Abstract
To evaluate the generalizability of artificial intelligence (AI) algorithms that use deep learning methods to identify middle ear disease from otoscopic images, between internal to external performance. 1842 otoscopic images were collected from three independent sources: (a) Van, Turkey, (b) Santiago, Chile, and (c) Ohio, USA. Diagnostic categories consisted of (i) normal or (ii) abnormal. Deep learning methods were used to develop models to evaluate internal and external performance, using area under the curve (AUC) estimates. A pooled assessment was performed by combining all cohorts together with fivefold cross validation. AI-otoscopy algorithms achieved high internal performance (mean AUC: 0.95, 95%CI: 0.80–1.00). However, performance was reduced when tested on external otoscopic images not used for training (mean AUC: 0.76, 95%CI: 0.61–0.91). Overall, external performance was significantly lower than internal performance (mean difference in AUC: −0.19, p ≤ 0.04). Combining cohorts achieved a substantial pooled performance (AUC: 0.96, standard error: 0.01). Internally applied algorithms for otoscopy performed well to identify middle ear disease from otoscopy images. However, external performance was reduced when applied to new test cohorts. Further efforts are required to explore data augmentation and pre-processing techniques that might improve external performance and develop a robust, generalizable algorithm for real-world clinical applications.
Similar content being viewed by others
Introduction
It estimated that over 1.5 billion people in the world live with hearing loss, representing nearly 20% of the global population1. The prevalence of hearing loss is expected to rise to over 2.5 billion by 20501. In the US, it is estimated that the overall cost of deafness and hearing loss amounts to US$980 billion annually2. Direct and indirect costs related to hearing loss are comprised of health sector costs, educational support, loss of productivity, and societal costs3,4. In the global pediatric population, 34 million children experience hearing loss, of which, 60% can be attributed to preventable causes5. The WHO estimates that over one-third of preventable childhood hearing loss is attributed to infections including mumps, rubella, meningitis, measles and middle ear infections5.
Otoscopy is an important component of ear health and hearing assessments to identify ear infections. Otoscopy is performed routinely by multiple healthcare workers including medical students, nurses, general practitioners, emergency medicine physicians, paediatricians, audiologists, and otolaryngologists. However, the ability to accurately interpret otoscopic findings varies by user experience6,7,8,9,10. Pichichero et al. (2005) demonstrated differences between general practitioners, paediatricians, and otolaryngologists to recognise tympanic membrane (TM) abnormalities8. Significant performance differences were found when non-otolaryngologists were asked to differentiate between acute otitis media (AOM), otitis media with effusion (OME), or retracted TMs8. These findings are consistent with recent studies that demonstrate that diagnostic accuracy is reduced when non-experts must differentiate between multiple ear disease sub-types6,11. Otolaryngologists significantly outperformed non-otolaryngologists to identify ear disease sub-types and important pathological characteristics (e.g. tympanic membrane perforations, attic retraction, or myringitis)6,8.
Accurately recognising middle ear diseases is important for early management, prevention of long-term hearing loss, can improve speech and language development and reduce healthcare costs12. However, the subjective and often inconsistent nature of traditional otoscopy makes accurate interpretation difficult. To address this challenge, deep learning -based image classification algorithms have been developed using machine learning techniques13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35. Traditional machine learning algorithms rely on statistical models and pre-defined features, whereas deep learning algorithms use neural networks to learn features and extract patterns directly from raw data13,36,37. Deep learning methods have the potential to improve early ear disease detection by bridging the gap between expert and non-expert performance and enhancing the performance of previously developed machine learning models for otoscopy38.
Previous studies have explored image classification algorithms for otoscopy using deep learning approaches13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35. In a recent systematic review and meta-analysis, a pooled analysis of 14 studies achieved 91% accuracy in differentiating between normal versus abnormal otoscopic images39. From a pooled analysis of three studies, multi-classification algorithms achieved 98% to differentiate AOM, OME, and normal aerated ears without middle ear pathology39. Despite high levels of accuracy reported, the binary and multi-classification analyses yielded substantial analyses heterogeneity between studies39. Variability between models to classify ear disease subtypes suggests performance may not be generalisable to new test environments.
The inability to generalize to environments beyond those used for training could limit the clinical utility and sustainability of deep learning algorithms for otoscopy. Variability in data source, capture devices, ear disease sub-type, and deep learning methods may impact classification performance in clinical encounters different to those used for training. To date, an exploration of the generalisability of deep learning algorithms for otoscopy has not been evaluated.
