Abstract
The emergence of antibiotic-resistant bacteria has limited treatment options and led to the untreatable infections, thereby necessitating the discovery of new antibiotics to battel against bacteria. Natural products from endophytic actinobacteria (EA) serve as a reservoir for discovery of new antibiotics. Therefore, the current study focused on the isolation and antibacterial properties of EA isolated from Luffa cylindrica. Six strains were identified using morphological characterization, SEM analyses and 16S rRNA gene sequencing from the roots and leaves of the plant. They were taxonomically classified as Streptomycetaceae family. This is the first report on EA form L. cylindrica. The strains produced a chain of oval, cubed or cylindrical shaped spores with spiny or smooth surfaces. Three strains; KUMS-B3, KUMS-B4 and KUMS-B6 were reported as endophytes for the first time. Fifty percent of isolates were isolated from leaves samples using YECD medium. Our results showed that the sampling time and seasons may affect the bacterial diversity. All six strains had antibacterial activity against at least one of the tested bacteria S. aureus, P. aeruginosa, and E. coli. Among the strains, KUMS-B6 isolate, closely related to S. praecox, exhibited the highest antibacterial activity against both gram-positive and negative bacteria. KUMS-B6, KUMS-B5 and KUMS-B4 isolates strongly inhibited the growth of P. aeruginosa. Interestingly, the strains, isolated from leaves exhibited stronger antagonist activities compared to those isolated from the roots. The study revealed that the isolated strains from Luffa produce a plethora of bioactive substances that are potential source of new drug candidates for the treatment of infections.
Similar content being viewed by others
Introduction
Despite the amazing therapeutic effects of antibiotics against infectious diseases, the emergence of drug-resistant microorganisms has become a serious problem1,2. The baseless prescribing of antibiotics in addition to the inappropriate and arbitrary use of antibiotics has led to antibiotic-resistant bacteria, thus posing a significant challenge to prevention and treatment of infectious diseases. In the United States, over 2 million people are infected with antibiotic-resistant bacteria every year, causing 35,000 deaths annually. It is estimated that the global annual mortality rate due to antibiotic resistance will exceed 10 million by 20503. Moreover, the emergence of pandrug resistance, a phenomenon characterized by bacteria becoming resistant to all classes of antibiotics, is also a growing concern4. Therefore, prevention and control of these microorganisms requires new strategies such as discovery and development of new drugs. Natural product screening is considered as the most promising line for novel antibiotic discovery, which is required to counteract the loss of effectiveness of the presently available antibiotics5. Plant-associated microorganisms have been widely used as sources of bioactive compounds for therapeutic purposes for many years6,7. Endophytic microorganisms reside inside plant tissues without causing obvious symptoms and damage and may include various groups such as fungi and bacteria, including actinobacteria8. Studies suggest a symbiotic interaction between host plants and endophytes in which the host provides shelter and nutrition, while endophytes act as chemical guards, promote plant growth, improve the host's tolerance for abiotic and biotic stresses, and increase disease resistance9. In addition, these endophytic actinobacteria may play crucial roles in maintenance and sustenance of specific natural habitats, but these endeavors call upon extensive scientific investigation to develop the necessary knowledge base8,10. Actinobacteria are generally free-living, Gram-positive filamentous bacteria with a high GC DNA content ranging from 50% to over 70%, found in aquatic and terrestrial habitats9. Over the last decades, actinobacteria, especially the genus Streptomyces, have been known to produce more than two-thirds of natural antibiotics (such as beta-lactams and tetracyclines). The discovery and use of antibacterials produced by actinomycetes is a new approach to deal with drug-resistant pathogens11. Fifty percent of 22,000 bioactive secondary metabolites of microbial origin is related to actinobacteria. Currently, about 160 antibiotics are used in the treatment of human infections and in agricultural industry12. Actinobacteria are known to be prolific producers of a wide range of bioactive natural products (NPs), and scientific evidence highlights their important protective role against diseases like colorectal cancer (CRC)13 and lung cancer14. Secondary metabolites of 17 Actinobacteria strains isolated from Antarctica had promising antitumor effects15. They also possess the potential to produce unique secondary metabolites, which can be exploited in pharmaceutical, agricultural and other industries16. Endophytic actinobacteria are critical resource of microbial biodiversity17,18. The taxonomic status of endophytic actinobacteria associated with medicinal plants has been investigated in several studies17,19. According to previous studies, almost all plants harbor endophytes. Endophytic bacteria can colonize various parts of the plant, including stems, roots, petioles, leaves, weed inflorescences, fruits, buds, seeds, along with dead and hollow hyaline cells. However, specificity and developmental stage of the host, geographical conditions and microbial diversity are variable factors that influence colonization of endophytes in different plants8. Search of particular ecological niches along with new methods of isolation of novel genera/species of actinobacteria may result in the identification of new gene clusters, and finally, new products8. Hence, the aim of present study was to explore the endophytic actinobacteria inhabiting special niches for discovering hitherto unexploited strains producing bioactive compounds. These strains can be used as an alternative source for the production of new antibiotics against pathogenic bacteria. For this purpose, Luffa cylindrica plant collected from different parts of Kerman, Iran. L. cylindrica is a medicinal plant utilized in traditional medicine for the treatment of cough, general body pain, asthma, tuberculosis, shingles, and boils. The fruits are anthelmentic, carminative, laxative, depurative, emollient, expectorant, tonic, and galactagogue and are useful in fever, syphilis, tumours, bronchitis, splenopathy and leprosy. This plant is mostly specific to the subtropical regions of Asia and Africa. Its fruits and leaves contain triterpenoid saponins20,21.
Many researchers have studied the antimicrobial properties of plant's endophytic actinobacteria22,23. There are plenty of reports on EA and their antibacterial activity against pathogenic bacteria. Their findings show a promisingly high probability of discovering a novel compound with different modes of action.
A few endophytic fungi including Phialemoniopsis endophytica sp have been reported from L. cylindrica24. However, to our knowledge, no study has been reported on the population of endophytic actinobacteria associated with L. cylindrica and this is the first report. Therefore, the aims of this study were to isolate and identify the endophytic actinobacteria from L. cylindrica, thus tapping into a still unexploited potential source of beneficial bacterial metabolites, and to determine their antimicrobial capability against the pathogenic bacteria; Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli.
Results
Isolation of endophytic actinobacteria
The surface sterilized samples were cultured on four different media and six strain having morphological characteristics of actinobacteria were isolated from different section of plant after 4–8 weeks of incubation onto TWYE medium at 28 °C. Figure 1 indicates actinobacterial hyphae rising from leaf fragments after 6 weeks of incubation onto YECD medium. The arrows show the growth of colonies from the leaves part of Luffa.
