Abstract
The Dmrt (Doublesex-mab3-related transcription factor) gene family is a class of crucial transcription factors characterized by one or several conserved DM (Doublesex/Mab-3) domains. Dmrt family genes can participate in various physiological developmental processes, especially in sex determination/differentiation. Echinoderms are extremely important research objects in various fields, such as sex determination/differentiation and neuroscience. However, to date, the genome-wide characterization and analysis of Dmrt genes in echinoderms have not been investigated. In this study, the identification and analysis of Dmrt genes in 11 representative echinoderms were performed using bioinformatics methods. A total of 43 Dmrt genes have been found in the studied echinoderms, and the number of Dmrt genes in different species ranges from 2 to 5. The phylogenetic tree showed that all Dmrt genes from echinoderms can be subdivided into 5 classes, the Dmrt2-like class, Dmrt3-like class, Dmrt4/5-like class, Dsx-like class, and a novel Dmrt (starfish-specific) class. Furthermore, selective pressure assessment suggested that the Dmrt genes underwent purifying selection pressure. In general, this study provides a molecular basis for echinoderm Dmrt genes and may serve as a reference for in-depth phylogenomics.
Similar content being viewed by others
Introduction
The Dmrt (Doublesex-mab3-related transcription factor) gene, including one or several DM (Doublesex/Mab-3) domains, has been widely studied due to various functions1,2,3,4, especially in sex determination/differentiation. For example, the Z-linked Dmrt1 gene is vital for sex determination/differentiation in birds5. In Xenopus laevis, a W-linked Dmrt gene can be involved in the development of the primary ovary6. A Y-specific Dmrt gene, DMY/Dmrt1bY, can determine the sex of Oryzias latipes7,8,9. In general, the Dmrt gene family is an important gene family involved in sex-related development during organism evolution.
To date, Dmrt family genes have been studied in mammals, teleosts, and insects. The members of the Dmrt gene family showed substantial differences in different organisms. For example, eight Dmrt members have been found in some mammals4, such as Homo sapiens and Mus musculus. In teleosts, a total of seven Dmrt members have been identified from Larimichthys crocea10, and five Dmrt genes have been found in Oreochromis niloticus11. In Drosophila melanogaster, only four Dmrt genes have been identified. However, to date, no genome-wide study has been conducted to identify Dmrt genes in echinoderms.
Echinoderms have usually been considered to be the closest invertebrate sister group of vertebrates12, with particular evolutionary classification and phylogeny. Meanwhile, as an ancient invertebrate group, echinoderms have diverse reproduction modes, including asexual multiplication, parthenogenesis, hermaphroditism, and dioecy13. Therefore, echinoderms are extremely important research objects in many fields, such as sex determination/differentiation13, neuroscience14, and regeneration biology15. In particular, recent studies have suggested that some biological processes in echinoderms are associated with Dmrt genes16,17. However, no research has focused on the systematic investigation of Dmrt family genes in echinoderms.
The main research objective of this study was to systematically analyze the abundance of Dmrt genes in echinoderms. With the decoding of many echinoderm genomes, including Acanthaster planci18, Anneissia japonica19, Apostichopus japonicus20, Asterias rubens (NCBI: PRJNA683060), Hemicentrotus pulcherrimus21, Holothuria glaberrima22, Lytechinus variegatus23, Patiria miniate (NCBI: PRJNA683060), Plazaster borealis24, Strongylocentrotus purpuratus25, and Temnopleurus reevesii26, genome-wide identification and analysis of the Dmrt gene family was carried out. Furthermore, the functional domains and sequence structures of the Dmrt genes were predicted, and phylogenetic analysis was carried out. The findings from this study can provide fundamental insights into the evolutionary and physiological aspects of Dmrt genes in invertebrates.