Deep learning techniques have been shown to excel in tasks such as image classification, with the ability to classify images with high accuracy and to generalize to new data40,41. Unlike traditional machine learning techniques, deep learning algorithms can automatically extract relevant features from images, reducing the need for manual feature engineering. This makes deep learning algorithms less sensitive to variations in data quality and pre-processing steps, and more robust to changes in data source, capture devices, and ear disease subtypes.
The purpose of this study was to construct a deep learning-based image classifier capable of differentiating between normal and abnormal otoscopic images using three independently collected otoscopy databases, evaluate the generalisability of the algorithm on images not used for training, and explore the optimal convolutional neural network (CNN) and deep learning approach to optimise performance.
Methods
Ethics
This research study was conducted in accordance with the Helsinki Declaration, the Standard for Reporting Diagnostic Accuracy Studies (STARD) and the Consolidated Standards of Reporting Trials for interventions involving artificial intelligence (CONSORT-AI) reporting guidelines42,43. This study utilised three open-access otoscopic image databases that were collected after obtaining Institutional Review Board (IRB) approval from each of the respective human research ethics committees and published online for public access, as indicated by each of the authorship groups (Bitlis Eren University Ethics Committee34,44, University of Chile Scientific and Research Ethics Committee18, Ohio State University Institutional Review Board45).
Data sources
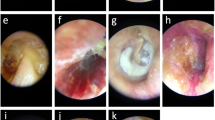
Otoscopic images ascertained for the purposes of this study can be found elsewhere for online public access, as per the authorship groups18,34,45. In Turkey (Cohort A), 848 otoscopic images were collected at the Özel Van Akdamar Hospital using digital video otoscopes (unspecified brand and model) with ground-truth classified by two otolaryngologists and one paediatrician as: normal, acute otitis media (AOM), and chronic otitis media (COM) consisting of chronic suppurative otitis media34. In Chile (Cohort B), 540 otoscopic images were collected at the University of Chile Clinical Hospital using the Firefly DE500 digital video otoscope (Firefly Global, Belmont, MA, US. 2022) with ground-truth classified by 1 otolaryngologist as: normal or COM18. In the United States (Cohort C), 454 otoscopic images were collected at Nationwide Children’s Hospital and Ohio State University using the Jedmed Horus+ (Jedmed, St Louis, MO, US, 2022) with ground-truth classified by 1 otolaryngologist as: normal or otitis media with effusion (OME)45. Otoscopic images where the TM could not be visualised were excluded (e.g., obstruction with wax or foreign body).
Algorithm development
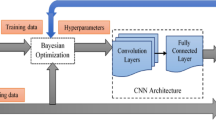
Deep learning-based binary class algorithms were developed to classify normal or abnormal otoscopic images using Cohort A, B and C independently. Abnormal sub-classes included AOM, OME, and COM. Pre-existing ground-truth labels were available for each otoscopic image from the online data sources18,34,45. We used four different neural net architectures including ResNet-50 (comprised of 50 layers and 23 million parameters)46, VGGNet-16 (comprised of 16 layers and 134 million parameters)47, DenseNet-161 (comprised of 16 layers and 26 million parameters)48, and Vision Transformer (comprised of 12 layers and 85 million parameters)49. Images were resized to 224 by 224 pixels. Each model was trained for 200 epochs using the Stochastic Gradient Descent (SGD) with a learning rate = 0.003 as the optimizer. The loss function is cross entropy. Training was completed using the Microsoft Azure Machine Learning Studio (Redmond, Washington, USA). All the models used the weights of models trained on the ImageNet dataset as their initial weights. Raw data was used for the purposes of developing the model and validation.
To assess the generalizability of a model trained on a single-source dataset, we first randomly split the dataset from one single source into training and validation and testing sets, allocating 70% for training, 10% for validation and 20% for testing (internal validation). We trained the model using the training set and selected the final model as the one at the best-performing epoch minimizing the loss on the validation set. Next, we tested the final model on the testing (internal validation) set. Then, we tested the final model on the two external validation cohorts. To ensure the robustness and reliability of the results, we ran each experiment for three times, and the mean and standard deviation of accuracy, area under the curve (AUC), sensitivity, and specificity were computed, as shown in Table 3. This was done to account for any potential variability or instability in the results due to the stochastic nature of deep learning algorithms. The mean and standard deviation values provided a more comprehensive understanding of the performance of the models, as well as the variability of their performance across multiple runs.
To evaluate the pooled performance, we first combined all the data from the three cohorts, and then used fivefold cross-validation as follows: (1) The data was randomly split into fivefolds; (2) a model was trained using the training set consisting of three of the five folds. We selected the final model as the one at the best-performing epoch minimizing the loss on the validation set consisting of one of the five folds. We tested the final model on the remaining fold. (3) Step 2 was repeated five times until each fold had been used as a test fold exactly once. All the metrics were averaged to estimate the pooled performance.
Statistical analysis
To compare the algorithm to the reference standard for Cohorts A, B, and C, contingency tables were generated to summarise Area Under the Curve (AUC), accuracy, sensitivity, and specificity. To evaluate internal performance, models were trained and tested using otoscopic images from the same cohort (e.g., split Cohort A otoscopic images into training and test sets). To evaluate external validation performance and generalizability, models were validated on external cohorts (e.g., trained using Cohort A and validated on Cohort B and Cohort C, Fig. 1)50. Mean difference in AUC were calculated to compare internal and external performance. We used bootstrapping to compare the Receiver Operating Characteristics (ROC) curves, and the null hypothesis is that the external validation AUC is greater or equal to the internal validation AUC50. Pooled assessment was performed by combining Cohort A, B, and C and fivefold cross-validation was applied (Fig. 1).
Results
In total, 1842 otoscopic images were collected from three online resources, comprised of 1117 normal and 725 abnormal images. Table 1 summarises the distribution of normal and abnormal otoscopic images by Cohort A, B and C.
Internal performance
Table 2 summarises internal performance for each cohort. Sensitivity and specificity results are provided in the Supplementary Appendix. Models trained using Cohort A otoscopic images achieved AUC between 0.82 and 0.86 (accuracy: 80 to 84%; sensitivity: 57 to 70%; specificity 90 to 91%). Models trained using Cohort B otoscopic images achieved an AUC of 1.00 (100% accuracy, 100% sensitivity, and 100% specificity). Models trained using Cohort C otoscopic images achieved AUC between 0.98 to 0.99 (accuracy: 91 to 95%; sensitivity: 91 to 96%; specificity: 89 to 96%).
Table 2 summarises external validation performance for each cohort. Models trained using Cohort A otoscopic images and tested on Cohort B achieved AUC between 0.80 and 0.87 (accuracy: 62 to 77%; sensitivity: 76 to 96%; specificity: 47 to 78%), while those tested on Cohort C achieved AUC between 0.61 and 0.82 (accuracy: 65 to 76%; sensitivity: 80 to 92%; specificity: 25 to 69%).
Models trained using Cohort B otoscopic images and tested on Cohort A achieved AUC between 0.60 to 0.67 (accuracy: 59 to 65%; sensitivity: 4 to 54%; specificity: 61 to 99%), while those tested on Cohort C achieved AUC between 0.87 and 0.91 (accuracy: 72 to 80%; sensitivity: 62 to 73%; specificity: 88 to 95%) (Table 2).
Models trained using Cohort C otoscopic images and tested on Cohort A achieved AUC: 0.54 to 0.68 (accuracy: 44 to 70%; sensitivity: 14 to 82%; specificity: 26 to 96%), while those tested on Cohort B achieved AUC: 0.85 to 0.94 (accuracy: 34 to 45%; sensitivity: 100%; specificity: 1 to 17%) (Table 2).
On average, external AUC was significantly lower than internal AUC (range of mean difference in AUC for DenseNet-161: 0.07–0.39). As shown in Table 3, Cohort A had the smallest difference between internal and external AUC between architectures (range of mean differences: −0.05 to 0.07).
Pooled performance
Models developed using a combination dataset of all cohorts achieved 90 to 91% accuracy (AUC: 0.96; sensitivity: 84 to 87%; specificity: 93 to 95%) (Table 4). DenseNet-161 had the highest AUC and smallest standard deviation for fivefold cross validation.
Discussion
Otoscopy is a common clinical task performed by multiple healthcare workers and practitioners with varying levels of expertise. Identifying normal anatomical landmarks and abnormal pathological processes is important in evaluating ear health. The diagnosis of ear disease relies on a combination of presenting symptoms, clinical history, tympanometry, audiometry, and otoscopic findings. AI-based algorithms have been explored in healthcare to augment existing clinical practices in order to enhance judgement and decision-making51. AI-based algorithms for otoscopy have achieved substantial accuracy to differentiate between normal or abnormal otoscopic images utilising pre-trained CNN architectures52,53,54,55,56. Despite achieving high performance in training and internal validation groups, substantial heterogeneity has been found between model results39. The differences may, in part, be attributed to the variability in image capture methods or devices, definition of diagnoses, ground truth determination, sample populations, and methodology used for algorithm development39. Translating performance achievements from training environments to new test cohorts is an important aspect for an AI-based model to be generalizable for routine clinical practice. Without achieving generalizable performance and real-world utility57, it is unclear if applying AI to otoscopy practices will contribute to improving clinical practice. The purpose of this study was to evaluate the generalisability of a binary classification AI-otoscopy algorithm to classify normal or abnormal otoscopic images, using three independently collected databases and trained using standardised deep learning methods.
This study demonstrates substantial internal performance within each cohort to differentiate between normal or abnormal otoscopic images. Between CNN architectures, models developed using DenseNet-161 achieved the highest internal performance for the pooled assessment. However, when the models were applied externally (i.e., to new validation cohorts not used for training), overall model performance was reduced. The extent of performance variability was dependent on the outcome measure assessed (e.g., accuracy, AUC, sensitivity, and specificity). Performance was reduced in most external comparisons, although the mean difference varied by outcome measure. Cohort A (Özel Van Akdamar Hospital, Van, Turkey) was found to have the smallest external performance drop in accuracy, AUC, and sensitivity. On the other hand, Cohort B (University of Chile’s Hospital, Santiago, Chile) was found to have the smallest performance drop in specificity, but the greatest performance drop in AUC and sensitivity. These findings demonstrate that measuring external model performance and evaluating generalizability to new cohorts need to be interpreted relative to the outcome measure of interest. The outcome variable ‘accuracy’ evaluated final predictions as discrete categories, where predictions could be considered either correct or incorrect. However, the outcome variable ‘AUC’ measured the degree of separability and as previously demonstrated, is a better measure of algorithm performance than accuracy58,59.
External validation performance differences between cohorts also may be attributed to intrinsic differences in the number of otoscopic images available for training, ear disease categories, and image capture devices. Cohort A had the lowest internal performance despite having both the greatest number of otoscopic images available for training and number of ear disease categories within overarching ‘abnormal’ classification labels. Although these factors may have contributed to less accurate internal performance due to greater heterogeneity in model learning across each category, these deficiencies were not translated to the model’s ability to generalize compared to Cohort B and C.
Interestingly, models naïve to OME during training were able to achieve an AUC between 0.72 to 0.90 and to differentiate OME from normal TMs in validation cohorts. This achievement may reflect the CNN’s architectural design and the feature extraction layers that were used to extract pertinent features common to normal TMs, ones that are translatable to new cohorts. However, the addition of AOM in Cohort A may have negatively impacted the CNN’s ability to extract pertinent features as, often, AOM, OME, and normal TMs are phenotypically comparable and challenging to differentiate, even among experts8,9,60,61.
In a pooled analysis of all three cohorts combined, the DenseNet-161 architecture achieved substantial performance (AUC 0.96) and exceeded the external performance from each individual cohort, respectively. This finding demonstrated that the reduction in performance between internal and external validation, could potentially and in part, be overcome by coming the data into a pooled cohort by sampling a broad collection of otoscopic images from multiple sources.
This study represents the first attempt to evaluate the generalisability of AI-otoscopy algorithms trained and tested on three unique and independently collected databases. Although the image capture devices and data collection protocols varied by each cohort, the deep learning methodology was standardised and uniform between groups. This approach revealed the potential for pre-trained CNNs to extract important features from otoscopic images used during training that could be applied universally to new images. Comparable to a healthcare worker’s evolving knowledge base that improves with otoscopy experience, CNNs demonstrate the potential to identify useful patterns in otoscopic images despite differences in otoscopes, image quality, and underlying ear disease diagnoses. The patterns and features extracted by pre-trained CNNs enable broad differentiation between normal versus abnormal TMs. However, the choice of architecture can impact model performance, with more complex architectures potentially achieving better performance to capture complex features in the otoscopic images. This was found as DenseNet-161, which has the largest number of layers compared to other architectures used, achieved the highest performance to generalise between cohorts. However, more complex architectures may also require more data and computing resources to train and may be more prone to overfitting. As these architectures were pre-trained on large datasets, they can extract features from images that are relevant for many different image classification tasks.
This study has several limitations. First, this study utilises secondary data published online for open-access use. As a result, the data collection protocols, image capture devices, ground-truth definitions, and labelling formats were not standardised across cohorts. Although the models developed for this study evaluated normal versus abnormal classifications, it is plausible that differences in the ear disease definitions and potential misclassifications could influence the models’ performance and overestimate its accuracy. Second, it is unclear if the ear disease diagnoses were based on otoscopic images alone or in concert with tympanometry, audiometry, pneumatic otoscopy, and clinical history. Inter-cohort variability and interpretation bias may exist in the ground-truth labels, potentially contributing to external measurement errors and misrepresenting the algorithm’s generalisability. Third, ground-truth labels were not established by expert consensus. Inter-rater variability and rater bias could impact the overall reliability of ground-truth labels and adversely impact external validation.
The potential for AI-based algorithms to augment otoscopy is promising. Autonomous AI-otoscopy tools have the potential to improve triage and clinical decision-making. In settings where the prevalence of ear disease is high and access to healthcare services is limited (e.g., rural and remote areas; Indigenous communities in North America, Australia, and New Zealand; under-resourced or marginalised populations in sub-Saharan Africa, India, south-east Asia)12,62,63, AI-otoscopy algorithms may enhance telehealth programs and empower local healthcare workers to perform otoscopy with more certainty. However, for this to be achieved, AI-otoscopy models must be accurate, reliable, generalizable, accessible in online/offline settings, sustainable, and ethnically constructed and deployed. Early studies have demonstrated that CNNs can achieve substantial performance to interpret otoscopy, although, as shown in this study, external validation performance and generalisability to new settings is limited. Further efforts are required to expand otoscopic image databases to include images from multiple countries, rural versus urban settings, various image capture devices, and include users from various experience levels to account for variability in image capture quality. Establishing standardised diagnostic definitions or consensus from expert panels is required to ensure the ground-truth labels are valid and subsequent models are trained with minimal classification bias. The consideration of data augmentation and pre-processing techniques may be explored to enhance external performance and overcome performance limitations. Otoscopy practices will inevitably vary between clinical settings and a robust, comprehensive, and generalizable AI-otoscopy algorithm is necessary for real-world applications.
Conclusion
Internal and pooled performance results are comparable to previously reported findings. However, external performance was reduced when applied to new test cohorts. Further efforts are required to explore data augmentation and pre-processing techniques to improve external performance and develop a robust, generalizable algorithm for real-world clinical applications.
Data availability
Data that support the findings of this study are available from the corresponding author on reasonable request.
References
World Health Organisation. World Report on Hearing [Internet]. Geneva; 2021. https://www.who.int/publications/i/item/world-report-on-hearing. Accessed 28 Aug 2022 (2021).
World Health Organization. Global Costs of Unaddressed Hearing Loss and Cost-Effectiveness of Interventions: A WHO Report [Internet]. Geneva; 2017. https://apps.who.int/iris/bitstream/handle/10665/254659/9789241512046-eng.pdf. Accessed 28 Aug 2022 (2017).
Deloitte Access Economics. The Social and Economic Costs of Hearing Loss in Australia [Internet]. https://apo.org.au/node/102776. Accessed 28 Aug 2022 (2017)
Shield, B. Evaluation of the Social and Economic Costs of Hearing Impairment: A Report for Hear-It [Internet]. https://www.hear-it.org/sites/default/files/multimedia/documents/Hear_It_Report_October_2006.pdf. Accessed 28 Aug 2022 (2006).
World Health Organization. Childhood Hearing Loss—Act Now, Here’s How! [Internet]. Geneva; 2016. https://apps.who.int/iris/handle/10665/204507. Accessed 28 Aug 2022 (2016).
Cha, D. et al. Differential biases and variabilities of deep learning-based artificial intelligence and human experts in clinical diagnosis: Retrospective cohort and survey study. JMIR Med. Inform. 9(12), 33049 (2021).
Kleinman, K. et al. Evaluation of digital otoscopy in pediatric patients: A prospective randomized controlled clinical trial. Am. J. Emerg. Med. 1(46), 150–155 (2021).
Pichichero, M. E. & Poole, M. D. Comparison of performance by otolaryngologists, pediatricians, and general practioners on an otoendoscopic diagnostic video examination. Int. J. Pediatr. Otorhinolaryngol. 69(3), 361–366 (2005).
Pichichero, M. E. Assessing diagnostic accuracy and tympanocentesis skills of South African physicians in management of otitis media [7]. S. Afr. Med. J. 92(2), 137–138 (2002).
Wormald, P. J., Browning, G. G. & Robinson, K. Is otoscopy reliable? A structured teaching method to improve otoscopic accuracy in trainees. Clin. Otolaryngol. Allied Sci. 20(1), 63–67 (1995).
Khan, M. A. et al. Automatic detection of tympanic membrane and middle ear infection from oto-endoscopic images via convolutional neural networks. Neural Netw. [Internet]. 126, 384–394. https://doi.org/10.1016/j.neunet.2020.03.023 (2020).
Graydon, K., Waterworth, C., Miller, H. & Gunasekera, H. Global burden of hearing impairment and ear disease. J. Laryngol. Otol. 133(1), 18–25 (2019).
Zhang, J., Li, C., Yin, Y., Zhang, J. & Grzegorzek, M. Applications of artificial neural networks in microorganism image analysis: A comprehensive review from conventional multilayer perceptron to popular convolutional neural network and potential visual transformer. Artif. Intell. Rev. 1, 1–58 (2022).
Sandström, J., Myburgh, H., Laurent, C., Swanepoel, D. W. & Lundberg, T. A machine learning approach to screen for otitis media using digital otoscope images labelled by an expert panel. Diagnostics 12(6), 1318 (2022).
Crowson, M. G. et al. Machine learning for accurate intraoperative pediatric middle ear effusion diagnosis. Pediatrics 147(4), e2020034546 (2021).
Byun, H. et al. An assistive role of a machine learning network in diagnosis of middle ear diseases. J. Clin. Med. 10(15), 3198 (2021).
Habib, A. R., Wong, E., Sacks, R. & Singh, N. Artificial intelligence to detect tympanic membrane perforations. J. Laryngol. Otol. 134(4), 311–315 (2020).
Viscaino, M. et al. Computer-aided diagnosis of external and middle ear conditions: A machine learning approach. PLoS One [Internet]. 15(3), 1–18. https://doi.org/10.1371/journal.pone.0229226 (2020).
Goshtasbi, K. Machine Learning Models to Predict Diagnosis and Surgical Outcomes in Otolaryngology [Internet]. https://escholarship.org/uc/item/1tr0c2p0 (University of California Irvine, 2020).
Livingstone, D., Talai, A. S., Chau, J. & Forkert, N. D. Building an otoscopic screening prototype tool using deep learning. J. Otolaryngol. Head Neck Surg. 48(66), 1–5 (2019).
Livingstone, D. & Chau, J. Otoscopic diagnosis using computer vision: An automated machine learning approach. Laryngoscope 13, 1–6 (2019).
Myburgh, H. C., Jose, S., Swanepoel, D. W. & Laurent, C. Towards low cost automated smartphone- and cloud-based otitis media diagnosis. Biomed. Signal Process. Control [Internet]. 39, 34–52. https://doi.org/10.1016/j.bspc.2017.07.015 (2018).
Myburgh, H. C., van Zijl, W. H., Swanepoel, D. W., Hellström, S. & Laurent, C. Otitis media diagnosis for developing countries using tympanic membrane image-analysis. EBioMedicine [Internet] 5, 156–160. https://doi.org/10.1016/j.ebiom.2016.02.017 (2016).
Başaran, E., Cömert, Z. & Çelik, Y. Neighbourhood component analysis and deep feature-based diagnosis model for middle ear otoscope images. Neural Comput. Appl. 34(8), 6027–6038 (2022).
Binol, H. et al. OtoXNet—Automated identification of eardrum diseases from otoscope videos: A deep learning study for video-representing images. medRxiv (Internet) https://doi.org/10.1101/2021.08.05.21261672 (2021).
Binol, H., Niazi, M.K.K., Elmaraghy, C., Moberly, A.C., & Gurcan, M.N. Automated video summarization and label assignment for otoscopy videos using deep learning and natural language processing. in Medical Imaging 2021: Imaging Informatics for Healthcare, Research, and Applications [Internet] (Park, B.J., Deserno, T.M. Eds.). 28. https://www.spiedigitallibrary.org/conference-proceedings-of-spie/11601/2582009/Automated-video-summarization-and-label-assignment-for-otoscopy-videos-using/https://doi.org/10.1117/12.2582009.full. Accessed 17 Mar 2021 (SPIE, 2021).
Binol, H. et al. SelectStitch: Automated frame segmentation and stitching to create composite images from otoscope video clips. Appl. Sci. (Switzerland) 10(17), 1–13 (2020).
Binol, H. et al. Digital otoscopy videos versus composite images: A reader study to compare the accuracy of ENT physicians. Laryngoscope 131(5), E1668–E1676 (2021).
Wang, W., Tamhane, A., Rzasa, J., Clark, J., Canares, T., & Unberath, M. Otoscopy video screening with deep anomaly detection. in (Drukker, K., Mazurowski, M.A. eds.) Medical Imaging 2021: Computer-Aided Diagnosis [Internet]. 50. https://www.spiedigitallibrary.org/conference-proceedings-of-spie/11597/2581902/Otoscopy-video-screening-with-deep-anomaly-detection/https://doi.org/10.1117/12.2581902.full. Accessed 17 Mar 2021. (SPIE, 2021)
Wang, W., Tamhane, A., Santos, C., Rzasa, J.R., Clark, J.H., Canares, T.L. et al. Pediatric Otoscopy Video Screening with Shift Contrastive Anomaly Detection. http://arxiv.org/abs/2110.13254. Accessed 25 Oct 2021 (2021).
Uçar, M., Akyol, K. & AtilaUçar, E. Classification of different tympanic membrane conditions using fused deep hypercolumn features and bidirectional LSTM. Irbm (Internet) 1, 1–11. https://doi.org/10.1016/j.irbm.2021.01.001 (2021).
Alhudhaif, A., Cömert, Z. & Polat, K. Otitis media detection using tympanic membrane images with a novel multi-class machine learning algorithm. PeerJ Comput. Sci. 7, e405 (2021).
Sundgaard, J. V. et al. Deep metric learning for otitis media classification. Med. Image Anal. 1, 71 (2021).
Cömert, Z. Fusing fine-tuned deep features for recognizing different tympanic membranes. Biocybern. Biomed. Eng. (Internet) 40(1), 40–51. https://doi.org/10.1016/j.bbe.2019.11.001 (2020).
Wu, Z. et al. Deep learning for classification of pediatric otitis media. Laryngoscope. 131, 1–8 (2020).
Liu, W. et al. Is the aspect ratio of cells important in deep learning? A robust comparison of deep learning methods for multi-scale cytopathology cell image classification: From convolutional neural networks to visual transformers. Comput. Biol. Med. 1, 141 (2022).
Li, X. et al. A comprehensive review of computer-aided whole-slide image analysis: From datasets to feature extraction, segmentation, classification and detection approaches. Artif. Intell. Rev. 55(6), 4809–4878 (2022).
Habib, A. R. et al. An artificial intelligence computer-vision algorithm to triage otoscopic images from Australian Aboriginal and Torres Strait Islander children. Otol. Neurotol. 43(4), 481–488 (2022).
Habib, A. R. et al. Artificial intelligence to classify ear disease from otoscopy: A systematic review and meta-analysis. Clin. Otolaryngol. 47(3), 401–413 (2022).
Chen, H. et al. IL-MCAM: An interactive learning and multi-channel attention mechanism-based weakly supervised colorectal histopathology image classification approach. Comput. Biol. Med. 1, 143 (2022).
Chen, H., Li, C., Wang, G., Li, X., Rahaman, M., Sun, H. et al. GasHis-Transformer: A Multi-scale Visual Transformer Approach for Gastric Histopathological Image Detection. http://arxiv.org/abs/2104.14528. Accessed 29 Apr 2021 (2021).
Cohen, J. F. et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: Explanation and elaboration. BMJ Open 6(11), 1–17 (2016).
Liu, X. et al. CONSORT-AI extension. Nat Med. 26, 1364–1374 (2020).
Başaran, E., Cömert, Z., Celik, Y., Velappan, S. & Togacar, M. Determination of tympanic membrane region in the middle ear otoscope images with convolutional neural network based YOLO method. Deu Muhendislik Fakultesi Fen ve Muhendislik. 22(66), 919–928 (2020).
Camalan, S. et al. OtoMatch: Content-based eardrum image retrieval using deep learning. PLoS One [Internet]. 15(5), 1–16. https://doi.org/10.1371/journal.pone.0232776 (2020).
He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. http://arxiv.org/abs/1512.03385. Accessed 10 Dec 2015 (2015).
Simonyan, K., & Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. http://arxiv.org/abs/1409.1556. Accessed 4 Sep 2014 (2014).
Huang, G., Liu, Z., van der Maaten, L., & Weinberger, K.Q. Densely Connected Convolutional Networks. http://arxiv.org/abs/1608.06993. Accessed 24 Aug 2016 (2016).
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T. et al. An Image is Worth 16 x 16 Words: Transformers for Image Recognition at Scale. arXiv [Internet]. 11929. http://arxiv.org/abs/2010.11929 (2020).
Ramspek, C. L., Jager, K. J., Dekker, F. W., Zoccali, C. & van Diepen, M. External validation of prognostic models: What, why, how, when and where?. Clin. Kidney J. (Oxford University Press) 14, 49–58 (2021).
Yin, J., Ngiam, K. Y. & Teo, H. H. Role of artificial intelligence applications in real-life clinical practice: Systematic review. J. Med. Internet Res. (JMIR Publications Inc.) 23, 25759 (2021).
Basaran, E., Comert, Z. & Celik, Y. Convolutional neural network approach for automatic tympanic membrane detection and classification. Biomed. Signal Process Control 56(1), 1–14 (2020).
Basaran, E., Sengur, A., Comert, Z., Budak, U., Celik, Y., & Velappan, S. Normal and acute tympanic membrane diagnosis based on gray level co-occurrence matrix and artificial neural networks. in 2019 International Conference on Artificial Intelligence and Data Processing Symposium, IDAP 2019. 5–10 (2019).
Basaran, E., Comert, Z., Sengur, A., Budak, U., Celik, Y., & Togacar, M. Chronic tympanic membrane diagnosis based on deep convolutional neural network. in UBMK 2019—Proceedings, 4th International Conference on Computer Science and Engineering. 635–638 (2019).
Lee, J. Y., Choi, S. H. & Chung, J. W. Automated classification of the tympanic membrane using a convolutional neural network. Appl. Sci. (Switzerland). 9(9), 1827 (2019).
Seok, J., Song, J.J., Koo, J.W., Kin, H.C., & Choi, B.Y. The semantic segmentation approach for normal and pathologic tympanic membrane using deep learning (internet). BioRxiv. https://doi.org/10.1101/515007v2.full. Accessed 3 Mar 2019 (2019).
Futoma, J., Simons, M., Panch, T., Doshi-Velez, F. & Celi, L. A. The myth of generalisability in clinical research and machine learning in health care. Lancet 1(2), e489–e492 (2020).
Bradley, A. P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Patter Recognit. 30(7), 1145–1159 (1997).
Huang, J. & Ling, C. X. Using AUC and accuracy in evaluating learning algorithms. IEEE Trans. Knowl. Data Eng. 17(3), 299–310 (2005).
Alenezi, E. M. A. et al. The reliability of video otoscopy recordings and still images in the asynchronous diagnosis of middle-ear disease. Int. J. Audiol. 61, 921 (2021).
Alenezi, E. M. A. et al. Clinician-rated quality of video otoscopy recordings and still images for the asynchronous assessment of middle-ear disease. J. Telemed. Telecare 26, 1357633X20987783 (2021).
Wahl, B., Cossy-Gantner, A., Germann, S. & Schwalbe, N. R. Artificial intelligence (AI) and global health: how can AI contribute to health in resource-poor settings?. BMJ Glob Health (Internet) 3(4), e000798. https://doi.org/10.1136/bmjgh-2018-000798 (2018).
Alami, H. et al. Artificial intelligence in health care: Laying the foundation for responsible, sustainable, and inclusive innovation in low- and middle-income countries. Global Health. 16(1), 1–6 (2020).
Acknowledgements
We acknowledge Microsoft’s AI for Humanitarian Action Grant, the Garnett Passe and Rodney Williams Memorial Foundation, the AI for Good Research Lab, and the Avant Foundation for providing research funding to support this project.
Funding
Microsoft AI for Humanitarian Action Program providing access to Azure and in-kind Data Science resources. (1) Microsoft AI for Humanitarian Action Grant, (2) Research Scholarship from the Garnett Passe and Rodney Williams Memorial Foundation, (3) Avant Foundation Doctor-in-Training and Early Career Research Grants.
Author information
Authors and Affiliations
Contributions
A.-R.H., Y.X., K.B., S.M., T.S., R.D., J.L.F., C.P., R.S., N.S. conceived the study idea. A.-R.H., Y.X., K.B., S.M. were involved in data collection. A.-R.H., Y.X. extracted data and performed the data analysis. A-RH wrote the first draft of the manuscript. A.-R.H., Y.X., K.B., S.M., T.S., W.B.W., R.D., J.L.F., C.P., R.S., interpreted the data analysis and critically revised the manuscript. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
A-RH discloses research funding from the Garnett Passe and Rodney Williams Memorial Foundation, Microsoft’s AI for Humanitarian Action grant, and the Avant Mutual Foundation Doctor-in-Training and Early Career research grants. CP discloses conflict of interest from Australian Medical Association (Queensland) and provides consulting services for the Deadly Ears Program. RS is a consultant for Medtronic. NS discloses research funding from Microsoft’s AI for Humanitarian Action grant, consultant for ResMed, Optinose, Nasus, GSK, and ENT Technologies, and has received conference funding from Medtronic, Karl Storz, and NeilMed. YX, KB, SM, TS, RD, and JLF are affiliated with the AI for Good Research Lab.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Habib, AR., Xu, Y., Bock, K. et al. Evaluating the generalizability of deep learning image classification algorithms to detect middle ear disease using otoscopy. Sci Rep 13, 5368 (2023). https://doi.org/10.1038/s41598-023-31921-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-31921-0
- Springer Nature Limited
This article is cited by
-
An ensemble approach for classification of tympanic membrane conditions using soft voting classifier
Multimedia Tools and Applications (2024)