Six strains of endophytic actinobacteria, based on the morphological characteristics, were isolated from L. cylindrica using four different media. Three isolates (KUMS-B1, KUMS-B2, KUMS-B3) were isolated from the roots using TWYE culture medium and the other three isolates (KUMS-B4, KUMS-B5 and KUMS-B6) were isolated from the leaves using YECD culture medium. However, no actinobacteria growth was discerned on PDA or HPDA media. This is the first time to reported KUMS-B6 isolate as an endophyte based on morphological, microscopical features and 16S rDNA gene sequencing (Table 1). The isolates were cultivated onto ISP2 and their cultural and physiological features were determined using International Streptomyces Project (ISP2)25,26, used for characterization of strains.
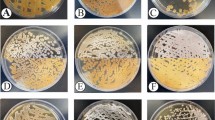
Six endophytic actinomycetes were isolated from either root or leaf tissues, whereas no actinobacteria was isolated from fruits, stems and flowers. They were described based on colonial morphology onto ISP2, ability to generate aerial hyphae and substrate mycelia, and pigmentation as summarized in Table 1. The presence of aerial mycelium, spore mass colour, distinctive reverse colony colour, diffusible pigment, and sporophore and spore chain shape were studied based on the ISCC-NBS centroid colour system27. The actinobacteria colonies have white, creamy, pale yellow or brown to white brown and reddish colors after two weeks of incubation onto ISP-2, as demonstrated in Fig. 2. The morphological and structural characteristics of the isolates are listed in Table 1.
Morphological characteristics of six endophytic actinobacteria isolates from Luffa cylindrica, grown onto ISP2 after 2 weeks of incubation at 28 °C. KUMS-B1 isolate; cream–brown colony (A): KUMS-B2 isolate; grey–creamy watery colony (B) KUMS-B3 isolate; creamy-brownish colony (C): KUMS-B4 isolate; creamy–light yellowish colony (D): KUMS-C5 isolate; creamy–yellowish colony (E): KUMS-B6 isolate; small white–brownish colony (F).
The morphology of spore shape, surface, and spore structure are important criteria for identifying actinobacteria. The morphology of spore chains and mycelia were also studied using SEM after two weeks growth on ISP2 medium at 28 °C. Scanning electron micrograph of mycelia and spores presenting the spore-chain morphology and spore-surface ornamentation of strains at different magnification are demonstrated in Fig. 3. The SEM results revealed that the aerial mycelia produced spore chains that were cuboid- spiral, cylindrical shaped, the branched and/or spiral or rod-shaped and/or long aerial mycelia and spore chains as demonstrated in Fig. 3.
Scanning electron micrograph of mycelia and spores representing the spore-chain morphology and spore-surface ornamentation of strains and mycelia grown on ISP2 medium for 14 days at 28 °C at different magnification. Streptomyces koyangensis KUMS-B1 (oval and ovoid shape with smooth surfaces) (A); S. griseorubens KUMS-B2 isolate (flexuous spiral spore chains, oval and ovoid shape with warty, hairy or rough and coarse surfaces) (B); S. intermedius KUMS-B3 (straight spore chains, oval and ovoid shape with a warty, spiny or hairy surfaces) (C); S. microflavus KUMS-B4 (spores are cylindrical cube-shapes with indenting surfaces, and fragmented) (D); S. cavourensis KUMS-B5 (spores have cylindrical shapes with indenting surfaces) (E); S. praecox KUMS-B6 (The spores have cylindrical shapes with smooth surfaces) (F).
Figure 3 shows spore-chain morphology and spore surface ornamentations of isolates after two weeks of incubation on ISP2 medium at 28 °C. They were clustered of single spores or spore chains. Spores of some actinobacteria had hairy surface and some had rugose or spiny surfaces. Generally, the spores were of oval and ovoid shape and had smooth surface (Fig. 3A), or coarse surfaces (Fig. 3B and C), but globes and rod-shaped spores, or cube shape such as KUMS-B4 (Fig. 3D) were also frequently found. The spores have cylindrical cube-shapes with indenting surfaces, which form in long straight branched chain ornamentations (Fig. 3D). Spores with spiny surface were observed with variable spine sizes (Fig. 3B and C). The spore chain of some strains such as KUMS-B2 and KUMS-B3 have hairy surface. Some are spiny and spiral chain such as KUMS-B2 (Fig. 3B). The spiral or flexuous spore chains were simple, open, loose coils of one to two turns, but some are hairy and in a long straight chain and branched ornamentations (Fig. 3C). Spore of some strains like KUMS-B1 have smooth surface. Strain KUMS-B2 showed a spore chain morphology of exhibiting spiral morphology.
Molecular identification
PCR results
The genomic DNA was extracted, and 16S rDNA amplification performed to identify endophyte actinobacterial isolates. The results of the amplification of 16S rDNA fragments on 1.5% agarose gel electrophoresis are shown in Fig. 4. The original image of PCR gel electrophoresis is shown in Supplementary Fig. 1.
Gel electrophoresis of PCR products using 27F and 1492R primers. The 1.5 Kb amplified 16S rDNA fragments of isolated actinobacteria monitored on a 1.5% agarose gel. RCR product of 16S rRNA sequence of isolates on a 1.5% agarose gel from left to right: KUMS-B4, KUMS-B5, KUMS-B6, positive control, negative control and Ladder.
16S rDNA gene sequencing
Endophytic actinobacteria strains were also identified by sequencing 16S rRNA gene segments. The sequences of isolates were aligned with those sequences of bacteria available in the datasets using the ClustalW and determined their similarity values. The presumptive relationships of these sequences were obtained from database comparison.
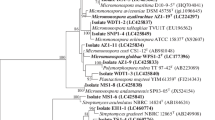
The phylogenetic tree of the isolates was constructed based on the neighbour-joining algorithm with MEGA. The phylogenetic relationship of isolates based on 16S rRNA gene sequences is shown in Fig. 5. Based on 16S rRNA gene alignments and phylogenetic studies of isolates, they were categorized into one main cluster (Fig. 5). All identified isolates belonged to the Streptomyces genus, Streptomycetaceae family. They all belong to one distinct family within the Phylum Actinobacteria. According to our analyses the similarity of all isolates' 16 s rDNA sequences was more than 99% when compared to those of data presented in the dataset. It was possible to assign all the isolates to a species. 16S rRNA gene sequencing revealed that endophytic actinobacteria represented six species; S. koyangensis, S. griseorubens, S. intermedius, S. microflavus, S. cavourensis, S. praecox (Fig. 5). The similarity, homology and accession numbers of the isolates with their closely related strains are summarized in Table 1. Among the examined isolates, KUMS-B4 had 100% similarity to S. microflavus NBRC 13062 in EzBioCloud and GenBank databases. The isolates had high percentage of similarity with their closely related strains, < 99% similarity to the existing database sequences of 16S rRNA genes. All the 16S rRNA gene sequences were submitted to GenBank and were assigned GenBank accession numbers ON626444, ON626445, ON626446, ON626447, ON626448, and ON626449.
Phylogenetic tree of endophytic actinobacteria isolated from Luffa cylindrica constructed by the neighbour-joining method demonstrating their closely related type strains based on partial 16S rRNA gene sequences generated by MEGA X. The numbers at branch nodes show levels of bootstrap support (%) from 1000 replicates and scale bar refers to a phylogenetic distance of 0.02 nucleotide substitutions per site. Kitasatospora setae KM -6054 was used as outgroup.