Materials and methods
Sequence identification
A set of DMRT protein sequences within different species was first obtained from NCBI databases, including H. sapiens, M. musculus, Macaca fascicularis, Balaenoptera musculus, Gallus gallus, O. latipes, O. niloticus, L. crocea, D. melanogaster, Bombyx mori, Aedes aegypti, Sagmariasus verreauxi and Cherax quadricarinatus (Supplementary Table S1). The Dmrt genes in 11 echinoderms were identified by combining BLAST and HMM search strategies. First, the genome and annotation files of 11 echinoderms were downloaded from different genome databases (Supplementary Table S2), and the DM domain query (accession: PF00751) was downloaded from Pfam (http://pfam.xfam.org/). Second, BLAST V2.11.027 and HMMER V3.2.128 were used simultaneously to search DMRT proteins in all genomes with the DM domain query. The initial E-values for both the BLAST and HMM searches were set to 1 × 10–5 and 1.0, respectively. Third, the candidate genes obtained by BLAST and HMM searches were merged, and then the redundant genes were removed. Finally, to confirm the existence of the DM domain, the nonredundant genes were further checked by using the NCBI CDD database according to an E-value of 10–5. When multiple transcripts were annotated for a gene, the longest transcript was selected. The properties of Dmrt proteins in echinoderms were predicted by using TBtools v1.09829.
Phylogenetic analyses of the Dmrt gene family
All retrieved Dmrt proteins from NCBI and those identified from 11 echinoderms were utilized to perform the phylogenetic analysis. Multiple sequence alignments of Dmrt proteins were first generated using MAFFT v7.31030. Then, the phylogenetic trees were constructed by IQTREE v2.1.231 with the following settings: -m MFP -bnni -B 2000 -T AUTO. The phylogenetic trees were displayed using iTOL (interactive tree of life) online tool32.
Sequence analyses and genomic distribution of Dmrts
The general feature format file was used to reveal the Dmrt gene structure and exon information. The conserved motifs of the Dmrt genes were predicted by using MEME33 with the following options: largest number, 25; minimum length, 6; maximum length, 50; and default values for other parameters. Conserved motifs and gene structure were both visualized by TBtools v1.09829. In addition, the conserved domains of all identified DMRT genes were analyzed using the Batch SMART plug-in in TBtools software (version 1.098)29 and visualized with the iTOL (interactive tree of life) online tool32. The genomic distribution was visualized with gene arrow maps generated by using the gggenes package in R34.
Selective pressure assessment
Selective pressure was assessed by using the branch and site model in EasyCodeML V1.035. The branch models assume that the ratios (ω) of nonsynonymous substitution sites (dN) and synonymous substitution sites (dS) vary among branches. Under the branch model, the comparison of two models (one ratio and free ratio) was calculated to test whether ω is different among different branches. The site models assume that the ω ratio varies among sites. Under the site models, the specific models (M0, M1a, M2a, M3, M7, and M8) were tested by adjusting the parameters. Among these models, comparison of M3/M0 is used to detect whether the ω ratio between different sites is consistent, while the comparisons of the M2a/M1a and M8/M7 model pairs test for positive selection.
Results
Identification and characterization of Dmrt genes
A total of 43 Dmrt genes have been identified in 11 representative echinoderms. The amino acid sequences of the identified Dmrt genes are provided in Supplementary Table S3. The number of Dmrt genes in each species ranges from 2 to 5, which are listed in Table 1. The characteristics of all the identified proteins in echinoderms were predicted and are listed in Table 1. The results showed that the biophysical properties of different Dmrt proteins were different. AA length varied from 110 to 794. The MW ranged from 12,660.77 to 88,920.97 Da, while the PI values varied from 5.44 to 10.37. Additionally, the vast majority of Dmrt proteins were considered unstable (instability index greater than 40).
Phylogenetic analysis of Dmrt genes
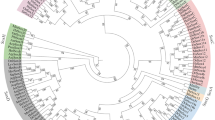
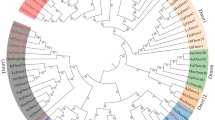
To understand the evolutionary relationships of Dmrt genes in echinoderms, a phylogenetic analysis was carried out using Dmrt protein sequences from vertebrates and invertebrates. As shown in Fig. 1, 43 Dmrt genes from echinoderms were divided into 5 classes: the Dmrt2-like class, Dmrt3-like class, Dmrt4/5-like class, Dsx-like class, and novel Dmrt class. The Dmrt2-like class contains 11 genes from 11 echinoderms. The Dmrt3-like class consists of 11 Dmrt genes from 10 echinoderms. Nine Dmrt genes from 8 echinoderms have formed a Dmrt4/5-like class. The Dmrt genes in the Dsx-like class come from sea urchins, sea cucumbers, and crinoids. The remaining genes from starfish were divided into a novel Dmrt class.
