as a grayscale, thematic, or contour map. As with all models discussed here, the values generated are not probability estimates but the degrees of classification confidence66.
Mahalanobis
The Mahal function implements SDMs based on Mahalanobis distance67. The Mahalanobis distance considers the correlations of the variables in the dataset, and it is not dependent on the scale of measurements.
where \({\alpha }_{\mu v}\) is the dispersion, \((\alpha {)}^{\mu }\) is the mean value, \(P\) is the independent variable, and \({D}^{2}\) is the sample value of the \({\Delta }^{2}\) statistic. Both the dispersion and mean values are subject to sampling fluctuations.
Generalized linear models (GLM)
In GLM, \({X}_{k}(k=1,\cdot \cdot \cdot ,r)\) is the vector of \(k\) predictor variables, which is related to the expected value \(\mu =E(Y)\) of the response variable \(Y\) (denote the response variable) through a link function \(g(\cdot )\) (Eq. 5). The use of GLM in species distribution modelling is provided in68.
where \(\alpha \) is a constant called the intercept, and \(\beta \) is the vector of \(k\) regression coefficients (one for each predictor). (Eq. (6)).
It is assumed that the topographic factor is normally distributed and is defined as X, and the species distribution probability as Y; Therefore in the response distribution of GLM, which uses the “family = gaussian”.
Generalized additive models (GAM)
GAM69,70 are extensions of GLMs. In GAMs, the linear predictor is the sum of the smoothing functions. This makes GAMs very flexible and they can fit complex functions. This also makes them very similar to machine-learning methods. The GAMs were implemented using the 'mgcv' package R 3.4.3.
where \(\mu =E(Y\left|{X}_{1},{X}_{2},\cdot \cdot \cdot {X}_{p}\right.)\), n is for the linear prediction, and \({s}_{i}(\cdot )\) is the nonparametric smooth function. This model does not require any assumptions. It consists of a random component, an additive component, and a link function connecting the two parts. The distribution of the response variables(Y) belonging to the family of exponential distributions can be binomial.
Random forest (RF)
The RF method is an extension of the regression and classification trees (CART)71. This method can be implemented using the “random forest” function in the package with the same package name in R 3.4.3. The function RF can take a formula, or in two separate arguments, a data frame with the predictor variables and a vector with the response. If the response variable is a factor (categorical) type, RF performs classification; otherwise, it performs regression. In species distribution modelling, because RF showed remarkable modelling results, it is considered a viable option.
-
(1)
Suppose there is a dataset \(D=\left\{{x}_{i1},{x}_{i2},\cdot \cdot \cdot {x}_{in},{y}_{i}\right\}(i\in \left[1,m\right])\) (\({{X}_{i}}_{m}\) is the vector of \(m\) predictor variables, \({y}_{i}\) are the response variables) with feature number n, sampling with replacement can generate a sampling space \((m*n{)}^{m*n}\).
-
(2)
Build a basic learner (a decision tree): \({d}_{i}=\left\{{x}_{i1},{x}_{i2},\cdot \cdot \cdot {x}_{in},{y}_{i}\right\}(i\in \left[1,m\right])\) generates a decision tree for each sample (where K < < M) and records the results of each decision tree \({h}_{j}(x)\).
-
(3)
Training times to T \(H(x)=\mathit{max}{\sum }_{t=1}^{T}\phi (h(x)=y)\), where \(\phi (x)\) is an algorithm (absolute majority voting, majority voting, weighted voting method, etc.).
RF requires two parameters: (1) mtry, the number of predictor variables performing data partitioning at each node, and (2) ntree, the total number of trees to be grown in the model run. In this study, ntree (number of trees to grow) was set to 500 and mtry to 10 after some initial tuning experiments.