Antibacterial effects of EA strains on pathogenic bacteria in solid-state culture
The set of test bacteria was used to determine the spectrum of antibiotic activity of strains. The antibacterial activity of ethyl acetate and methanolic extracts of the isolates, inoculated onto the solid culture was tested against Gram-positive and Gram-negative bacteria including S. aureus 25923, P. aeruginosa 27853, and E. coli 25922 using well diffusion assay. The results showed that the crude extract of the endophytic actinobacteria isolated from L. cylindrica were able to inhibit the growth of at least one of the test bacteria (Table 2). The test isolates displayed antibacterial activity against at least one of the target bacteria. In general, all strains had an inhibitory effect on at least one test bacteria. On the other hand, inhibitory activity was observed against all tested pathogens. Some strains showed antagonist activity against both gram-positive and gram-negative bacteria. The KUMS-B4, KUMS-B5 and KUMS-B6 strains showed a broad spectrum of antibiotic properties against all test bacteria, and demonstrated the largest inhibition zone against P. aeruginosa as compared to all isolated strains. Interestingly, all these strains were isolated from leaves. Ethyl acetate and methanolic extracts of KUMS-B4, KUMS-B5 and KUMS-B6 strains had inhibitory effects on the three tested pathogens. Ethyl acetate extracts of these three isolates showed the same inhibitory activity against S. aureus. Ethyl acetate extract of KUMS-B4 strain also showed a moderate effect on E. coli 25922, while KUMS-B5 and KUMS-B6 showed a stronger activity as compared to this strain. Methanolic extract of KUMS-B5 had a strong effect on P. aeruginosa 27853.
The KUMS-B6 strain showed the strongest activity against target bacteria as compared to other isolates. Interestingly, the ethyl acetate and methanolic extracts of KUMS-B5 and KUMS-B6 strains had strong inhibitory effects against P. aeruginosa 27853. Ethyl acetate extract of KUMS-B4 was highly potent against P. aeruginosa 27853 and S. aureus 25923, while its methanolic extract had no a significant inhibitory activity against these bacteria. KUMS-B6 was active against all target pathogens making it as a promising source of broad-spectrum antibiotics that should be investigated further, while KUMS-B1 strain showed the weakest antimicrobial property so that only methanolic crude extract of this strain demonstrated a weak antimicrobial property against only one bacterium tested (E. coli 25922). Methanolic extract of KUMS-B1 strain was the only effective extract from this strain against E. coli 25922 and had no effect against other test pathogens. KUMS-B1 strain was shown the weakest antibacterial activity among 6 endophytic strains. From our findings, it seems that S. aureus was the most frequently inhibited target bacterium. No test bacteria showed an inhibitory zone with ethyl acetate extract of KUMS-B1 and KUMS-B2. This might indicate that there are no bioactive compounds in the extracts, having biocidal activities against test bacteria.
Discussion
The antibiotic resistance is a crucial public health concern. Natural products have always been an important course of bioactive compounds namely antibiotics. They can be used to counteract the antibiotic resistance. Due to the increasing prevalence of multiple drug resistant pathogens (MDR) and widespread drug resistance (XDR) and problems related to the treatment and management of diseases caused by "ESKAPE" pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pnumoniae, Acinetobacter baumanni, Pseudomonas aeruginosa, and Enterobacter species), the development of new drugs is increasingly urgent8,28,29. Therefore, the use of diverse and newly endophytic actinobacteria isolated from particular habitats offers unique opportunities to extract new drugs and therapies. It is fascinating to explore and understand the biology and chemistry of endophytic actinobacteria and their host plants in order to develop new, non-destructive applications for human health, agriculture, and the environment8,9. Although the genus Streptomyces has been reported more frequently, the non-Streptomyces genus has also been reported as endophytes in different plants from harsh habitats8. Actinobacteria are one of the most important reservoirs of secondary metabolites with beneficial biological activities and also, one of the main manufacturers of common antibiotics30. The therapeutic properties of some of these secondary metabolites strongly suggest the abundance of potential new drug molecules in the vast chemical repertoire of endophytic actinobacteria that demonstrate plant protection and growth and, consequently, their importance in agriculture8,19. Sivalingam and colleagues (2019) reported the prominence of Actinobacteria, especially Streptomyces species derived from extreme sources, as a remarkable sources of new biosynthetic gene clusters with potential for anticancer drug development31. Actinomycetes are considered as a rich source of natural products, which have numerous bioactive functions, including phytotoxic, antimicrobial, insecticidal, and mainly antiproliferative and antitumor activities32,33,34,35,36,37. This is, to our knowledge, the first report on the diversity and antimicrobial activity of EA isolated from L. cylindrica. As with other reports, the present study also found that high-G + C Gram-positive endophytic bacteria were dominant in the luffa with streptomyces being the most abundant group. These observations were similar to the study conducted by Govindasamy et al. They isolated and identified endophyte actinobacteria from cactus (Opuntia ficus-indica) for the first time38.
The present study for the first time addressed that L. cylindrica's root and leaves are rich in endophytic actinobacteria, which six endophytic actinobacteria were isolated from these samples using cultivar-dependent method. Many studies performed on endophytic actinobacteria populations using culture-independent and culture-dependent methods. For example, Sessitsch et al. identified several species of Streptomyces in potatoes using PCR for16S rRNA gene followed by denaturing gradient gel electrophoresis (DGGE)39. There are many studies on endophytic actinobacteria isolation by which several genera such as Streptomyces, Microbispora, Micromonospora and Nocardioides have been isolated and reported from different plants40. In the present study based on the morphological characteristics and also partial 16S rRNA gene sequencing, six endophytic actinobacteria strains were isolated from L. cylindrica. Four different selection media were consumed to isolate actinobacteria. Three isolates were isolated from the roots (KUMS-B1, KUMS-B2, KUMS-B3) sampling from November 2020 and the other three isolates were isolated from the leaves (KUMS-B4, KUMS-B5 and KUMS-B6) sampling from September. The results showed that the sampling time and seasons may affect the results and bacterial diversity. The result also revealed that the leaves and roots are good habitats for EA. Fifty percentage of strains were isolated from leaves and the other 50% isolated from roots of the plant. We were unable to identify any EA from the fruits, stems and flowers. This might result from the intrinsic feature of plant species which are herbaceous, not woody plants. They are more vulnerable to the surface sterilization reagents and thus suggested that these substances might have penetrated to the samples and perished EA. The soft internal tissue and having a fibrous spongy skeleton network inside the fruits allow the penetration of sterilization reagents. In addition, decreasing the incubation time of surface sterilization reagents might lead to the isolation of EA from these tissues. Three isolates were isolated from TWYE culture medium (KUMS-B1, KUMS-B2, KUMS-B3) and three isolates (KUMS-B4, KUMS-B5 and KUMS-B6) from YECD culture medium. No growth of actinobacteria was detected on other selected media used. The results unravel the importance of using a specific culture medium as well as the plant sampling site to isolate different species of endophytic actinobacteria. According to morphological analysis, all isolates belonged to Streptomycetaceae family. Fragmented mycelium was observed for all isolates. The appearance of the KUMS-B1 colonies on ISP-2 medium was spherical, wrinkled, and protruding with a cream-colored bald surface, which was surrounded by a white layer of spores. The KUMS-B2 colonies into ISP-2 medium was spherical and protruding with cinereous, dove-grey color and watery surface, but smaller than KUMS-B1 colonies. The KUMS-B3 colonies was also spherical and protruding with the creamy-white and fuzzy surface. White spores covered all colonies after two weeks. The KUMS-B4 colonies into ISP-2 medium was small spherical with cream-yellowish color. In this strain, spore production was faster. The KUMS-B5 colonies was full of colorful creamy- light yellowish colour, while the KUMS-B6 produced small colonies with white-brownish color into ISP-2 medium. Spore production was done in this strain at a lower speed than other strains. Similar to our results, in a study conducted by Chromkaew's Shutsrirung et al. the most frequent isolates recovered were members of Streptomyces41.