Sequence analyses and genomic distribution
The exon‒intron diversification among echinoderm Dmrt genes is also displayed in Fig. 2. The exon numbers of Dmrt genes in 11 echinoderms varied from 1 to 6. Genes in the same class have more similar exon‒intron structures. In addition, although all the predicted Dmrt proteins contain motif 1, the proteins in the same class have more similar motif structural features. By using the Batch SMART search, it was found that all Dmrt genes include a DM domain, and some Dmrt genes contain Pfam:DMA (Fig. 3). The genomic locations of Dmrt genes in different species are shown in Fig. 4. The Dmrt gene cluster can be found in several species. The Dmrt2-like/Dmrt3-like/Dmrt4-like cluster can be found in A. rubens, L. variegatus, and S. purpuratus, while the Dmrt3-like/Dmrt4-like cluster can be found in A. planci. In addition, a Dmrt3-like/Dmrt4-like/Dmrt4-like cluster was identified in P. miniata. Dmrt genes in other echinoderms were randomly distributed on separate scaffolds, which may be due to incomplete genome assemblies.
Selective pressure analysis
In the branch model, Model = 0 (M0) and 1 (M1) were chosen to test whether the ω values across different branches were significantly different (Table 2). The ω for the M0 model was 0.016. Comparison of the M1 model with the M0 model revealed that each branch has similar ω values (PLRT = 0.976). Subsequently, the site model was used to identify the positive selection sites of the Dmrt gene of echinoderms. In the site model, the M3 model significantly outperformed the M0 model, suggesting that variable alternative pressure existed among different sites. However, no significantly positively selected sites were found in Dmrt by comparisons (M2a/M1a and M8/M7). Considering differences in terms of protein sequence, these results suggest that the homologous (or conserved) domain region of Dmrt genes underwent purification selection.
Discussion
The Dmrt gene family has been identified genome-wide in various animal groups, including mammals, insects, and teleosts10,36,37. However, little is known about aquatic invertebrates. In particular, a comprehensive survey of Dmrt genes has not been carried out in echinoderms, although some echinoderm genome sequences have been available for several years. In the current study, a systematic analysis of Dmrt family genes was performed in 11 echinoderm genomes. Two to five Dmrt genes have been identified in different echinoderms. According to previous studies, the difference in the number of Dmrt genes may be related to genome size and genomic duplication rounds37. Although Dmrt genes are widely represented across the animal kingdom, they present a certain degree of species specificity. For example, Dmrt1 is found only in vertebrates, and Dmrt 6–8 is only present in mammals. This pattern was confirmed in this study. No Dmrt1-like gene or Dmrt6/7/8-like gene was identified in echinoderms, while Dmrt2-like, Dmrt3-like, Dmrt4/5-like and possibly Dsx-like genes were found in this study. Similar Dmrt members can also be found in Panarthropoda38. These results imply that Dmrt2, Dmrt3, Dmrt4/5, and Dsx-like genes may be widely present in invertebrates.
Unlike some fish that harbored two paralogs of Dmrt2 (Dmrt2a and Dmrt2b), all the studied echinoderms carried one Dmrt2-like gene, suggesting that the Dmrt2-like gene may be conserved in echinoderms. Dmrt2 has very important roles in sex reversal, testicular development, and embryonic development. For example, in humans, Dmrt2 is associated with XY sex reversal and gonadal dysgenesis39. Dmrt11E was proven to be a crucial factor for gametic formation in domesticated silkworm40. Analogous functions of Dmrt2 have also been reported in several aquatic invertebrates. In Penaeus monodon, Dmrt11E was proposed to affect muscle development, testis development, spermatogenesis, and somites41. In Chlamys nobilis, Dmrt2 is expressed exclusively in gonads, implying that it may be involved in the maintenance of gonadal function or gonadal development42. In addition, in zebrafish, Dmrt2 was found to have a function in regulating the left–right patterning of the mesoderm43. In summary, given that Dmrt2 has diverse functions, Dmrt2-like genes in echinoderms should be further studied.
In this study, both Dmrt2-like genes and Dmrt3-like genes were found in all species except P. borealis. Furthermore, the Dmrt2-like/Dmrt3-like cluster can be found in numerous echinoderms. These results may support the previous conclusion that Dmrt3 may have emerged through a gene duplication event of Dmrt2 during deuterostome evolution44. In addition, functional investigations on Dmrt3 have only been performed in vertebrates, showing that this gene can play pivotal roles in configuring the spinal circuits controlling stride45. Consistent with previous findings, the current phylogenetic analysis showed that Dmrt4 and Dmrt5 were clustered into a major branch, suggesting that these two types of genes originated from the same ancestor of Dmrt. In this study, the Dmrt4/5-like gene was identified in 8 echinoderms with the exception of H. pulcherrimus, T. reevesii, and P. borealis. In particular, the Dmrt4/5-like gene was duplicated in P. miniata. To date, Dmrt4 and Dmrt5 have been found to be closely related to neurogenesis. For instance, in Xenopus, Dmrt4 and Dmrt5 are important regulators of olfactory placode neurogenesis46. During the development of the hippocampus in mice, Dmrt5 was shown to be involved in the regulation of the neuron–glia cell-fate switch47. A similar function was also observed in invertebrates. In Drosophila, Dmrt99B plays an essential role in initiating temporal patterning in medulla neuroblasts. Thus, it will be interesting to investigate whether Dmrt4/5 play similar functions in echinoderms.
Dsx was found to have a pivotal role in sexual dimorphism in genetic sex-determining animals, including insects and nematodes. In Drosophila, Dsx has male- and female-specific isoforms (DsxM and DsxF), which can regulate different target genes, resulting in sex-specific morphology48. In B. mori, two Dsx isoforms (BmDsxF and BmDsxM) can enhance male and female differentiation in gonads and external genitalia, respectively49. DapmaDsx1 (Dsx ortholog) in Daphnia magna was confirmed as a critical regulator of the male phenotype50. In this study, a possible Dsx-like gene class was found in the phylogenetic tree. However, whether these genes have similar functions is unclear. In addition, it should be noted that the Dsx gene class in the phylogenetic tree was backed by low bootstrap values. In particular, these sequences from sea urchins, sea cucumbers, crinoids, and two Dsx sequences from shrimp seem to be unrelated to the Dsx from A. aegypti, B. mori, and D. melanogaster. This result may be caused by relatively few informative characters outside of the DM domains. In particular, it is worth noting that the members in the Dsx-like cluster present different protein characteristics. For example, the AA and MW of the Dsx-like gene from sea urchins were significantly higher than those of the Dsx-like gene from other species. These results may imply that the function of Dsx-like genes in sea urchins may be different from that of Dsx-like genes in other species.
Moreover, a starfish-specific Dmrt class was identified in the current study. These genes were phylogenetically distant from the other Dmrt members. Their exon‒intron structure is also unique. Similar results can be learned in other aquatic invertebrates. Comparative phylotranscriptomics revealed that DMRT1L is a mollusk-specific gene51, and a novel Dmrt gene (EsDmrt-like) was identified in Eriocheir sinensis52. These results indicate that there may be more members of the Dmrt gene family, especially in aquatic invertebrates. Therefore, it is necessary to conduct systematic identification and analysis of the Dmrt gene family in other classes of invertebrates.
Conclusion
In this study, a systematic analysis of Dmrt family genes in 11 representative echinoderms was performed. A total of 43 Dmrt genes have been found, and the number of Dmrt genes in different echinoderms ranges from 2 to 5. The phylogenetic tree showed that all Dmrts from echinoderms were classified into 5 classes: the Dmrt2-like class, Dmrt3-like class, Dmrt4/5-like class, Dsx-like class, and novel Dmrt class. Furthermore, selective pressure assessment suggested that the Dmrt genes underwent purifying selection pressure. In general, this study provides a molecular basis for echinoderm Dmrt and may serve as a reference for in-depth phylogenomics.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Kopp, A. Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28, 175–184 (2012).
Zarkower, D. DMRT genes in vertebrate gametogenesis. Curr. Top. Dev. Biol. 102, 327–356 (2013).
Zhang, T. & Zarkower, D. DMRT proteins and coordination of mammalian spermatogenesis. Stem Cell Res. 24, 195–202 (2017).
Hong, C. S., Park, B. Y. & Saint-Jeannet, J. P. The function of Dmrt genes in vertebrate development: it is not just about sex. Dev. Biol. 310, 1–9 (2007).
Hirst, C. E. et al. Sex reversal and comparative data undermine the W chromosome and support Z-linked DMRT1 as the regulator of gonadal sex differentiation in birds. Endocrinology 158, 2970–2987 (2017).