Support vector machines (SVM)
SVM76 applies a simple linear method to data; however, in a high-dimensional feature space, nonlinearly exists in the input space. However, in practice, this does not involve any computations in a high-dimensional space. This simplicity, combined with state-of-the-art performance on many learning problems (classification, regression, and novelty detection), has contributed to the popularity of SVM77. They were first used in species distribution modelling by Guo et al.78. There are several implementations of SVM in R 3.4.3. The most useful implementations in our context are the function 'ksvm' in the package 'kernlab' and the 'svm' function in the package 'e1071'. The 'ksvm' includes many different SVM formulations and kernels, provides useful options, and features a method for plotting, but it lacks a proper model selection tool. The 'svm' function in package 'e1071' includes a model selection tool: the 'tune' function77.
Given the training examples, \({x}_{i}(i=\mathrm{1,2},\cdot \cdot \cdot m)\) indicates the samples to be classified, \(\widehat{y}\in \left\{+1,-1\right\}\) indicates the marked label value for the samples, + 1 denotes the point above the separating hyperplane, and -1 denotes the point under the separating hyperplane, where \(K\left(x,{x}_{i}\right)\) is the Gaussian kernel function, \(K\left(x,{x}_{i}\right)\) is nonlinearity, and \({\lambda }_{i}^{*}\) is the optimal solution to the primal problem obtained by solving the dual problem. Note that if a sample \({x}_{i}\) is not a support vector, to maximize the Lagrangian function, there must be a λ* = 0 non-support vector corresponding to \({\lambda }^{*}\) equal to zero, which theoretically shows that the decision boundary of the SVM is only related to the support vector.
Evaluation and comparison of SDMs
Accuracy assessment
Different measures may be used to evaluate the prediction ability of each model79,80,81. Many measures for evaluating models based on presence-absence or presence-only data are threshold dependent. This implies that a threshold must first be set. However, many statistics are threshold independent, such as the correlation coefficients and the area under the receiver operator curve (AUROC, generally abbreviated as AUC). The AUC value is the area of ROC and the X-axis under the ROC curve; the larger the area, the higher the relationship, and the greater the creditability. When the AUC value is greater than 0.7, the result is fair, and when the AUC is higher than 0.9, the result is excellent82. We applied this measure to evaluate the SDMs.
In this framework, an algorithm is supposed to predict “positive” or “negative”. However, some concepts are confusing, as summarized in Table 2.
Suitable habitat index (SHI)
The probability of species occurrence was extracted using ArcGIS. The lowest probability value was selected to distinguish the suitable zone from the non-suitable zone83. The habitats of the entire study area were divided into three categories using the frequency statistical method84: unsuitable habitat (SHI ≤ 0.3), low suitable habitat (0.3 < SHI ≤ 0.5), and suitable habitat (SHI > 0.5).
Conclusion
This is the first attempt at modelling the relationships between the spatial distribution of desert plant species and topographical factors extracted from the LiDAR data. The results indicated a general relationship between model performance and modelled species distribution on a small scale. Topographical factors should be contingent on the modelled species distribution. The LiDAR technique has certain applications in species distribution simulations. The RF produced the highest AUC when AUC was integrated into TNR + TPR (false positive rate plus true positive rate); Manhalanobis showed the best SDMs to predict the distribution of Nitraria tangutorum Bobr. in the desert on a small scale. The suitable habitat distribution area of Nitraria tangutorum Bobr. predicted in the study was larger than the actual area. A flat land with an aspect of the south at an altitude near 1010 m was found to be the driving topography of the Nitraria tangutorum Bobr. distribution. Thus, this study provides a reliable reference for the restoration and management of desert vegetation.
Data availability
Data used in this study are available from the National Forestry and Grassland Science Data Center (http://www.forestdata.cn/). Data is available when needed, and Huoyan Zhou should be contacted.
References
Zhang, X. et al. Discussion on the method of high-resolution remote sensor data interpretation of haloxylon ammodendron in Ulan Buh Desert. Remote Sens. Technol. Appl. 25(6), 828–835 (2010).
Zhang, W., Lu, T. & Ma, K. Analysis of environmental and spatial factors of plant community distribution in arid valley of the upper Minjiang River. Ecology 3, 552–558 (2004).
Guisan, A. et al. Predicting species distributions for conservation decisions. Ecol. Lett. 16, 1424–1435 (2013).
Walther, G. R. et al. Alien species in a warmer world: risks and opportunities. Trends Ecol. Evol. 24(12), 86–93 (2009).
Willis, K. J. & Bhagwat, S. A. Biodiversity and climate change. Science 326(5954), 806–807 (2009).
Hao, T., Guillera-Arroita, G., May, T. W., Lahoz-Monfort, J. J. & Elith, J. Using species distribution models for fungi. Fungal Biol. Rev. (2020)
Buonincontri, M. P. et al. Shedding light on the effects of climate and anthropogenic pressures on the disappearance of Fagus sylvatica in the Italian lowlands: Evidence from archaeo-anthracology and spatial analyses. Sci. Total Environ. 877, 162893 (2023).
Elith, J. & Leathwick, J. R. Species Distribution Models: Ecological Explanation and Prediction Across Space and Time. 677–697 (2009)
Safaei, M., Rezayan, H., ZeaieanFirouzabadi, P. & Sadidi, J. Optimization of species distribution models using a genetic algorithm for simulating climate change effects on Zagros forests in Iran. Ecol. Inform. 63, 101–288 (2021).
Benkendorf, D. J. & Hawkins, C. P. Effects of sample size and network depth on a deep learning approach to species distribution modeling. Ecol. Inform. 60, 101–137 (2020).
Kaky, E., Nolan, V., Alatawi, A. & Gilbert, F. A comparison between Ensemble and MaxEnt species distribution modelling approaches for conservation: A case study with Egyptian medicinal plants. Ecol. Inform. 60, 101–150 (2020).
Walker, P. O. Modelling wildlife distribution using a geographic information system: kangaroos in relation to climate. J. Biogeogr. 17, 279–289 (1990).
Bio, A. M. F., Alkemande, R. & Barendregt, A. Determining alternative models for vegetation response analysis—A non-parametric approach. J. Veg. Sci. 9, 5–16 (1998).
Huiller, W. T. BIOMOD—Optimising predictions of species distributions and projecting potential future shifts under global change. Glob. Change Biol. 9, 1353–1362 (2003).
Pereira, J. M. C. & Itami, R. M. GIS-based habitat modeling using logistic multiple regression: a study of the Mt. Graham Red Squirrel. Photogram. Eng. Remote Sens. 57, 1474–1486 (1991).
Pedro, S. & Araújo, M. B. An evaluation of methods for modelling species distributions. J. Biogeogr. 31, 1555–1568 (2004).
Lopatin, J., Dolos, K., Hernández, H. J., Galleguillos, M. & Fassnacht, FEta. Comparing Generalized Linear Models and random forest to model vascular plant species richness using LiDAR data in a natural forest in central Chile. Remote Sens. Environ. 173, 200–210 (2016).
Randin, C. F. et al. Monitoring biodiversity in the Anthropocene using remote sensing in species distribution models. Remote Sens. Environ. 239, 111626 (2020).
Milanesi, P., Rocca, F. D. & Robinson, R. A. Integrating dynamic environmental predictors and species occurrences: Toward true dynamic species distribution models. Ecol. Evol. 10(2), 1087 (2020).
Isaac, N. J. B. et al. Data integration for large-scale models of species distributions. Trends Ecol. Evol. 35(1), 56–67 (2020).
Pradervand, J.-N., Anne, D., Loïc, P., Antoine, G. & Christophe, R. Very high resolution environmental predictors in species distribution models: Moving beyond topography?. Prog. Phys. Geogr.: Earth Environ. 38(1), 79–96 (2014).
Karger, D. N., Conrad, O., Böhner, J., Kawohl, T., Kreft, H. et al. Climatologies at high resolution for the earth’s land surface areas. Sci. Data. 4(1) (2017)
Dixon, P. P. M. Small-scale environmental heterogeneity and the analysis of species distributions along gradients. J. Veg. 1(1), 57–65 (2010).
Mujuru, L. & Kundhlande, A. Small-scale vegetation structure and composition of Chirinda Forest, southeast Zimbabwe. Afr. J. Ecol. 45, 624 (2010).
Punchi-Manage, R. et al. Effects of topography on structuring local species assemblages in a Sri Lankan mixed dipterocarp forest. J. Ecol. 101, 149 (2012).
Swanson, F. J., Kratz, T. K., Caine, N. & Woodmansee, R. G. Landform effects on ecosystem patterns and processes. Bioscience 38(2), 92–98 (1988).
Noy-Meir, I. Desert ecosystems: Environment and producers. Annu. Rev. Ecol. Syst. 4(1), 25–51 (1973).
Zhang, X.
Runquist, R. D. B., Lake, T., Tiffin, P. & David, A. M. Species distribution models throughout the invasion history of Palmer amaranth predict regions at risk of future invasion and reveal challenges with modeling rapidly shifting geographic ranges. Sci. Rep. 9, 2426 (2019).
Dempsey, S. J., Gese, E. M., Kluever, B. M., Lonsinger, R. C. & Waits, L. P. Evaluation of scat deposition transects versus radio telemetry for developing a species distribution model for a Rare Desert Carnivore, The Kit Fox. PLoS ONE 10(10), 17 (2015).
Nussear, K. E. et al. Modeling habitat of the desert tortoise (Gopherus agassizii) in the Mojave and parts of the Sonoran Deserts of California, Nevada, Utah, and Arizona. U.S. Geol. Surv. Open-File Report. 1102, 18 (2009).
Saatchi, S., Buermann, W., Steege, H. T., Mori, S. & Smith, T. B. Modeling distribution of Amazonian tree species and diversity using remote sensing measurements. Remote Sens. Environ. 112(5), 2000–2017 (2008).
Normand, S. et al. Importance of abiotic stress as a range-limit determinant for European plants: insights from species responses to climatic gradients. Glob. Ecol. Biogeogr. 18(4), 437–449 (2009).
Lembrechts, J. J. et al. Comparing temperature data sources for use in species distribution models: From in-situ logging to remote sensing. Glob. Ecol. Biogeogr. 28(11), 1578–1596 (2019).
Austin, M. P. & Van Niel, K. P. Improving species distribution models for climate change studies: Variable selection and scale. J. Biogeogr. 38, 1–8 (2011).
Zellweger, F., De Frenne, P., Lenoir, J., Rocchini, D. & Coomes, D. Advances in microclimate ecology arising from remote sensing. Trends Ecol. Evol. 34(4), 327–341 (2019).
Park, J., Choi, B. & Lee, J. Spatial distribution characteristics of species diversity using geographically weighted regression model. Sens. Mater. 31(10), 3197–3213 (2019).
Leempoel, K. et al. Data from: Very high resolution digital elevation models: are multi-scale derived variables ecologically relevant?. Methods Ecol. Evol. 6, 1373 (2016).
Mathys, L., Zimmermann, N. E. & Guisan, A. Spatial pattern of forest resources in a multifunctional landscape. Photogramm. Remote. Sens. 36, 340–342 (2004).
Hagar, J. C., Yost, A. & Haggerty, P. K. Incorporating LiDAR metrics into a structure-based habitat model for a canopy-dwelling species. Remote Sens. Environ. 236, 11–49 (2020).
Wüest, R. O., Bergamini, A., Bollmann, K. & Baltensweiler, A. LiDAR data as a proxy for light availability improve distribution modelling of woody species. For. Ecol. Manage. 456, 117–644 (2020).
Zhou, Z. et al. Ecological niche modeling with LiDAR data: A case study of modeling the distribution of fisher in the southern Sierra Nevada Mountains, California. Biodiver. Sci. 26(08), 878–891 (2018).
Zellweger, F., De Frenne, P., Lenoir, J., Rocchini, D. & Coomes, D. Advances in microclimate ecology arising from remote sensing. Trends Ecol Evol. 34(4), 327–341 (2019).
Lefsky, M. A., Cohen, W. B. & Parker, G. G. Lidar remote sesing for ecosystem studies. Bioscience 52, 19–30 (2002).
Westoby, M. J., Brasington, J., Glasser, N. F., Hambrey, M. J. & Reynolds, J. M. Structure-from-motion photogrammetry: A low-cost, effective tool for geoscience applications. Geomorphology 179, 300–314 (2012).
Randin, C. F. et al. Monitoring biodiversity in the Anthropocene using remote sensing in species distribution models. Remote Sens. Environ. 239, 11–62 (2020).
Lenoir, J., Hattab, T. & Parker, G. Climatic mocrorefugia under anthropogenic climata change:implications for species redistribution. Ecography 40, 253 (2017).
Manfreda, S. et al. On the use of unmanned aerial systems for environmental monitoring. Remote Sensing. 10, 641 (2018).
Elith*, J. et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29(2), 129–151 (2010).
Marmion, M., Parviainen, M., Luoto, M., Heikkinen, R. K. & Thuiller, W. Evaluation of consensus methods in predictive species distribution modelling. Divers. Distrib. 15(1), 59–69 (2010).
Bonneu, M., Delecroix, M. & Malin, E. Semiparametric versus nonparametric estimation in single index models, a computational approach. Comput. Statistics 8(3), 207–222 (1993).
Zurell, D. et al. Testing species assemblage predictions from stacked and joint species distribution models. J. Biogeogr. 47(1), 101–113 (2019).
Fernandes, R. F., Daniel, S. & Guisan, A. How much should one sample to accurately predict the distribution of species assemblages? A virtual community approach. Ecol. Inform. 48, 125–134 (2018).
Ali, H. et al. Expanding or shrinking? Range shifts in wild ungulates under climate change in Pamir-Karakoram mountains, Pakistan. PLoS ONE 16(12), e0260031 (2021).
Ma, S., Li, X., Luo, C. & Sun, F. The impact of climate change of Haloxylon ammodendron plants suitable for distribution. Ecology Magazine. 36(05), 1243–1250 (2017).
Salinas-Ramos, V. B. et al. Artificial illumination influences niche segregation in bats. Environ. Pollut. 284, 117187 (2021).
Qiu-Yan, L. I., Zhao, W. Z. & Fang, H. Y. Adaptation of Nitraria sphaerocarpa to wind-blown sand environments at the edge of a desert oasis. J. Environ. Sci. 19(4), 482–487 (2007).
Tang, G.-a., X. Liu & G. Yan, The principle and method of geological analysis and Digital elevation model. 2005: Science Press.
Li, Z. & Li, F. Interspecific associations of natural nitraria tangtorum population and its main companion species in Ulanbuh Desert. Bull. Bot. Res. 28, 99 (2008).
Luo, F. et al. Characteristics of soil temperature variation and influence factors at Northeastern Margin Region of Ulan Buh Desert. J. Desert Res. 39(01), 179–186 (2019).
Kramerschadt, S. et al. The importance of correcting for sampling bias in MaxEnt species distribution models. Divers. Distrib. 19(11), 1366–1379 (2013).
Zheng, J. Statistical Big Dictionary Vol. 1 (China Statistical Press, 1995).
Bosso, L. et al. The rise and fall of an alien: Why the successful colonizer Littorina saxatilis failed to invade the Mediterranean Sea. Biol. Invasions 24(10), 3169–3187 (2022).
Valavi, R., Guillera-Arroita, G., Lahoz-Monfort, J. J. & Elith, J. Predictive performance of presence-only species distribution models: a benchmark study with reproducible code. Ecol. Monogr. 92(1), e1486 (2022).
Booth, T. H., Nix, H. A., Busby, J. R. & Hutchinson, M. F. BIOCLIM: the first species distribution modelling package, its early applications and relevance to most current MAXENT studies. Divers. Distrib. 20(1), 1–9 (2014).
Carpenter, G., Gillison, A. N. & Winter, J. Domain: a flexible modelling procedure for mapping potential distributions of plants and animals. Biodiver. Conserv. Biol. 2, 667–680 (1993).