The spore-chain morphology and spore surface ornamentations are crucial for identification of actinobacteria. Actinomycete spore chain length, shape, position, color are the important basis for classification5. The growth rate and sporulation rate of each isolate varied on ISP2 medium. The strains had clear and unique pattern of spores’ arrangement. The spore surface of the isolates varied from smooth and fluffy to rough and flattened and indented. Some isolates from root had oval and ovoid -shaped spores with spiny and hairy surfaces, while isolated strains from leaves had a chain of cylindrical or cubed shape spores with smooth and/or indented surfaces. Our results are in agreement with findings of Riquelme5 and Ali et al42. We used inclined glass coverslip technique and acquired the good result, as describe by Kurtboke43 as the best technique. The concept of smooth, warty, spiny, and hairy spore surfaces has been used for characterization of actinobacteria.
16S rRNA gene sequencing revealed that endophytic actinobacteria represented six species; S. koyangensis, S. griseorubens, S. intermedius, S. microflavus, S. cavourensis, S. praecox. and exhibited 99% similarity with their closely related actinobacterial strains. The present study indicated all the six isolates belong to same phylum Actinomycetota and one genus, Streptomyces, which is consistent with other studies, reported mainly from Actinomycetota phylum22,41.
Three strains, closely related to S. microflavus, S. intermedius and S.praecox were reported as endophytes for the first time and they were isolated from leaf and root samples using YECD and TWYE culture media.
Numerous factors can affect the diversity of endophytes, such as plant age, plant sampling site, soil type, seasonal collection, geographical location, and other environmental conditions44,45. The similarity of strains with their closely related stains, deposited in GenBank and other databases is more that 99%. The closely related strain to KUMS-B4 strain in the GenBank and EZbiocloud databases was found to be S. microflavus strain NBRC 13062, which had 100% similarity in sequence. This is first to report S. microflavuas as an endophyte. It was originally isolated from soil and recently from marine environment46. According to the phylogenetic tree, KUMS-B5 was closely related to S. cavourensis NBRC 13026. S. cavourensis YBQ59, and S. cavourensis strain 1AS2a reported as endophytes47,48. Based on the phylogenetic tree, KUMS-B6 strain was placed next to S. praecox strain NBRC 13073, which had 99.80% similarity in sequence. Besides, the KUMS-B1 strain was placed next to S. koyangensis VK-A60, which had 99.04% sequence similarity. The S. koyangensis strain B025 reported as endophytes49. The 16S rRNA gene sequence of strain KUMS-B3 showed the highest sequence similarity with S. intermedius NBRC 13049, reported from soil. Nevertheless, further investigations of strains should be carried out to determine their correct phylogenetic placement.
The present study demonstrated the antibacterial effect and inhibitory potential of endophytic bacteria isolated from L. cylindrica against test bacteria. The results revealed that the crude extracts of the isolates were able to inhibit the growth of at least one of the test bacteria due to the presence of natural bioactive compounds with growth-inhibitory properties8,50. This finding is consistent with other studies, which reported the endophytic actinobacteria as a potential source of secondary metabolites containing bioactive compounds50,51. Some strains such as KUMS-B6 isolate had antagonist activity against both gram-positive and gram-negative bacteria. The KUMS-B6, isolated for the first time as endophyte, exhibited the highest antibacterial activity against all test bacteria, highlighting this strain as new potential source for a wide range of antibacterial agents. Zarandi, et.al. studied the antagonist activity of S. praecox strain R7 to control Rhizoctonia solani in tomatoes. They stated that S. praecox enhances the expression of lipoxygenase and phenylalanine ammonia lyase by triggering jasmonic acid and phenyl propanoid signaling pathways in plants; therefore, resulting an induced defense status in tomatoes against R. solani52. Interestingly, KUMS-B6, KUMS-B5 and KUMS-B4 isolates also strongly inhibited the growth of P. aeruginosa. A few compounds; chromomycin A9, chromomycin Ap, chromomycin A2, and chromomycin A3 were reported from the ethyl acetate extract of S. microflavus which showed potent antibacterial activities against Gram-positive bacteria46 which are consistent with our findings. Four metabolites; (2E, 4E) -5- (3-hydroxyphenyl) -penta-2,4-dienamide, Ergosterol, Ergosterol peroxide and Halolithuralin B, were documented from Streptomyces sp. YIM65484 to have antibacterial activity53. In addition, eight compounds; 1-monolinolein, bafilomycin D, nonactic acid, daidzein, 3'-hydroxydaidzein, 5,11-epoxy-10-cadinanol, prelactone B, and daucosterol were identified from S. cavourensisYBQ59 exhibited inhibitory activity against Gram positive bacteria47 which is in agreement with our findings. Mondal and Rai et al., also reported antibacterial activity of endophytic S. cavourensis MH16 isolated from M. hortensis against Gram-positive and negative bacteria54. Among the isolates, KUMS-B1 strain showed the weakest antimicrobial properties. This strain had no antibacterial activity on test bacteria or showed a weak antimicrobial property against E. coli 25922. The antifungal activity of 4-phenyl-3-butenoic acid from S. koyangensis VK-A60, isolated from soil55 was reported by Lee et al.56. The KUMS-B2 strain had 99.86% similarity to S. griseorubens strain NBRC 12780. S. griseorubens MPT42 has already been isolated as an endophyte from Litsea cubeba57. This strain reported to expresses all three biosynthetic genes associated with the antibiotics PKS-I, PKS-II and NRPS and has shown significant antimicrobial activity57. The antimicrobial properties of this strain have been demonstrated in many studies58,59. Methanolic extract of KUMS-B2 strain had a great inhibitory effect against S. aureus 25923. The antibacterial compounds generated by this strain have probably polar structures and the Gram-positive bacteria generally were more susceptible to extracts from this strain as compared to gram-negative bacteria. The influential difference may be due to the structure of their cell walls60. Similarly, Ameen, et al.61 reported the antibacterial activity of thirty endophytic actinobacterial strains against antibiotic-resistant bacteria. Ethyl acetate extracts of actinobacteria inhibited the growth of Gram positive and negative bacteria62,63. The methanolic extract of KUMS-B3 had a moderate inhibitory activity against S. aureus. This is the first antibacterial report on closely related strain to S. intermedius. The present study for the first time indicated that L. cylindrica is rich in endophytic bacteria where six endophytic bacteria were isolated from roots and leaves samples and all the isolates belonged to same phylum Actinomycetota and represented one genera Streptomyces. In general, the strains, isolated from leaves, exhibited stronger antagonist activities against test bacteria as compared with those, isolated from roots. Therefore, they are more promising sources for discovery of novel antibacterial compounds and industrial applications. In this study, S. praecox was isolated as endophyte for the first time, exhibited the highest antibacterial activity against all three tested bacterial pathogens.
Methods
Samples collection and authentication
Symptomless and mature L. cylindrica plant collected from different parts of Kerman, Iran (southeast of Iran, 28.6751° N, 57.7/372° E). The samples collected in September and November 2021 to cover seasonal variation. All the procedure related to plants were complied with relevant institutional, national, and international guidelines and legislations. The collection of specimens was done randomly under the Research Center of Agriculture and Natural Resources of Kermanshah (RANK) permit, provided by the governments. They were then identified and authenticated by an expert, Dr. Nastaran Jalilian at the Kermanshah Agricultural and Natural Resources Research and Education Centre, Kermanshah, Iran according to the Flora of Iran and Flora Iranica. The voucher specimen (10395) was deposited in the Herbarium of RANK. The plants were dissected to stems, leaves, roots, and, fruits, and surface-sterilized and cultured within 48 h of sampling.
Surface sterilization
The surface sterilization of plant samples (roots, stems, leaves and fruits) was performed according to procedure described by Kaewkla and Franco64 and Ali et al42. Breifly, the plant tissues were washed with running tap water to remove all contamination and any physical debris. The samples were cut into pieces using a sterile saw/knife and tweezers and dried in natural air after washing three times with distilled water. The segments were cut to create small pieces of two to five cm length and their surfaces were sterilized with various surface disinfectants to prevent the growth of bacteria and non-endophytic fungi. Each sample was immersed in sterile 0.1% Tween 20 for five minutes. The samples were then treated with 70% ethanol for five minutes, and then the samples were rinsed with freshly prepared 6% sodium hypochlorite (NaOCl) solution. Samples were washed several times with DDW to remove chemical residues and immersed in sterile 10% (w/v) sodium bicarbonate (NaHCO3) for 10 min to delay the growth of endophytic fungi65 and were washed three times with sterile distilled water and dried in a laminar flow. In order to ensure the efficacy of the surface sterilization, 100 µL of the last double distilled washed water was cultivated onto ISP-2 media and incubated at 28 °C for 1 week. The absence of microbial growth on the culture media after plating the last washing water demonstrated the effectiveness of surface sterilization66.
Isolation of endophytic actinobacteria
To isolate the endophytic actinobacteria, HPDA, PDA (Merck, Germany)67, TWYE (Merck, Germany)68, and YECD (Merck, Germany)69 media supplemented with nalidixic acid (25 µg/mL) and benomyl (10 µg/mL) to inhibit the growth of gram negative bacteria and fungi were used70,71. Then the surface sterilized samples were aseptically cut to 1–2 cm, placed directly on the culture medium (in triplicate), and incubated at 28 °C for 16 weeks. The plates checked periodically and the growth of endophytic actinobacteria colonies from the tissues monitored daily, up to 16 weeks. Different colonies with putative actinobacteria characteristics were introduced and sub-cultured onto international Streptomyces Project-2 medium (ISP2)72. Pure cultures obtained after two to three consecutive subcultures and transferred to fresh isolation media, considering the possibility that each colony resembles a distinct strain73. Finally, the stock culture was prepared using Tryptic Soybean Broth (TSB) (Merck, Germany) containing 30% glycerol from the strains cultured on the ISP2 medium and stored at − 20 °C for further analysis.
Identification of endophytic actinobacteria
The isolates were identified according to their morphological characteristics19 such as, colony features, the presence of vegetative and reproductive mycelium, fragmentation of the mycelium, spore production, and pigments, following the general guidelines of the International Streptomyces Project. In addition, Gram staining and Scanning Electron Microscope (SEM) analysis (FEI, Quanta 450, America) were used to confirm the isolates74,75.
Scanning electron microscopic (SEM) examination
The morphology of isolates was also studied using SEM (FEI, Quanta 450, America). The isolates were cultured onto ISP2, the aerial mycelium and spores were obtained on a cover slip /slide. Cover slides were cut into one cm pieces and sterilized. The Sterile coverslips were inserted into solidified ISP2 at an angle of 45°. The inoculum was spread along with the glass-agar medium interface and incubated at 28 °C for 14 days. During incubation, the organisms grew over the surface of the glass pieces. The cover slip containing organisms was then coated with a film of gold about 150–200 A° thickness and observed under SEM at an accelerating voltage of 25.000–30.000 V for spore surface ornamentation74. The spore chain morphology and ornamentation of the potential strains were analyzed.
DNA extraction
Molecular identification of isolates was carried out using 16S rRNA gene sequencing to identify selected endophytic actinobacteria species. The genomic DNA was extracted from the endophytic actinobacteria according to Coombs et al. protocol69 using TSB broth medium. For this purpose, first, under sterile conditions the bacterial colony was inoculated into Erlen Meyers containing 25 mL TSB and incubated for 7 days in a shaker incubator at 28 °C (HYSC, Korea) at 160 rpm. After about 7 days, the purity of cultures was monitored using wet mount and Gram staining methods by a light microscope (Olympus). The supernatant was then removed with a Hettich 320R, Germany (4,000 rpm, 10 min, 4 °C) to separate the mycelia, and 50–100 mg of bacterial pellets were used to DNA extraction and poured into 2 mL microtubes and added 500 µL TE (Tris 10 mm, EDTA 1 mm), vortexed and then centrifuged, (3 min, 5000 rpm). Then, 10 µL of lysosome (10 mg/mL) was added to each microtube and incubated for 60 min at 37 °C. Ten µL of proteinase K (10 mg/mL) and 32.5 µL of SDS 10% were added to the microtubes and incubated for 60 min at 55 °C. After that 100 µL NaCl 5 M and 65 µL CTAB/NaCl were added to the microtubes and incubated ten minutes at 55 °C. Then, 600 µL chloroform-alcohol alcohol (24:1) was added to the microtubes and after a short vertex, incubated 30 min at room temperature, and then Centrifuged, at 12,000 rpm for 15 min. The supernatant was transferred to the new microtubes and 500 µL chloroform was added, and incubated at room temperature for 15 min and then centrifuged for 15 min at 12,000 rpm. The supernatant was transferred to the new microtubes and added 10 µL RNAase (10 mg/mL), incubated for 60 min at 37 °C and then 55 °C for 5 min to inactivate the RNase enzyme. After that, washed with chloroform. The supernatant was treated with 1 × volume of 3 M sodium acetate (50 µL) and 3× volume of 100 percent cold ethanol (1 mL) and kept in a freezer at – 20 ℃ overnight. Following centrifugation for 15 min at 15,000 RPM, the supernatant was transferred to the new microtube and 500 µL ethanol 70% was added and centrifuged 15 min at 8000 rpm. The supernatant was discarded and the pellet was dried using a heat block. Finally, 50 µL of deionized distilled water was added to the sediment and the microtube was preserved in – 20 ℃. Agarose gel electrophoresis and NanoDrop 2000c spectrophotometer (Thermo Scientific, USA) were used to evaluate the quality of the extracted DNA.
Molecular identification of isolates
PCR
The 16S rRNA genes were amplified by PCR as described by Coombs and Franco using Mastercycler® nexus, Eppendorf and primers namely 27F (AGAGGTTGATCMTGGCTCAG) and 1492R (TACGGTACTTTTTACTACTTT)40. Briefly, the amplifications were carried out in a total volume of 25 µL reaction mixture, including 12.5 µ of master mix (Sinaclon Cat.No.MM2062), 0.3 µL of each primer (10 pmol/μL), 1 µL of DNA template (50–100 ng), and 10.9 µL of D.W. Amplification was performed in a thermocycler (Mastercycler® nexus, Eppendorf). The thermocycle program was 94 °C for 4 min, followed by 30 cycles of 94 °C for 1 min, 50 °C for 2 min and 15 s and 72 °C for 2 min, and finally an extension step at 72 °C for 5 min. The PCR product was then confirmed by gel electrophoresis and UV-translummant device (Gel Doc device (Vilber, France). PCR products were sequenced by a commercial company, Macrogen, South Korea.
Molecular phylogenetic analysis
The Partial 16S rRNA gene sequences were compared with the sequences retrieved from EzBioCloud and GenBank, NCBI databases using BLASTn tool (Basic Local Alignment Search Tool) (http://www.ncbi.nlm.nih.gov/BLAST/Blast.cgi)76. Multiple sequence alignment was performed using the CLUSTAL W 1.83 program, and a phylogenetic tree was reconstructed via the bootstrap test of neighbor-joining method using MEGA X (Molecular Evolutionary Genetics Analysis) software19. The Kimura two-parameter model was used to calculate pairwise distances for the neighbour-joining procedure (Kimura 1980). Strength and reliability of internal branches (topologies of the neighbor- joining) of the resultant tree was assessed after a 1000 bootstrap -replicate analysis77. Kitasatospora setae KM -6054 was used as an outgroup strain for tree constructions78. The partial 16S rRNA gene sequences, determined in the present study, were deposited in GenBank under accession numbers listed in Table 1.
Metabolites extraction
To test the antibacterial activity of endophytic isolated strains, the secondary metabolites from the isolates were extracted using ethyl acetate and methanol, respectively. Briefly, Isolates were grown for 7 days on plates (10 cm in diameter) containing TSA medium at 28 ± 2 °C. After that, they were cut into small pieces with a razor blade and then poured into a 100 mL pre-sterilized Erlenmeyer. Then 50 mL of ethyl acetate was added to the Erlenmeyer flakes and place on a shaker (180 rpm) for 24 h at room temperature. The ethyl acetate was then filtered and transferred to the Falcon 50 mL. Then we added 50 mL of methanol to the pellet and allowed the device to operate for another 24 h under the same conditions. After that, the methanolic extracts were filtered. A rotary evaporator (Rotary Evaporator RE100, Heidolph, Germany) (70 RPM, 38 °C) was used to concentrate the extracts. The extracts were stored at − 20 °C79 until used.
Antimicrobial activity of isolated strains
The antibacterial activity of the isolates (KUMS-B1, KUMS-B2, KUMS-B3, KUMS-B4, KUMS-B5, and KUMS-B6) was screened against three pathogenic bacteria (Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 25923 and Pseudomonas aeruginosa ATCC 27853), purchased from microbial Bank of Pasteur Institute of Iran, using agar well diffusion method. Initially, the test bacteria were inoculated in TSB medium at 37 °C for 24 h. The growth was adjusted to half the McFarland's dilution (OD = 0.2) using Spekol 1500 spectrophotometer at 600 nm (OD600 nm). Then, the antibiotic agar medium (AAM-MERCK) was seeded with the tested culture (1% V/V) and dispensed into petri dish plates. After that, 10 wells with a diameter of 6 mm were prepared using a sterile cork borer with specified intervals in each plate. Each well was than loaded with 50 µL of extracts, allowing the wells to dry completely. Finally, the plates incubated at 37 °C for 24 h. Vancomycin (20 µg/mL) and Imipenem (15 µg/mL P. aeruginosa ATCC 27853 and 1.5 µg/mL E. coli ATCC 25,922) were used as positive controls for Gram-positive and Gram-negative bacteria, respectively. Wells containing methanol or ethyl acetate (50 mL) were used as negative controls. Each test was repeated thrice and the zone of growth inhibition around each well was measured and its average was recorded in mm.
Conclusion and future prospective and directions
This is the first study screening the EA from Luffa cylindrica. The study revealed that L. cylindrica harbors untapped diversity of endophytic actinobacteria with valuable antimicrobial potential. All isolates belong to the family Streptomycetaceae in which three of those reported as endophytes for the first time. We analyzed limited numbers of strains based on 16S rRNA gene sequences which need further analysis regarding their PKI and NRPs. To understand the chemical structure of antimicrobial compounds, further analytical purification of extracts is required to decipher their mechanism of action.
Interestingly, some isolates were able to inhibit the growth of both gram-positive and gram-negative pathogens and reveal the possible presence of broad-spectrum of antibacterial compounds. These bioactive substances can be considered as a potential source for the synthesis of new antibacterial drugs for the treatment of infectious diseases, but more studies are required to isolate and identify these bioactive substances.
Our finding along with literature highlights that actinobacteria are able to produce a diverse range of secondary metabolites, which should be explored further for new antimicrobial drug candidates. These data may have significant implications for the antibiotic resistance, health and pharmaceutical applications and are particularly relevant to the development of antibiotic agents.
Data availability
The following accession numbers have been assigned to the nucleotide sequences generated during the current study, deposited in the GenBank databases: ON626444, ON626445, ON626446, ON626447, ON626448, and ON626449.
References
Shahbazi, S., Karam, M. R. A., Habibi, M., Talebi, A. & Bouzari, S. Distribution of extended-spectrum β-lactam, quinolone and carbapenem resistance genes, and genetic diversity among uropathogenic Escherichia coli isolates in Tehran, Iran. J. Glob. Antimicrob. Resist. 14, 118–125 (2018).
Shahbazi, S. et al. Identification of novel putative immunogenic targets and construction of a multi-epitope vaccine against multidrug-resistant Corynebacterium jeikeium using reverse vaccinology approach. Microb. Pathog. 164, 105425 (2022).
Opoku-Temeng, C., Kobayashi, S. D. & DeLeo, F. R. Klebsiella pneumoniae capsule polysaccharide as a target for therapeutics and vaccines. Comput. Struct. Biotechnol. J. 17, 1360–1366 (2019).
Falagas, M. E. & Karageorgopoulos, D. E. Pandrug resistance (PDR), extensive drug resistance (XDR), and multidrug resistance (MDR) among Gram-negative bacilli: Need for international harmonization in terminology. Clin. Infect. Dis. 46, 1121–1122 (2008).
Riquelme, C. et al. Biotechnological potential of Actinobacteria from Canadian and Azorean volcanic caves. Appl. Microbiol. Biotechnol. 101, 843–857. https://doi.org/10.1007/s00253-016-7932-7 (2017).
Subbulakshmi, G., Thalavaipandian, A., Ramesh, V. & Rajendran, A. Bioactive endophytic fungal isolates of Biota orientalis (L) Endl., Pinus excelsa Wall and Thuja occidentalis L.. Int. J. Adv. Life Sci. 4, 9–15 (2012).
Abedinlou, H., Bahrami, Y., Mohammadi, S. & Kakaei, E. Rare actinobacteria and their potential biotechnological applications. Sci. J. Kurdistan Univ. Med. Sci. 26, 108–131 (2022).
Singh, R. & Dubey, A. K. Diversity and applications of endophytic actinobacteria of plants in special and other ecological niches. Front. Microbiol. 9, 1767. https://doi.org/10.3389/fmicb.2018.01767 (2018).
Bahrami, Y. et al. In Natural Products from Actinomycetes: Diversity, Ecology and Drug Discovery (eds Ravishankar V. Rai & Jamuna A. Bai) 119–150 (Springer Singapore, 2022).
Zhongming, Z., Linong, L., Xiaona, Y., Wangqiang, Z. & Wei, L. Low quality healthcare is increasing the burden of illness and health costs globally. World Health Organization, Geneva, Switzarland. http://www.who.int/servicedeliverysafety/quality-report/en/ (2018).
Baltz, R. H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 8, 557–563 (2008).
Bérdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 65, 385–395 (2012).
Bahrami, Y., Bouk, S., Kakaei, E. & Taheri, M. Natural products from Actinobacteria as a potential source of new therapies against colorectal cancer: A review. Front Pharmacol. 2022, 5 (2022).
Rajivgandhi, G. et al. Antibacterial and anticancer potential of marine endophytic actinomycetes Streptomyces coeruleorubidus GRG 4 (KY457708) compound against colistin resistant uropathogens and A549 lung cancer cells. Microb. Pathog. 125, 325–335. https://doi.org/10.1016/j.micpath.2018.09.025 (2018).
Silva, L. J. et al. Actinobacteria from Antarctica as a source for anticancer discovery. Sci. Rep. 10, 1–15 (2020).
Golinska, P. et al. Endophytic actinobacteria of medicinal plants: Diversity and bioactivity. Antonie Van Leeuwenhoek 108, 267–289. https://doi.org/10.1007/s10482-015-0502-7 (2015).
Matsumoto, A. & Takahashi, Y. Endophytic actinomycetes: Promising source of novel bioactive compounds. J. Antibiot. 70, 514–519 (2017).
Vurukonda, S. S. K. P., Giovanardi, D. & Stefani, E. Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 19, 952 (2018).
Musa, Z. et al. Diversity and antimicrobial potential of cultivable endophytic actinobacteria associated with the medicinal plant Thymus roseus. Front. Microbiol. 11, 191 (2020).
Khan, M., Bhatia, S., Javed, K. & Khan, M. Evaluation of anti-stress (adaptogenic) activity of luffa cylindrica fruit extract. Ind. J. Pharm. Sci 54, 75–76 (1992).
Partap, S., Kumar, A., Sharma, N. K. & Jha, K. Luffa cylindrica: An important medicinal plant. J. Natural Product Plant Resour. 2, 127–134 (2012).
Shan, W., Zhou, Y., Liu, H. & Yu, X. Endophytic actinomycetes from tea plants (Camellia sinensis): Isolation, abundance, antimicrobial, and plant-growth-promoting activities. BioMed. Res. Int. 2018, 6 (2018).
Gos, F. M. et al. Antibacterial activity of endophytic actinomycetes isolated from the medicinal plant Vochysia divergens (Pantanal, Brazil). Front. Microbiol. 8, 1642 (2017).
Su, L., Deng, H. & Niu, Y.-C. Phialemoniopsis endophytica sp. nov., a new species of endophytic fungi from Luffa cylindrica in Henan, China. Mycol. Progress 15, 48. https://doi.org/10.1007/s11557-016-1189-5 (2016).
Meklat, A. et al. Actinopolyspora algeriensis sp. nov., a novel halophilic actinomycete isolated from a Saharan soil. Extremophiles 16, 771–776 (2012).
Shirling, E. B., Gottlieb, D. J. I. J. O. S. & Microbiology, E. Cooperative description of type cultures of Streptomyces. II. Species descriptions from first study. Int. J. Syst. Evol. Microbiol. 18, 69–189 (1968).
Kelly, K. L. & Judd, D. B. Color: Universal Language and Dictionary of Names. Vol. 13 (US Department of Commerce, National Bureau of Standards, 1976).
Goodarzi, N. N., Fereshteh, S., Sabzi, S., Shahbazi, S. & Badmasti, F. Construction of a chimeric FliC including epitopes of OmpA and OmpK36 as a multi-epitope vaccine against Klebsiella pneumonia. (2021).
Taati-Moghadam, M. et al. The challenge of global emergence of novel colistin-resistant Escherichia coli ST131. Microb. Drug Resist. 27, 1513–1524 (2021).
Delbari, Y., Mohassel, Y., Bahrami, Y., Kakaie, E. & Mostafaie, A. A review on isolation and identification of endophytic actinobacteria, their chemical structure, bioactive compounds, and potential medical-pharmaceutical applications. J. Mazandaran Univ. Med. Sci. 30, 195–217 (2020).
Sivalingam, P., Hong, K., Pote, J. & Prabakar, K. Extreme environment Streptomyces: Potential sources for new antibacterial and anticancer drug leads?. Int. J. Microbiol. 2019, 5 (2019).
Almeida, L. C. D. et al. Pradimicin-IRD exhibits antineoplastic effects by inducing DNA damage in colon cancer cells. Biochem. Pharmacol. 168, 38–47 (2019).
Martinez, A. F. C., de Almeida, L. G., Moraes, L. A. B. & Cônsoli, F. L. Tapping the biotechnological potential of insect microbial symbionts: New insecticidal porphyrins. BMC Microbiol. 17, 1–10 (2017).
Bauermeister, A., Zucchi, T. D. & Moraes, L. A. B. Mass spectrometric approaches for the identification of anthracycline analogs produced by actinobacteria. J. Mass Spectrom. 51, 437–445 (2016).
Crevelin, E. J. et al. Isolation and characterization of phytotoxic compounds produced by Streptomyces sp. AMC 23 from red mangrove (Rhizophora mangle). Appl. Biochem. Biotechnol. 171, 1602–1616 (2013).
Rodrigues, J. P. et al. Bioguided isolation, characterization and media optimization for production of Lysolipins by actinomycete as antimicrobial compound against Xanthomonas citri subsp. citri.. Mol. Biol. Rep. 45, 2455–2467 (2018).
Bauermeister, A. et al. Pradimicin-IRD from Amycolatopsis sp. IRD-009 and its antimicrobial and cytotoxic activities. Natural Prod. Res. 33, 1713–1720 (2019).
Govindasamy, V. et al. Characterization of root-endophytic actinobacteria from cactus (Opuntia ficus-indica) for plant growth promoting traits. Arch. Microbiol. 204, 1–14 (2022).
Sessitsch, A., Reiter, B., Pfeifer, U. & Wilhelm, E. Cultivation-independent population analysis of bacterial endophytes in three potato varieties based on eubacterial and Actinomycetes-specific PCR of 16S rRNA genes. FEMS Microbiol. Ecol. 39, 23–32 (2002).
Coombs, J. T. & Franco, C. M. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 69, 5603–5608 (2003).
Shutsrirung, A., Chromkaew, Y., Pathom-Aree, W., Choonluchanon, S. & Boonkerd, N. Diversity of endophytic actinomycetes in mandarin grown in northern Thailand, their phytohormone production potential and plant growth promoting activity. Soil Sci. Plant Nutr. 59, 322–330 (2013).
Ali, A.R., Bahrami, Y., Kakaei, E. et al. Isolation and identification of endophytic actinobacteria from Citrullus colocynthis (L.) Schrad and their antibacterial properties. Microb Cell Fact 21(1), 206. https://doi.org/10.1186/s12934-022-01936-9 (2022).
Kurtböke, D. I. Correct interpretation of actinomycete imagery using scanning electron microscopy. Microbiol. Australia 43, 28–31. https://doi.org/10.1071/MA22009 (2022).
Fernandes, E. G., Pereira, O. L., da Silva, C. C., Bento, C. B. P. & de Queiroz, M. V. Diversity of endophytic fungi in Glycine max. Microbiol. Res. 181, 84–92 (2015).
Martín-García, J., Espiga, E., Pando, V. & Diez, J. J. Factors influencing endophytic communities in poplar plantations. Silva Fennica 45, 169–180 (2011).
Cho, E. et al. Antibacterial activity of chromomycins from a marine-derived Streptomyces microflavus. Mar. Drugs https://doi.org/10.3390/md18100522 (2020).
Vu, H.-N.T. et al. Antimicrobial and cytotoxic properties of bioactive metabolites produced by Streptomyces cavourensis YBQ59 isolated from Cinnamomum cassia Prels in Yen Bai Province of Vietnam. Curr. Microbiol. 75, 1247–1255 (2018).
Vargas Hoyos, H. A., Santos, S. N., Padilla, G. & Melo, I. S. Genome sequence of Streptomyces cavourensis 1AS2a, a rhizobacterium isolated from the Brazilian Cerrado Biome. Microbiol. Resour. Announc. https://doi.org/10.1128/mra.00065-19 (2019).
Saini, P. & Gangwar, M. Inhibition spectrum, purification and kinetic study of alpha-amylase inhibitor from Murraya koenigii endophytic actinobacterium (Streptomyces koyangensis strain B025). Curr. Sci. 115, 393–395 (2018).
Sarmiento-Vizcaíno, A., Martín, J., Reyes, F., García, L. A. & Blanco, G. bioactive natural products in actinobacteria isolated in rainwater from storm clouds transported by Western Winds in Spain. Front. Microbiol. https://doi.org/10.3389/fmicb.2021.773095 (2021).
Manivasagan, P. et al. Marine actinobacteria: An important source of bioactive natural products. Environ. Toxicol. Pharmacol. 38, 172–188 (2014).
Ebrahimi-Zarandi, M. et al. Exploring two Streptomyces species to control Rhizoctonia solani in tomato. Agronomy 11, 1384 (2021).
Zhou, H. et al. Anti-mycobacterium tuberculosis active metabolites from an endophytic Streptomyces sp. YIM65484. Records Natural Prod. 9, 196 (2015).
Mondal, S. & Rai, V. R. Molecular profiling of endophytic Streptomyces cavourensis MH16 inhabiting Millingtonia hortensis Linn. and influence of different culture media on biosynthesis of antimicrobial metabolites. Sci. Nature 106, 1–10 (2019).
Lee, J. Y., Lee, J. Y., Jung, H. W. & Hwang, B. K. Streptomyces koyangensis sp. nov., a novel actinomycete that produces 4-phenyl-3-butenoic acid. Int. J. Syst. Evol. Microbiol. 55, 257–262 (2005).
Lee, J. Y., Lee, J. Y., Moon, S. S. & Hwang, B. K. Isolation and antifungal activity of 4-phenyl-3-butenoic acid from Streptomyces koyangensis strain VK-A60. J. Agric. Food Chem. 53, 7696–7700 (2005).
Nguyen, Q. H. et al. Characterization of endophytic Streptomyces griseorubens MPT42 and assessment of antimicrobial synergistic interactions of its extract and essential oil from host plant Litsea cubeba. J. Fungi. 8, 197 (2019).
Rashad, Y. M., Al-Askar, A. A., Ghoneem, K. M., Saber, W. I. & Hafez, E. E. Chitinolytic Streptomyces griseorubens E44G enhances the biocontrol efficacy against Fusarium wilt disease of tomato. Phytoparasitica 45, 227–237 (2017).
Al-Hamad, B. M. et al. Effectiveness of augmentative biological control of Streptomyces griseorubens UAE2 depends on 1-aminocyclopropane-1-carboxylic acid deaminase activity against Neoscytalidium dimidiatum. J. Fungi 7, 885 (2021).
Nikaido, H. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178, 5853–5859 (1996).
Ameen, F. et al. Prevalence of antibiotic resistant mastitis pathogens in dairy cows in Egypt and potential biological control agents produced from plant endophytic actinobacteria. Saudi J. Biol. Sci. 26, 1492–1498 (2019).
Bhakyashree, K. & Kannabiran, K. Actinomycetes mediated targeting of drug resistant MRSA pathogens. J. King Saud Univ.-Sci. 32, 260–264 (2020).
Pandey, A., Ali, I., Butola, K. S., Chatterji, T. & Singh, V. Isolation and characterization of Actinomycetes from soil and evaluation of antibacterial activities of Actinomycetes against pathogens. Int. J. Appl. Biol. Pharm. Technol. 2, 384–392 (2011).
Kaewkla, O. & Franco, C. M. J. M. E. Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microb. Ecol. 65, 384–393 (2013).
Ponpandian, L. N. et al. