Abstract
Vibrio species are classified as potent hazards because of their tendency to effect serious diseases like cholera and other gastrointestinal ailments in humans, as well as vibriosis in fish. A total of 144 freshwater samples were aseptically collected monthly across four rivers (Asejire, Ona, Dandaru and Erinle rivers) over a 12-month period from which Vibrio spp. were isolated using culture procedures, confirmed by means of biochemical test as well as Polymerase Chain Reaction (PCR) assay and further characterized for their phenotypic antibiotic susceptibilities and relevant antimicrobial resistant determinants by PCR. Three hundred and fifteen (58%) isolates confirmed across the sampled sites (Asejire = 75, Dandaru = 87, Eleyele = 72, Erinle = 81) showed high resistance against erythromycin—95%, Sulphamethoxazole—94%, rifampicin—92%, doxycycline—82%, tetracycline—75%, amoxicillin—45%, cephalothin—43% and varied susceptibilities to other antibiotics. The multiple antibiotic resistance indices of 97% of the Vibrio isolates were above the 0.2 threshold limit with MAR phenotype pattern E-SUL-RF-TET-DOX (0.38) found to be the most prevalent pattern among the isolates. The distributions of resistance determinant of the tested antibiotics were revealed as follows: sulII 33%, sulI 19% (sulfonamides); blaOXA 27%, ampC 39%, blapse 11% (beta-lactams); tetA 28%, tetE 20%, tet39 8%, (tetracyclines) and strA 39%. aacC2 24%, aphA1 14% (aminoglycosides). Strong positive associations were observed among tetA, sulI, tetE and sulII. This study raises concerns as these selected rivers may contribute to the environmental spread of waterborne diseases and antibiotic resistance genes. Therefore, we recommend environmental context-tailored strategies for monitoring and surveillance of resistance genes so as to safeguard the environment from becoming reservoirs of virulent and infectious Vibrio species.
Similar content being viewed by others
Introduction
Vibrio species are aquatic bacteria of ecological significance belonging to Vibrionaceae family. It is phenotypically characterized as Gram-positive, oxidase positive, motile rods, halophilic and facultatively anaerobic in its metabolism. They are commonly regarded as autochthonous to marine environment and its detection in freshwater ecosystem has been documented globally. There are over 140 different species of Vibrios documented currently1, however, 12 Vibrio species are classified as infectious to humans with V. cholerae recognized as the causative agent of cholera disease in humans. Other non-cholerae Vibrio spp. are common causative agents of foodborne related infection in humans arising from contaminated water, undercooked or raw seafood consumption2. The common symptom of Vibrio infection is gastroenteritis and diarrhea which may sometimes be associated with virulence factors like shiga-like cytotoxin, toxin-coregulated pilus, cytotoxins, siderophores, hemolysins, proteases and heat-stable enterotoxin3,4.
The incidence of Vibrio associated diseases in human and animal is increasingly being reported globally. This bacterium has developed to inhabit diverse settings within ecosystems due to their complex mode of living5. They have been isolated from diverse environments including crustacean guts, fishes in sea hydrothermal vent, rivers, wastewater, plants etc. and they carry out important role of nutrient cycling in aquatic milieus6,7,8.
Vibrio infections are commonly treated with tetracyclines, third-generation cephalosporin, fluoroquinolone, trimethoprim-sulfamethoxazole as well as aminoglycoside. Tetracyclines and fluoroquinolones are mostly used in protracted or severe cases of Vibrio illnesses as they are known to be associated with lesser mortality rates4. However, in recent decades, many bacteria including Vibrio have emerged with unprecedented resistance to many antimicrobials owing to abusive use in agricultural and human systems9.
Antibiotics is among a prominent member of the list of “contaminants of emerging concerns” circulating in different environments. Bacteria that are exposed to antibiotics in aquatic environments can become antibiotic resistance10 and their resistance genes can be transferred to other pristine microorganisms by means of mobile genetic elements and horizontal gene transfer thereby causing changes in the development and adaptation of microbial residents of aquatic environment11.
Evidence has shown that anthropogenic activities resulting in agricultural, municipal and aquaculture wastes as well as water run-off can introduce multiple antibiotic resistance organism and their resistance factors into the environment and water bodies12,13. Likewise unregulated and ease of access to antibiotics in developing nations like Nigeria can contribute to persistent exposure of humans to antibiotics via consumption of water and food. Thus, polluted aquatic environments have now been identified as significant reservoirs of antibiotic resistance genes requiring more investigation14. Some resistance genes contribute to the virulence expression in Vibrio infections in human and animal setting and this includes beta lactam genes (blaOXA, ampC), sulfonamides genes (sul1 and sul2), quinoline genes (tetA, tetE) etc.
In most developing countries, identification of new and emerging threats, practices of good treatment of patients, and effective control of antibiotic resistance is not effective due to non-existence of national antimicrobial resistance surveillance programs. Even where surveillance programs do exist, implementation strategies are lacking due to poor infrastructural and equipment support15. Hence, monitoring antibiotic resistance patterns of pathogenic bacteria in anthropogenically impacted environment is a relatively cost effective substitute where a national surveillance activity is absent16.
Moreover, if the dynamics of antibiotic resistance in pathogenic organisms and diversity of their resistance genes are to be accurately evaluated; there is a need to assess the freshwater environment for contamination with pathogens such as Vibrio spp. and clinically relevant antibiotic resistance genes. Therefore, we document the first assessment of the prevalence of 11 resistance determinants of Vibrio isolates obtained from four selected freshwater bodies in South-western region of Nigeria.
Methodology
The study area
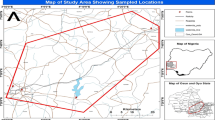
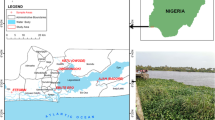
The sampled rivers selected for this study includes: Asejire—SS1 (N7° 21′ 46.9″ E4° 07′ 51.5″), Dandaru—SS2 (N7° 26′ 25.0″ E3° 49′ 57.5″), Eleyele—SS3 (N7° 26′ 17.1″ E3° 52′ 42.2″), Erinle—SS4 (N7° 46′ 23.7″ E4° 27′ 58.3″); located in three towns in Southwest Nigeria (Ibadan, Asejire and Ede) (Fig. 1). The climate is tropical with rain forest vegetation and atmospheric temperature average of 32 °C. These rivers function as key water source particularly to the rural neighborhoods that borders the cities. The river Ona at Eleyele and River Dandaru at Agodi were selected in Ibadan, for Oyo State. Asejire river, the river at the boundary of Osun and Oyo states (abstracted by Oyo State) and Erinle river in Ede were selected for Osun State. These rivers which serve as collector of effluents from animal, human, hospitals, industries, markets, are often contaminated with organic and inorganic substances from anthropogenic activities with higher concentration during dry seasons. There are a couple of activities around these rivers which includes: markets, small scale farming, animal rearing, active fishing and people were also seen washing clothes and bathing during visit.