Yoshimoto, S. et al. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. 105, 2469–2474 (2008).
Matsuda, M. et al. DMY gene induces male development in genetically female (XX) medaka fish. Proc. Natl. Acad. Sci. 104, 3865–3870 (2007).
Matsuda, M. et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417, 559–563 (2002).
Nanda, I. et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. 99, 11778–11783 (2002).
Wan, H.-F. et al. Genome-wide investigation of Dmrt gene family in large yellow croaker (Larimichthys crocea). Theriogenology 156, 272–282 (2020).
Rather, M. A. & Dhandare, B. C. Genome-Wide identification of doublesex and Mab-3-Related transcription factor (DMRT) genes in nile tilapia (Oreochromis niloticus). Biotechnol. Rep. 24, e00398 (2019).
Satoh, N., Rokhsar, D. & Nishikawa, T. Chordate evolution and the three-phylum system. Proc. R. Soc. B Biol. Sci. 281, 20141729 (2014).
Wang, Y., Yang, Y., Li, Y. & Chen, M. Identification of sex determination locus in sea cucumber Apostichopus japonicus using genome-wide association study. BMC Genomics 23, 1–14 (2022).
Ortega, A. & Olivares-Bañuelos, T. N. Neurons and glia cells in marine invertebrates: An update. Front. Neurosci. 14, 121 (2020).
Byrne, M. The link between autotomy and CNS regeneration: Echinoderms as non-model species for regenerative biology. BioEssays 42, 1900219 (2020).
Cui, Z. et al. Testis-specific expression pattern of dmrt1 and its putative regulatory region in the sea urchin (Mesocentrotus nudus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 257, 110668 (2022).
Slota, L. A., Miranda, E. M. & McClay, D. R. Spatial and temporal patterns of gene expression during neurogenesis in the sea urchin Lytechinus variegatus. EvoDevo 10, 2 (2019).
Hall, M. R. et al. The crown-of-thorns starfish genome as a guide for biocontrol of this coral reef pest. Nature 544, 231–234 (2017).
Li, Y. et al. Genomic insights of body plan transitions from bilateral to pentameral symmetry in Echinoderms. Commun. Biol. 3, 371 (2020).
Zhang, X. et al. The sea cucumber genome provides insights into morphological evolution and visceral regeneration. PLoS Biol. 15, e2003790 (2017).
Kinjo, S., Kiyomoto, M., Yamamoto, T., Ikeo, K. & Yaguchi, S. HpBase: A genome database of a sea urchin, Hemicentrotus pulcherrimus. Dev. Growth Differ. 60, 174–182 (2018).
Medina-Feliciano, J. G., Pirro, S., García-Arrarás, J. E., Mashanov, V. & Ryan, J. F. Draft genome of the sea cucumber Holothuria glaberrima, a model for the study of regeneration. Front. Mar. Sci. 8, 603410 (2021).
Davidson, P. L. et al. Chromosomal-level genome assembly of the sea urchin Lytechinus variegatus substantially improves functional genomic analyses. Genome Biol. Evol. 12, 1080–1086 (2020).
Lee, Y. et al. Chromosome-level genome assembly of Plazaster borealis sheds light on the morphogenesis of multiarmed starfish and its regenerative capacity. GigaScience 11 (2022).
Sodergren, E. et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science 314, 941–952 (2006).
Kinjo, S. et al. TrBase: A genome and transcriptome database of Temnopleurus reevesii. Dev. Growth Differ. 64, 210–218 (2022).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Eddy, S. R. Profile hidden Markov models. Bioinformatics (Oxford, England) 14, 755–763 (1998).
Chen, C. et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202 (2020).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Bailey, T. L., Johnson, J., Grant, C. E. & Noble, W. S. The MEME suite. Nucleic Acids Res. 43, W39–W49 (2015).
Wilkins, D. & Kurtz, Z. gggenes: draw gene arrow maps in ‘ggplot2’. R package version 0.4. 0 342 (2019).
Gao, F. & Chen, J. EasyCodeML: an interactive visual tool for CodeML analysis. Ecol. Evol. (2016).
zafar, I., Rather, M. A. & Dhandare, B. C. Genome-Wide identification of doublesex and Mab-3-Related transcription factor (DMRT) genes in nile tilapia (Oreochromis niloticus). Biotechnol. Rep. 24, e00398 (2019).