Mahalanobis, P. C. On the generalised distance in statistics. Proc. Natl. Inst. Sci. India. 2, 49–55 (1936).
Guisan, A., Edwards, T. C. Jr. & Hastie, T. Generalized linear and generalized additive models in studies of species distributions: Setting the scene. Ecol. Model. 157(2–3), 89–100 (2002).
Hastie, T. J. & Robert, T. Generalized Additive Models, in Statistical Science 249–307 (Routledge, 1986).
Wood, S. N. Generalized additive models: an introduction with R (CRC Press, 2006).
Breiman, L., Friedman, J., Olshen, R. A. & Charles, J. S. Classification and Regression Trees (CRC Press, 1984).
Hijmans, R. J. & Graham, C. Testing the ability of climate envelope models to predict the effect of climate change on species distributions. Glob. Change Biol. 12, 2272–2281 (2006).
Elith, J. et al. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 17(1), 43–57 (2011).
Elith, J., Graham, C. H. Anderson, R. P., Ferrier, S., Guisan, A., et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography (2010)
Gower, JCta. A general coefficient of similarity and some of its properties. Biometrics 27, 57–71 (1971).
Fan, R. E., Chen, P. H. & Lin, C. J. Working set selection using second order information for training support vector machines. J. Mach. Learn. Res. 6, 1889–1918 (2005).
Karatzoglou, A., Meyer, D. & Hornik, K. Support vector machines in R. J. Stat. Softw. 15(9), 1–28 (2006).
Guo, Q., Kelly, M. & Graham, C. H. Support vector machines for predicting distribution of Sudden Oak Death in California. Ecol. Model. 182(1), 75–90 (2005).
Friedman, J. H. Greedy function approximation: a gradient boosting machine. Ann. Stat. 29, 1189–1232 (2001).
Liu, C., White, M. & Newell, G. Measuring and comparing the accuracy of species distribution models with presence-absence data. Ecography 34, 232–243 (2011).
Potts, J. M. & Elith, J. Comparing species abundance models. Modelling. 199, 153–163 (2006).
Guo, C., Li, X., Zhao, Z. & Nawaz, Z. Predicting the impacts of climate change, soils and vegetation types on the geographic distribution of Polyporus umbellatus in China. Sci. Total Environ. 648, 1–11 (2019).
Pearson, R. G., Raxworthy, C. J., Nakamura, M. & Peterson, A. T. ORIGINAL ARTICLE: Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. J. Biogeogr. 34(1), 102 (2007).
Chen, H., Chen, L. & Albright, T. P. Predicting the potential distribution of invasive exotic species using GIS and information-theoretic approaches: A case of ragweed (Ambrosia artemisiifolia L.) distribution in China. Chin. Sci. Bull. 52(9), 1223–1230 (2007).
Acknowledgements
We thank the 14th Five-Year Plan Pioneering Project of High Technology Plan of the National Department of Technology under Grant(Grant No. 2021YFD2200405), and the National Natural Science Foundation of China (Grant Nos. 31470641, 31300534, and 31570628) for financial support.
Funding
This research was funded by the 14th Five-Year Plan Pioneering Project of High Technology Plan of the National Department of Technology under Grant (Grant No. 2021YFD2200405), and National Natural Science Foundation of China (Grant Nos. 31470641, 31300534, and 31570628).
Author information
Authors and Affiliations
Contributions
H.Z., X.Z., and L.F. collected data, H.Z., X.Z., and X.Z. undertook the formal identification of the plant material; H.Z. and X.Z. analyzed the data; H.Z., X.Z., L.F., and R.P.S. wrote the manuscript, contributed critically to improving the manuscript, and gave final approval for publication. All the authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, H., Feng, L., Fu, L. et al. Modelling the effects of topographic heterogeneity on distribution of Nitraria tangutorum Bobr. species in deserts using LiDAR-data. Sci Rep 13, 13673 (2023). https://doi.org/10.1038/s41598-023-40678-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-40678-5
- Springer Nature Limited