Sampling, enumeration and isolation of presumptive Vibrio
Pre-sterilized 1 L plastic bottles were used for water sample collection. After the bottles were triple rinsed with sample water, actual samplings were done at approximately 20 cm below the water surface. Samples were subsequently transported in ice coolers and processed according to the recommendation of the American Public Health Association16. By means of standard membrane filtration method, 100 ml each of sampled was filtered through 90 mm, 0.45 μm pore-size filter papers (Millipore, Ireland) and were thereafter transferred into agar plates of TCBS (Thiosulphate Citrate Bile Salts, Conda, Pronasida) and then incubated overnight at 37 °C. Yellow and green characteristic colonies were quantified as colony forming units (CFU) per 100 mL of samples and 3–5 colonies each were randomly selected, Gram-stained and subjected to oxidase test. Afterwards, 315 presumptive Vibrio species that were Gram-negative and oxidase positive were purified on a non-selective agar and stored for molecular analysis.
Extraction of genomic DNA
The isolates' genomic DNA extraction was carried out using the boiling technique6. Specifically, 18–24 h old pure colonies of presumptive Vibrio spp. were suspended in sterile 200 μl distilled water, then lysed by heating for 15 min at 100 °C temperature using an AccuBlock (Labnet). A Mini Spin microcentrifuge was used to decant cell debris by centrifugation for 10 min at rate of 15,000 rpm. The DNA template from the recovered supernatants was employed in subsequent PCR tests.
Molecular confirmation of Vibrio genus
Genotypic confirmation of Vibrio was established by PCR using the Vibrio genus primers (F: CGG GAAATGCGTAGAGAT R: TACTAGCGATTCCGAGTTC)17 to amplify a 663 bp sequence of the 16s RNA genes of Vibrio.
The reaction mixture contains PCR Master Mix (12.5 μl), (Thermo Scientific), oligonucleotide primer (1 μl each) (Inqaba Biotech, SA), DNA template (5 μl), and, nuclease free water (5.5 μl), to make up 25 μl. reaction volume. The PCR protocols used comprises: Initial denaturation for 15 min at 93 °C followed by 35 cycles of denaturation—annealing—elongation steps at 92 °C for 40 s; 57 °C for 1 min and 72 °C for 1.5 min respectively followed by 7 min final extension at 72 °C. Electrophoresis was performed by loading 5 μl amplicons, 3 ul 100-bp ladder (Thermo Scientific), on 2% (w/v) agarose gel (Merck) pre-stained with 5 μl ethidium bromide (Sigma-Aldrich). The electrophoresis process was carried out at 90 V for 50 min and visualized under an ultra-violet trans-illuminator system (Alliance 4.7).
Antibiotic susceptibility profiling of Vibrio isolates
Standards disc diffusion technique with Mueller–Hinton agar (MHA) was used for antimicrobial susceptibility testing18, Fresh cultures (18–22 h old) of all isolates that have been positively identified were introduced into 5 mL of 0.85% sterile saline with the suspension turbidity set to 0.5 McFarland standards. MH agar plates were inoculated uniformly with sterile swabs after which discs were aseptically added and incubated for 18–24 h at 37 °C. Measured Zones of inhibition were interpreted as resistant (R), susceptible (S), intermediate (I), using the zone diameter interpretation according to CLSI guideline18.
Eighteen (18) antibiotics which comprise the Centre for disease Control (CDC) recommended antibiotics for Vibrio infection treatment selected for this assay are: Amikacin—AMK (30 μg), Streptomycin—S (300 μg), Gentamycin—G (10 μg), Cefotaxime—CEF (30 μg), Imipenem—IMI (10 μg), Ciprofloxacin—CIP (5 μg), Meropenem—MEM (10 μg), Ampicillin—AP (10 μg), Sulphamethoxazole—SUL (25 μg), Chloramphenicol—C (30 μg), Tetracycline—TET (30 μg), Erythromycin—E (15 μg), Trimethoprim + Sulphamethoxazole—TS (25 μg), Amoxycillin—AMC (25 μg), Norfloxacin—NOR (30 μg), Doxycycline—DOX (30 μg), Cephalothin—CEP (30 μg) and Rifampin—RF (5 µg).
Antibiotics resistance phenotyping and indexing
The antibiotic resistance profile of the confirmed Vibrio spp. was obtained and those that presented resistance to three and more antibiotics were “phenotyped” to indicate the multiple antibiotic resistances (MAR) phenotypes expressed as:
a = total antibiotics that showed resistant, b = total antibiotics selected for the assay19,20.
MAR index value ≥ 0.2 threshold suggests high-risk pollution, possibly by uncontrolled use of antimicrobials around the sample sites, which may lead to development of antibiotic resistance21. Pattern of antibiotic resistance (ARPA) was likewise evaluated22.
Resistance quotients (RQs) determination
Possible variations in the phenotypes of antimicrobial-resistant Vibrio isolates in the selected freshwaters were determined by calculating their resistance quotients23. This determines probable ecological-risk presented by antibiotics tested on the confirmed Vibrio isolates. The RQs were determined according to the equation24:
PCR detection antibiotic resistance genes of Vibrio isolates
Phenotypically resistance Vibrio isolates were evaluated for their antibiotic resistant genes using specific primer pairs and PCR conditions (Supplementary Table 1). Resistance genes assayed includes; sulfonamide genes (sulI, sulII), tetracycline genes (tetA, tet39, tetE), β-lactamases genes (blaOXA, ampC, blaP,SE), and aminoglycoside genes (aacC2, aphA2, strA). The PCR assay and electrophoresis procedure is as described in “Extraction of genomic DNA” section. Likewise multiple antibiotic resistance genes patterns of all Vibrio isolates harboring ≥ 2 resistance genes were determined.
Statistical analysis
The data obtained were statistically analyzed using IBM (SPSS) version 22.
The associations between the resistance genes found in Vibrio isolates were evaluated with Pearson’s chi-square exact test at P = 0.05. A positive relationship between two genes indicates that the genes were associated together, while a negative correlation shows there is no association between the genes.
Results
Vibrio genus confirmation
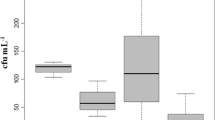
The counts of Vibrio confirmed isolates across the samples freshwaters ranges between 72 CFU/100 ml at SS3 and 87 CFU/100ML at SS2 (Table 1). In total, 58% (315) isolates were positively confirmed by PCR as Vibrio (Fig. 2) out of the 548 presumptive isolates confirmed by Gram staining and oxidase test.
Profile of antibiotic resistance phenotypes
In total, 315 confirmed Vibrio isolates were subjected to susceptibility test with 18 antibiotics belonging to 10 antimicrobial classes (supplementary Table 2). The antibiogram profile of the Vibrio isolates across the sampling sites varies. However, notable resistance to Sulphamethoxazole—83%, Tetracycline—70%, Erythromycin—82%, Rifampicin—80% were recorded in SS2 (Dandaru) whereas resistance to doxycycline were recorded in 72% Vibrio isolates in Erinle.
None of the sample sites presented 100% susceptible or resistance of isolates to any classes of antibiotics. The total susceptibility of all Vibrio tested varied and follows the order: ciprofloxacin—93%, norfloxacin—98%, meropenem—91%, amikacin—76%, cefotaxime—88%, Trimethoprim + Sulphamethoxazole—75%, gentamicin—74%, ampicillin—54%. Although, high resistance to erythromycin—95%, sulfamethoxazole—94%, rifampicin—92%, doxycycline—82%, tetracycline—75% were observed as well as mild resistance to amoxicillin—45% and cephalothin—43% respectively (Fig. 3).
Antibiotic resistance profile of Vibrio isolates across the selected sampling sites. CIP ciprofloxacin, AMK amikacin, MEM meropenem, E erythromycin, AMC amoxicillin, IMP imipenem, TS trimethoprim–sulphamethoxazole, CEF cefotaxime, C chloramphenicol, G gentamicin, SUL sulphamethoxazole, RF rifampin, CEP cephalothin, AP ampicillin, NOR norfloxacin, TET tetracycline, DOX doxycycline, S Streptomycin.
The antibiotic resistance and susceptibility characteristics across the sampling sites were analyzed by heatmap clusters (supplementary Fig. 1A–D). Two major clusters were formed column wise by the Vibrio isolates. The hetero-site isolates showed cluster characteristics indicating that all sampled rivers had 3 antibiogram clusters except river Eleyele with 4 hetero-site antibiogram clusters of the isolates.
Phenotypes and indexes of multiple antibiotic resistance (MAR)
The multiple antibiotic resistance phenotype result indicated that all the isolates were resistance to three or more antibiotics except one (1) isolate at SS1 (Erinle).
MAR indexes of Vibrio isolates across the sampling sites ranged between 0.11 and 0.72 with MAR indexes less than 0.2 threshold limit obtained in SS2 (n = 2), SS3 (n = 4) and SS4 (n = 3). Highest (0.72) MAR index was recorded in SS3 with resistance to 13 of the tested antibiotics. Abundance of resistance pattern observed across the sample sites showed that SS1 had the lowest resistance pattern with ARPA of 0.106 while the remaining 3 sampled sites had 0.126 ARPA respectively. MAR phenotype pattern E-SUL-RF-TET-DOX with index of 0.38 was found to be the most common pattern to all the isolates across the sampling sites having being recorded in 8, 4,10 and 5 isolates in SS1 (Asejire), SS2 (Dandaru), SS3 (Eleyele) and SS4 (Erinle) respectively.
The MARindex of 97% of Vibrios from all the four rivers sampled were above 0.2 threshold value (Supplementary Table 3).
Antibiotics resistance genes profile among Vibrio isolates
The profiling of antibiotic resistance genes was carried out on all resistance phenotypes of Vibrio isolates25,26. The profile of resistance genes of Vibrios from the selected river water samples is as shown in Table 2. Among the 297 sulfonamide-resistant isolates assayed for resistance determinant genes, only 19% possessed the sulI gene, whereas 33% harbored sulII gene. However, 20 of the sulfonamide-resistant Vibrio isolates expressed dual sulI-sulII genes pattern (Table 3). Likewise, 237 tetracycline-resistant Vibrio isolates were examined for probable detection of three tetracycline resistance genes in which 28% were detected positive for tetA gene, 20% positive for tetE gene and 8% harbored tet39 gene respectively. Also, dual and multiple resistant patterns were observed in the order: tetA-tetE (16), tetE-tet39 (3), tetA-tet39 (1) and tetA-tetE-tet39 (1) (Table 3). Regarding the Beta-lactam antibiotics, the PCR amplification of 94-ampicillin-resistant Vibrio isolates revealed that 39% were ampC gene positive. Similarly, amoxicillin-resistant Vibrio isolates (141) were examined for probable detection of blaOXA and blaPSE genes. Both genes were spotted in 27% and 11% of the isolates respectively with dual blaPSE-blaOXA resistant pattern observed in 8 of the isolates (Table 3).
Among the 3 resistance genes encoding aminoglycosides, aacC2 genes account for 24% of the 25 gentamicin-resistant Vibrio isolates whereas 14% of the amikacin-resistant isolates carried the aphA1 gene while 39% of the streptomycin-resistant Vibrio isolates were identified to possess strA gene (Table 2). Overall, 391 resistance gene fingerprints were recovered across all sampled sites, with highest frequency recorded at SS1 (Asejire) with a total of 117 prints while SS4 had the lowest with 85 prints. Also, the occurrence of multiple antibiotics resistance genes ranged between 2 and 3 combinations with tet genes most occurring combinations followed by sul combination (Table 3). Generally, occurrence of resistance determinant follows the sequence: sulII > tetA > sulI > tetE > ampC > strA > blaOXA > blaPSE > tet39 > aacC2 > aphA1. The representative gel electrophoresis profile of antibiotic resistance determinant encoding the genes detected are as shown in Fig. 4 (A and B).
(A,B) Gel electrophoresis characteristic of antibiotics resistance genes in Vibrio Isolates. Lanes 1: Molecular marker (100 bp), lane 2: −ve control; lane 3: sulII (722 bp); lane 4: tetE (246 bp); lane 5: aphA1 (600 bp); lane 6: tetA (209 bp); lane 7: ampC (550 bp); lane 8: blaOXA (590); lane 9: sulI (822 bp); Lane 10: tet39 (711 bp); lane 11: strA (548 bp); lane 12: blaPSE (420 bp); lane 13: aacC2 (428).