Xu, S. et al. Genome-wide identification, phylogeny, and expression profile of the Dmrt (Doublesex and Mab-3 related transcription factor) gene family in channel catfish (Ictalurus punctatus). Front. Genet. 13, 891204 (2022).
Panara, V., Budd, G. E. & Janssen, R. Phylogenetic analysis and embryonic expression of panarthropod Dmrt genes. Front. Zool. 16, 23 (2019).
Raymond, C. S. et al. A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum. Mol. Genet. 8, 989–996 (1999).
Kasahara, R., Yuzawa, T., Fujii, T., Aoki, F. & Suzuki, M. G. dmrt11E ortholog is a crucial factor for oogenesis of the domesticated silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 129, 103517 (2021).
Wei, W. Y. et al. Molecular characterization and functional analysis of DMRT11E in black tiger shrimp (Penaeus monodon). Aquacult. Rep. 22, 100982 (2022).
Shi, Y., Wang, Q. & He, M. Molecular identification of dmrt2 and dmrt5 and effect of sex steroids on their expressions in Chlamys nobilis. Aquaculture 426–427, 21–30 (2014).
Saúde, L., Lourenço, R., Gonçalves, A. & Palmeirim, I. terra is a left–right asymmetry gene required for left–right synchronization of the segmentation clock. Nat. Cell Biol. 7, 918–920 (2005).
Mawaribuchi, S., Ito, Y. & Ito, M. Independent evolution for sex determination and differentiation in the DMRT family in animals. Biol. Open 8, bio041962 (2019).
Andersson, L. S. et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 488, 642–646 (2012).
Huang, X., Hong, C.-S., O’Donnell, M. & Saint-Jeannet, J.-P. The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system. Proc. Natl. Acad. Sci. 102, 11349–11354 (2005).
Muralidharan, B. et al. Dmrt5, a novel neurogenic factor, reciprocally regulates Lhx2 to control the neuron-glia cell-fate switch in the developing hippocampus. J. Neurosci. 37, 11245–11254 (2017).
Verhulst, E. C. & van de Zande, L. Double nexus—Doublesex is the connecting element in sex determination. Brief. Funct. Genom. 14, 396–406 (2015).
Yuzawa, T. et al. Transgenic and knockout analyses of Masculinizer and doublesex illuminated the unique functions of doublesex in germ cell sexual development of the silkworm, Bombyx mori. BMC Dev. Biol. 20, 1–15 (2020).
Kato, Y., Kobayashi, K., Watanabe, H. & Iguchi, T. Environmental sex determination in the branchiopod crustacean Daphnia magna: deep conservation of a Doublesex gene in the sex-determining pathway. PLoS Genet. 7, e1001345 (2011).
Evensen, K. G., Robinson, W. E., Krick, K., Murray, H. M. & Poynton, H. C. Comparative phylotranscriptomics reveals putative sex differentiating genes across eight diverse bivalve species. Comp. Biochem. Physiol. D-Genom. Proteom. 41, 100952 (2022).
Zhang, E. F. & Qiu, G. F. A novel Dmrt gene is specifically expressed in the testis of Chinese mitten crab, Eriocheir sinensis. Dev. Genes Evol. 220, 151–159 (2010).
Acknowledgements
This work was supported by the High-Performance Computing Platform of Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences.
Funding
This work was supported by the Open Project Program of Key Laboratory of Ecological Warning, Protection and Restoration for Bohai Sea, Ministry of Natural Resources [2022204], Natural Science Foundation of Shandong Province [ZR2021MC151, ZR2021QD158, ZR2022QC234], and Yantai Science and Technology Planning Project [2020MSGY074].
Author information
Authors and Affiliations
Contributions
Q.C.W. and T.G.C. conducted the experiment and data processing. Q.C.W. and Y.W. conceived and supervised the project. Y.X.W. and X.J.L. contributed to the data collection. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, Q., Cao, T., Wang, Y. et al. Genome-wide identification and comparative analysis of Dmrt genes in echinoderms. Sci Rep 13, 7664 (2023). https://doi.org/10.1038/s41598-023-34819-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-34819-z
- Springer Nature Limited
This article is cited by
-
Genome-wide identification and expression analysis of Dmrt genes in bivalves
BMC Genomics (2023)