Association between resistance determinants in Vibrio isolates
Statistical association was performed in order to determine probable relationship among resistance encrypting genes detected in Vibrio isolates and likewise to confirm if co-occurrence of a particular resistance genes could be established statistically. Significant associations were therefore established at P = 0.05. the following positive associations were observed: tetA was more strongly associated with sulI and tetE but less strongly with blaOXA and blaPSE. The tetE gene was also strongly associated sulII, sulI and blaOXA, but less strongly associated with tet39. Likewise, strA and aacC2 showed strong association with aphA1, while tet39 and sulI were less strongly associated with blaOXA (Table 4). There was no negative correlation between any of the genes.
Discussion
Pollution of water bodies also influence the microbial population and species of organisms found in them. Vibrio species are freshwater-associated organisms implicated in both human and animal infections. In this study, the incidence of Vibrio species was examined and 58% of the presumptive Vibrio spp. were confirmed after molecular confirmation, with Eleyele river (SS3) and Dandaru river (SS2) exhibiting the highest prevalence. Occurrence of high Vibrio prevalence in Erinle river suggested the presence of healthy Vibrio carriers who shed Vibrio into the environment on a regular basis. Although severe rainfall modifies the physical and chemical parameters in the aquatic environment, the subsequent influx of flood water together with organic waste (such as food, sewage, fertilizer and green waste as well as human and animal feces.) can aid Vibrio species proliferation in the chosen rivers. This finding is consistent with earlier studies that recorded high incidence of Vibrio species27,28. Furthermore, prior research has confirmed the link between Vibrio species and the aquatic environment6,25. The incidence of Vibrio spp. did not differ significantly between the sampled sites, according to statistical analysis.
Water ecosystems have become a well-known hotspot for environmental diffusion of both Vibrio species and antimicrobials29. Although, Vibrios are largely considered to be very susceptible to most antimicrobials recommended for medical treatment30, resistance among environmental isolates of Vibrio is increasingly being recorded in many countries.
Eighteen antibiotics from 9 different classes were examined in this investigation. Multiple antibiotics resistance was observed in the recovered Vibrio isolates against some endorsed and frequently used antibiotics in Vibrio-associated infection treatment which correlated with previous reports31. Remarkable resistance against erythromycin and sulfamethoxazole observed in this study is comparable with previous reports on clinical and environmental Vibrio isolates32,33. Likewise, relative resistance against the two tested β-lactams drugs (Amoxicillin 45% and amikacin 30%) recorded in this study is similar to that documented in environmental Vibrio isolates elsewhere34,35,36. The observed resistance of Vibrios to tested β-lactams in this study maybe related with the isolation source as bacteria such as Vibrio, recovered from water environment have been documented to show high resistance towards β-lactams class of antibiotics37,38. The resistance to amoxicillin and amikacin observed in this study is not surprising as these drugs enjoys widespread usage across different settings. Moreover, the frequency of antibiotics use over time has been hypothesized by researchers to contribute to development of resistance to antibiotics among bacteria36,39. Resistance to imipenem (18%) was also recorded in this study which corresponds to studies on Vibrio spp. recovered from shellfish and fish in Egypt33 and marine environment in Norway40. The recorded resistance to imipenem in this present study raises public health concerns as this drug is one of the major drugs of last resort for medical treatment of multiple drug-resistance Vibrio infections. Moreso, carbapenemases responsible for resistance to carbapenems has been associated with mobile genetic elements which can perhaps be transferred to humans through food interface41. Resistance recorded against tetracycline and cephalothin showed comparable profile with those stated in various studies across other countries42,43,44 and signifies public health threat as these are prescribed drugs in the treatment of humans infections42,45. High susceptibility to norfloxacin (98%), ciprofloxacin (93%), meropenem (91%), cefotaxime (88%), amikacin (76%), trimethoprim + sulfamethoxazole (75%), gentamicin (74%) recorded in this study is comparable to studies elsewhere46. Generally, varied resistance among antibiotics belonging to the sulfonamides, β-lactams, aminoglycosides, macrolides and tetracycline classes obtained in this study is similar to other findings22,35,47 in which similar findings of multiple antibiotic resistance of Vibrio spp. recovered from aquatic habitat have also been reported.
The high level of Vibrio isolate resistance to many of the antibiotics examined across all sample sites in this present study suggests a probable misuse or overuse of antimicrobials around the study area for a variety of purposes beyond what the environment can absorb, thereby resulting in the maintenance of unabsorbable residue in the water bodies. Such practices have been suggested to impact the likelihood of bacteria to develop resistance to commonly used antibiotics26. Furthermore, it has been hypothesized that the ability of microbes to share their genetic material in fresh and aquatic environments is a contributing factor to the rise in antibiotic resistance48. In light of the possible influence on treatment outcomes, the continued development of antimicrobial drug resistance to the standard oral treatments for Vibrio-associated diseases raises public health concerns6.
The degree of exposure of all the four rivers to contamination by antibiotics and its possible human health risk was assessed by the multiple antibiotic resistance index (MARI) which shows that the indexes range from 0.06 to 0.72 across all the sampled sites.
MARI of 0.06, 0.11 and 0.16 recorded in few of the Vibrio isolates recovered from SS2 (n = 2), SS3 (n = 4) and SS4 (n = 3) indicates low risk of environmental contamination. However, higher MARI values obtained across the sampling sites with highest (0.72) recorded in SS3 having exhibited resistance to 13 of the examined antibiotics indicates the Vibrio isolates have a high ability to contaminate the water sources; this is in accord with results documented in other findings25,42,43. MAR phenotype pattern E-SMX-RP-T-DXT with index of 0.38 was found to be the most common pattern among the isolates concurs with a study on Vibrio parahaemolyticus from shellfish in Selangor Malaysia in which resistance to five antibiotics was found prevalent among the isolates49. it was observed in this study that MARindex of 98% of Vibrio isolates from all the four rivers sampled greater than the threshold value. Similar findings of high MAR index of Vibrio spp. has been reported in aquatic environment elsewhere50,51,52. The high multiple resistance observed signifies a possible treat public health in the area of bacterial infection control and managements as these antibiotics are still the drug of choice in managing Vibrio related infection in both human and animal. According to many studies, the role of wastewater run-off in diffusion of antibiotic resistance in intestinal bacteria harboring resistant plasmids cannot be over-emphasized53.
Different samples recovered from environmental sources have been found to harbor diverse antimicrobial genes that encodes resistance to antibiotics such as aminoglycosides, β-lactams, tetracycline, chloramphenicol sulfonamides and trimethoprim25,48,54. In this present study, 11 different resistant genes were assayed on phenotypically resistant Vibrio isolates. Among the sulfonamide and tetracycline resistance genes tested, tetA (28%) and sulII (33%) had the highest frequency of occurrence. This outcome is similar to reports by55 in which tetA (64.28%) was among the highest detected genes in Vibrio parahaemolyticus isolated from Malaysian seawater and fishes56 and with high detection of sulII (53.8%) genes in Vibrio cholerae recovered from Hilsha fish.
Specifically, sulI gene is linked with class1 integron while sulll are typically found on large infectious multi-resistant or small non-conjugative plasmids which may contribute to the commonness of sulll detection in water environment. The detection of tetA and sulII genes advocates that they are extensively used antibiotics in animal and human therapy in this region. This is because tetracyclines and sulfonamides have broad spectrum of activities and are relatively cheap57. They are commonly incorporated into livestock feed as growth promoters in aquaculture. Likewise, the two classes of antibiotics are used in treatment of Vibrio associated infections in humans, as sulfonamide drugs are used as combination drug for cholera treatment in both children and adults57,58. This findings match reports of other studies where sulII gene detection was reported as the most profuse sulfonamide resistance conferring gene in freshwater environments25,59,60,61,62. Likewise, high tetA genes prevalence is consistent with63,64 reports in which the ribosomal efflux gene, tetA, have been found to be the most commonly detected resistance genes in Vibrio spp. from fresh environment attesting that tetA gene can be easily harbored by different class of bacteria due to their affinity for diverse host range.
Beta-lactams, an extensively used antibiotics in the management of several infections are usually distributed in freshwater environment and consequently pose substantial risk to environmental health. Three β-lactams genes blaOXA, blaPSE and ampC were examined. However, ampC had the highest frequency of recovery among all tested isolates. As stated in earlier studies, diverse pools of β-lactamases are existing in many environmental isolates of Vibrio species signifying that such acquirement of β-lactamase genes is not primarily due to β-lactamase treatment's selective pressure65. Some gram-negative environmental isolates have been found to naturally have low levels of β-lactam antibiotic resistance. It has been stated also that some intrinsic ampC resistance gene found in Enterobacteriaceae are positioned on mobile genetic element with the ability to transmit between diverse bacteria. Some other studies have also recorded high prevalence of ampC gene in gram negative bacteria from freshwater environment66.
Aminoglycosides are a type of antibiotic that has become increasingly significant in the management and prevention of severe bacterial infections in both animals and humans since resistance to diverse classes of antibiotics are increasing in gram negative bacteria. Among the different aminoglycosides genes that were examined, the str genes that bestows streptomycin phosphoryl transferase resistance, had more strA prevalence than others. These findings agree with similar studies in which high occurrence of strA detection were reported in aquatic bacteria67,68. Several means of acquiring resistance to aminoglycosides has been described; modification of enzymes such as neomycin phosphor-transferase, alteration of chromosome binding sites and altered aminoglycoside uptake/efflux pumps which bestows cross- resistance and change in cell membrane.
The correlation between the emergence of aminoglycoside resistance and the widespread use of antibiotics for cholera prevention and treatment as well as other usage is well known. The expression of multiple drug resistant by non-cholera Vibrio isolates to some of the antibiotics conventionally used in treatment of cholera calls for concern as this could have direct influence on the treatment of cholera cases in Southwestern Nigeria and other parts of the country where the organism could spread.
This study reiterated that antibiotic susceptibility and PCR assays are complementary. The PCR assay shows the existence of genes linked with resistance in many of the phenotypically resistant strains and absence of resistance conferring genes in some other strains that shows phenotypic resistance. As observed, tetA and sulII had the highest frequency of genotypic resistance occurrence while erythromycin, rifampin and sulphamethoxazole showed the most prevalent phenotypic resistance. Reasons for the observed discrepancy between genotypic and the phenotypic antibiotic resistance might be presence of expression profiles that were not subjected to testing due to limitations in selecting genes in this study as reported elsewhere69. Also, there is possibility that some of the ARGs in these Vibrios are unexpressed genes61. Consequently, these genes might be possible source of risk in the environment because apart from contributing to the resistance reservoir, they could also be horizontally transferred under suitable conditions. This suggests complexity of the linkage between phenotypes and genetic constructions. The results obtained were put through a statistical test to see if there was any correlation between the various resistance genes investigated, and high correlations were found among some of the resistance determinants. Notable comparable association between the resistance genes indicates tetA was strongly associated with sulI and both genes were the most detected among the three tetracycline and sulfonamide resistance genes tested. High proliferation of sulII than sulI genes in freshwater bacterial isolates is well documented26,70. This observation further affirms that resistance to antibiotics is triggered by more than two genes.
Antibiotic resistance development and persistent in the freshwater environment has been established to be initiated by various resistance mechanism such as selective pressure of antimicrobial substances71 horizontal gene transfer predispositions for genetic mutations, recombination events66 and the dissemination of antibiotic-resistant bacteria from human and veterinary medicine72. These mechanisms assist microorganisms to acclimatize to varying environments and contribute to the widely reported antibiotic resistance patterns observed in aquatic habitats. Furthermore, the genotypic resistance information can be conveyed via mobile genetic elements (plasmid, integron, transposon) from a particular species to different species (horizontal gene transfer)73,74. Also, antibiotics in the aquatic environment can lead to transformation in the ecological roles of microbes75 apart from encouraging the development and spreading of antibiotic resistant microorganisms.
Conclusion
The high concentrations of antibiotic resistant organisms and antibiotic resistant genes in the rivers examined indicated the rivers are polluted and highly impacted by waste discharge and other anthropogenic activities around the rivers. It is anticipated that unregulated waste disposal into the rivers and usage of antimicrobials in agriculture, clinical and domestic setting will keep on generating pools of resistance genes within the surface waters. Also, this can create a selective pressure in the immediate environment that would lead to genetic mutations in both allochthonous and autochthonous microorganisms thereby enabling them to persistently thrive and multiply. These observations therefore buttress the importance of environmental context-tailored strategies for continuous monitoring and surveillance of the aquatic environments for the presence of emerging contaminants such as, antibiotic resistance bacteria and their resistance determinant in order to evaluate their potential effects on environmental and human health.
Data availability
All data are listed in the main manuscript and other associated data are listed supplementary file.
References
Parte, A. C. LPSN—List of prokaryotic names with standing in nomenclature (Bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 68(6), 1825–1829. https://doi.org/10.1099/ijsem.0.002786 (2018).
Valáriková, J. et al. Potential pathogenicity and antibiotic resistance of aquatic Vibrio isolates from freshwater in Slovakia. Folia Microbiol. 65(3), 545–555. https://doi.org/10.1007/s12223-019-00760-w (2020).
Ceccarelli, D. et al. Non-O1/Non-O139 Vibrio cholerae carrying multiple virulence factors and V. cholerae O1 in the Chesapeake Bay, Maryland. Appl. Environ. Microbiol. 81(6), 1909–1918. https://doi.org/10.1128/AEM.03540-14 (2015).
da Silva, L. V., Ossai, S., Chigbu, P. & Parveen, S. Antimicrobial and genetic profiles of Vibrio vulnificus and Vibrio parahaemolyticus isolated from the Maryland Coastal Bays, United States. Front Microbiol. 12(May), 1–9. https://doi.org/10.3389/fmicb.2021.676249 (2021).
Reen, F. J., Almagro-Moreno, S., Ussery, D. & Boyd, E. F. The genomic code: Inferring Vibrionaceae niche specialization. Nat. Rev. Microbiol. 4(9), 697–704 (2006).
Adesiyan, I. M., Bisi-Johnson, M. A., Ogunfowokan, A. O. & Okoh, A. I. Occurrence and antibiogram signatures of some Vibrio species recovered from selected rivers in South West Nigeria. Environ. Sci. Pollut. Res. 28(31), 42458–42476. https://doi.org/10.1007/s11356-021-13603-4 (2021).
Bier, N., Schwartz, K., Guerra, B. & Strauch, E. Survey on antimicrobial resistance patterns in Vibrio vulnificus and Vibrio cholerae non-O1/non-O139 in Germany reveals carbapenemase-producing Vibrio cholerae in coastal waters. Front. Microbiol. 6, 1179. https://doi.org/10.3389/fmicb.2015.01179 (2015).
Hasan, N. A. et al. Deep-sea hydrothermal vent bacteria related to human pathogenic Vibrio species. Proc. Natl. Acad. Sci. 112(21), E2813–E2819 (2015).
Park, K. et al. Food-borne outbreaks, distributions, virulence, and antibiotic resistance profiles of Vibrio parahaemolyticus in Korea from 2003 to 2016: A review. Fish Aquat. Sci. https://doi.org/10.1186/s41240-018-0081-4 (2018).
Labella, A. et al. High incidence of antibiotic multi-resistant bacteria in coastal areas dedicated to fish farming. Mar. Pollut. Bull. 70(1–2), 197–203 (2013).
Christaki, E., Marcou, M. & Tofarides, A. Antimicrobial resistance in bacteria: Mechanisms, evolution, and persistence. J. Mol. Evol. 88(1), 26–40 (2020).
Adesiyan, I. M., Bisi-johnson, M. A., Ogunfowokan, A. O. & Okoh, A. I. Science of the total environment incidence and antimicrobial susceptibility fingerprints of Plesiomonas shigelliodes isolates in water samples collected from some freshwater resources in Southwest Nigeria. Sci. Total Environ. 665, 632–640. https://doi.org/10.1016/j.scitotenv.2019.02.062 (2019).
Jeamsripong, S., Khant, W. & Chuanchuen, R. Distribution of phenotypic and genotypic antimicrobial resistance and virulence genes in Vibrio parahaemolyticus isolated from cultivated oysters and estuarine water. FEMS Microbiol. Ecol. 96(8), 1–10. https://doi.org/10.1093/femsec/fiaa081 (2020).
Bergeron, S., Raj, B., Nathaniel, R., Corbin, A. & Lafleur, G. Presence of antibiotic resistance genes in raw source water of a drinking water treatment plant in a rural community of USA. Int. Biodeterior. Biodegrad. https://doi.org/10.1016/j.ibiod.2017.05.024 (2017).
Saeed, D. K. et al. Readiness for antimicrobial resistance (AMR) surveillance in Pakistan; a model for laboratory strengthening. Antimicrob. Resist. Infect. Control. 6(1), 1–7 (2017).
Hendriksen, R. S. et al. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat. Commun. https://doi.org/10.1038/s41467-019-08853-3 (2019).
Kwok, A. Y. C. et al. Phylogenetic study and identification of human pathogenic Vibrio species based on partial hsp60 gone sequences. Can. J. Microbiol. 48(10), 903–910. https://doi.org/10.1139/W02-089 (2002).
Testing S. Clsi. (2019) https://doi.org/10.1007/978-3-662-48986-4_300418.
Krumperman, P. H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46(1), 165–170 (1983).
Mok, J. S. et al. Distribution and antimicrobial resistance of Vibrio parahaemolyticus isolated from fish and shrimp aquaculture farms along the Korean coast. Mar. Pollut. Bull. https://doi.org/10.1016/j.marpolbul.2021.112785 (2021).
Titilawo, Y., Sibanda, T. & Okoh, A. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of faecal contamination of water. Environ Sci. Pollut. Res. 22(14), 10969–10980. https://doi.org/10.1007/s11356-014-3887-3 (2015).
Deng, Y. et al. Prevalence, virulence genes, and antimicrobial resistance of Vibrio species isolated from diseased marine fish in South China. Sci. Rep. 10(1), 1–8. https://doi.org/10.1038/s41598-020-71288-0 (2020).
Ekundayo, T. C. & Okoh, A. I. Antimicrobial resistance in freshwater Plesiomonas shigelloides isolates: Implications for environmental pollution and risk assessment. Environ. Pollut. https://doi.org/10.1016/j.envpol.2019.113493 (2019).
Amos, G. C. A. et al. The widespread dissemination of integrons throughout bacterial communities in a riverine system. ISME J. 12(3), 681–691. https://doi.org/10.1038/s41396-017-0030-8 (2018).
Onohuean, H., Okoh, A. I. & Nwodo, U. U. Antibiogram signatures of Vibrio species recovered from surface waters in South Western districts of Uganda: Implications for environmental pollution and infection control. Sci. Total Environ. 807, 150706. https://doi.org/10.1016/j.scitotenv.2021.150706 (2022).
Titilawo, Y., Obi, L. & Okoh, A. Antimicrobial resistance determinants of Escherichia coli isolates recovered from some rivers in Osun State, South-Western Nigeria: Implications for public health. Sci. Total Environ. 523, 82–94. https://doi.org/10.1016/j.scitotenv.2015.03.095 (2015).
Mookerjee, S., Batabyal, P., Sarkar, M. H. & Palit, A. Seasonal prevalence of enteropathogenic Vibrio and their phages in the riverine estuarine ecosystem of south Bengal. PLoS ONE https://doi.org/10.1371/JOURNAL.PONE.0137338 (2015).
Okoh, A. I. & Sibanda, T. Prevalence and characterisation of non-cholerae Vibrio spp. in final effluents of wastewater treatment facilities in two districts of the Eastern Cape Province of South Africa: Implications for public health. Environ. Sci. Pollut. Res. https://doi.org/10.1007/s11356-014-3461-z (2014).
Huijbers, P. M. C., Flach, C. F. & Larsson, D. G. J. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 130, 104880. https://doi.org/10.1016/j.envint.2019.05.074 (2019).
Letchumanan, V., Yin, W., Lee, L. & Chan, K. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6, 1–12. https://doi.org/10.3389/fmicb.2015.00033 (2015).
Shaw, K. S. et al. Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Chesapeake Bay and Maryland Coastal Bays. PLoS ONE 9(2), 1–11. https://doi.org/10.1371/journal.pone.0089616 (2014).
Abana, D. et al. Investigating the virulence genes and antibiotic susceptibility patterns of Vibrio cholerae O1 in environmental and clinical isolates in Accra, Ghana. BMC Infect Dis. 19(1), 1–10. https://doi.org/10.1186/s12879-019-3714-z (2019).
Sadat, A., El-Sherbiny, H., Zakaria, A., Ramadan, H. & Awad, A. Prevalence, antibiogram and virulence characterization of Vibrio isolates from fish and shellfish in Egypt: A possible zoonotic hazard to humans. J. Appl. Microbiol. https://doi.org/10.1111/jam.14929 (2021).
Jiang, Y. et al. Antimicrobial resistance, virulence and genetic relationship of Vibrio parahaemolyticus in seafood from coasts of Bohai Sea and Yellow Sea, China. Int. J. Food Microbiol. 290, 116–124. https://doi.org/10.1016/j.ijfoodmicro.2018.10.005 (2019).
Xie, T., Yu, Q., Tang, X., Zhao, J. & He, X. Prevalence, antibiotic susceptibility and characterization of Vibrio parahaemolyticus isolates in China. FEMS Microbiol. Lett. 367(16), fnaa136 (2020).
Sony, M. et al. Antimicrobial resistance and virulence characteristics of Vibrio vulnificus, Vibrio parahaemolyticus and Vibrio harveyi from natural disease outbreaks of marine/estuarine fishes. Aquaculture 539, 736608. https://doi.org/10.1016/j.aquaculture.2021.736608 (2021).
Elmahdi, S., Dasilva, L. V. & Parveen, S. Antibiotic resistance of vibrio parahaemolyticus and Vibrio vulnificus in various countries: A review. Food Microbiol. https://doi.org/10.1016/j.fm.2016.02.008 (2016).
Mohamad, N. et al. Virulence-associated genes and antibiotic resistance patterns of Vibrio spp. isolated from cultured marine fishes in Malaysia. BMC Vet. Res. 15(1), 1–13. https://doi.org/10.1186/s12917-019-1907-8 (2019).
Baker-Austin, C. et al. Vibrio spp. infections. Nat. Rev. Dis. Prim. https://doi.org/10.1038/S41572-018-0005-8 (2018).
Håkonsholm, F. et al. Vibrios from the Norwegian marine environment: Characterization of associated antibiotic resistance and virulence genes. Microbiologyopen. 9(9), 1–19. https://doi.org/10.1002/mbo3.1093 (2020).
Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Ther. Adv. Infect. Dis. 3(1), 15–21 (2016).
Ahmed, H. A. et al. Molecular characterization, antibiotic resistance pattern and biofilm formation of Vibrio parahaemolyticus and V. cholerae isolated from crustaceans and humans. Int. J. Food Microbiol. 274, 31–37. https://doi.org/10.1016/j.ijfoodmicro.2018.03.013 (2018).
Ashrafudoulla, M. et al. Genetic relationship, virulence factors, drug resistance profile and biofilm formation ability of Vibrio parahaemolyticus isolated from mussel. Front. Microbiol. https://doi.org/10.3389/fmicb.2019.00513 (2019).
Xu, X., Cheng, J., Wu, Q., Zhang, J. & Xie, T. Prevalence, characterization, and antibiotic susceptibility of Vibrio parahaemolyticus isolated from retail aquatic products in. BMC Microbiol. 100, 1–9. https://doi.org/10.1186/s12866-016-0650-6 (2016).
Xie, T., Wu, Q., Zhang, J., Xu, X. & Cheng, J. Comparison of Vibrio parahaemolyticus isolates from aquatic products and clinical by antibiotic susceptibility, virulence, and molecular characterisation. Food Control 71, 315–321. https://doi.org/10.1016/j.foodcont.2016.06.046 (2017).
Mok, J. S., Ryu, A., Kwon, J. Y., Kim, B. & Park, K. Distribution of Vibrio species isolated from bivalves and bivalve culture environments along the Gyeongnam coast in Korea: Virulence and antimicrobial resistance of Vibrio parahaemolyticus isolates. Food Control 106, 106697. https://doi.org/10.1016/j.foodcont.2019.06.023 (2019).
Okoh, A. I. & Igbinosa, E. O. Antibiotic susceptibility profiles of some Vibrio strains isolated from wastewater final effluents in a rural community of the Eastern Cape Province of South Africa. BMC Microbiol. 10, 143. https://doi.org/10.1186/1471-2180-10-143 (2010).
Beshiru, A., Okareh, O. T., Okoh, A. I. & Igbinosa, E. O. Detection of antibiotic resistance and virulence genes of Vibrio strains isolated from ready-to-eat shrimps in Delta and Edo States, Nigeria. J. Appl. Microbiol. https://doi.org/10.1111/jam.14590 (2020).
Letchumanan, V., Yin, W. F., Lee, L. H. & Chan, K. G. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. https://doi.org/10.3389/FMICB.2015.00033 (2015).
Igbinosa, E. O. & Odjadjare, E. E. O. Characterization of antibiogram susceptibility profile of Vibrio species isolated from fresh vegetables. Afr. Sci. 17(2), 147–152 (2016).
Silvester, R. & Alexander, D. Prevalence, antibiotic resistance, virulence and plasmid profiles of Vibrio parahaemolyticus from a tropical estuary and adjoining traditional prawn farm along the southwest coast of India. Ann. Microbiol. https://doi.org/10.1007/s13213-015-1053-x (2015).
You, K. G., Bong, C. W. & Lee, C. W. Antibiotic resistance and plasmid profiling of Vibrio spp. in tropical waters of Peninsular Malaysia. Environ. Monit. Assess. https://doi.org/10.1007/s10661-016-5163-0 (2016).
Akhter, A., Imran, M. & Akhter, F. Determination of multiple antibiotic resistance patterns and indexing among metal tolerant β-lactamase-producing Escherichia coli. Afr. J. Microbiol. Res. 8(7), 619–627. https://doi.org/10.5897/AJMR2013.6417 (2014).
Hossain, S., Wickramanayake, M. V. K. S., Dahanayake, P. S. & Heo, G. J. Occurrence of virulence and extended-spectrum β-lactamase determinants in Vibrio spp. isolated from marketed hard-shelled mussel (Mytilus coruscus). Microb. Drug Resist. 26(4), 391–401. https://doi.org/10.1089/mdr.2019.0131 (2020).
Faja, O. M. et al. Isolation, detection of virulence genes, antibiotic resistance genes, plasmid profile, and molecular typing among Vibrio parahaemolyticus isolated in Malaysian seawater from recreational beaches and fish. Vet. World. 12(7), 1140–1149. https://doi.org/10.14202/vetworld.2019.1140-1149 (2019).
Hossain, Z. Z., Farhana, I., Tulsiani, S. M., Begum, A. & Jensen, P. K. M. Transmission and toxigenic potential of Vibrio cholerae in hilsha fish (Tenualosa ilisha) for human consumption in Bangladesh. Front. Microbiol. 9, 222 (2018).
Granados-Chinchilla, F. & Rodríguez, C. Tetracyclines in food and feedingstuffs: from regulation to analytical methods, bacterial resistance, and environmental and health implications. J. Anal. Methods Chem. 2017 (2017).
Gao, P., Munir, M. & Xagoraraki, I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 421–422, 173–183. https://doi.org/10.1016/j.scitotenv.2012.01.061 (2012).
Titilawo, Y., Obi, L. & Okoh, A. Science of the total environment antimicrobial resistance determinants of Escherichia coli isolates recovered from some rivers in Osun State, South-Western Nigeria: Implications for public health. Sci. Total Environ. 523, 82–94. https://doi.org/10.1016/j.scitotenv.2015.03.095 (2015).
Zhang, S. H., Lv, X., Han, B., Gu, X. & Wang, P. F. Prevalence of antibiotic resistance genes in antibiotic-resistant Escherichia coli isolates in surface water of Taihu Lake. Environ. Sci. Pollut. Res. https://doi.org/10.1007/s11356-015-4371-4 (2015).
Xu, Y. et al. Occurrence and distribution of antibiotics, antibiotic resistance genes in the urban rivers in Beijing, China. Environ. Pollut. 213, 833–840. https://doi.org/10.1016/j.envpol.2016.03.054 (2016).
Yuan, Y. et al. Isolation, identification, and resistance gene detection of Vibrio harveyi from Scophthalmus maximus. Aquac. Int. 29(5), 2357–2368. https://doi.org/10.1007/s10499-021-00752-z (2021).
Tan, C. W. et al. Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from short mackerels (Rastrelliger brachysoma) in Malaysia. Front. Microbiol. 8, 1087 (2017).
Hu, Y., Li, F., Zheng, Y., Jiao, X. & Guo, L. Isolation, molecular characterization and antibiotic susceptibility pattern of Vibrio parahaemolyticus from aquatic products in the southern Fujian Coast, China. J. Microbiol. Biotechnol. 30(6), 856–867. https://doi.org/10.4014/jmb.2001.01005 (2020).
Avison, M. B., Bennett, P. M. & Walsh, T. R. β-Lactamase expression in Plesiomonas shigelloides. J. Antimicrob. Chemother. 45, 877–880 (2000).
Alexander, J., Bollmann, A., Seitz, W. & Schwartz, T. Science of the total environment microbiological characterization of aquatic microbiomes targeting taxonomical marker genes and antibiotic resistance genes of opportunistic bacteria. Sci. Total Environ. 512–513, 316–325. https://doi.org/10.1016/j.scitotenv.2015.01.046 (2015).
Okoh, A. I. & Igbinosa, E. O. Antibiotic susceptibility profiles of some Vibrio strains isolated from wastewater final effluents in a rural community of the Eastern Cape Province of South Africa. BMC Microbiol. 10, 1–6 (2010).
Baron, S. et al. Antimicrobial susceptibility of autochthonous aquatic Vibrio cholerae in Haiti. Front. Microbiol. 7, 1671 (2016).
Chen, S. et al. A DNA microarray for identification of virulence and antimicrobial resistance genes in Salmonella serovars and Escherichia coli. Mol. Cell Probes. 19(3), 195–201. https://doi.org/10.1016/j.mcp.2004.11.008 (2005).
Gao, P. et al. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 46(7), 2355–2364. https://doi.org/10.1016/j.watres.2012.02.004 (2012).
Seiler, C. & Berendonk, T. U. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 3, 1–10. https://doi.org/10.3389/fmicb.2012.00399 (2012).
Cantas, L. et al. A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front Microbiol. 4, 1–14. https://doi.org/10.3389/fmicb.2013.00096 (2013).
Martinez, J. L. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc. R. Soc. B Biol. Sci. https://doi.org/10.1098/rspb.2009.0320 (2009).
Jechalke, S. et al. Widespread dissemination of class 1 integron components in soils and related ecosystems as revealed by cultivation-independent analysis. Front. Microbiol. https://doi.org/10.3389/FMICB.2013.00420/ABSTRACT (2013).
Ding, C. & He, J. Effect of antibiotics in the environment on microbial populations. Appl. Microbiol. Biotechnol. 87(3), 925–941. https://doi.org/10.1007/S00253-010-2649-5 (2010).
Acknowledgements
We are grateful to the South African Medical Research Council for financial support. Organization for Women in Science for the Developing World (OWSD) and Swedish International Development Cooperation Agency (SIDA) provided research funding through the award of PhD fellowship. University of Fort Hare appreciated for providing laboratory access, equipment and financial support.
Author information
Authors and Affiliations
Contributions
Study design and coordination: I.A., A.O., and M.B. Sampling and Molecular analysis: I.A. Manuscript draft: A.I. Review and editing: A.O. and M.B. Final approval of manuscript: I.A., A.O., and M.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Adesiyan, I.M., Bisi-Johnson, M.A. & Okoh, A.I. Incidence of antibiotic resistance genotypes of Vibrio species recovered from selected freshwaters in Southwest Nigeria. Sci Rep 12, 18912 (2022). https://doi.org/10.1038/s41598-022-23479-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-23479-0
- Springer Nature Limited
This article is cited by
-
The prevalence, antimicrobial susceptibility, virulence, and antimicrobial resistance genes of multidrug-resistant Vibrio parahaemolyticus recovered from Oreochromis niloticus
Aquaculture International (2024)
-
Investigation and detection of multiple antibiotic-resistant pathogenic bacteria in municipal wastewater of Dhaka city
Discover Water (2024)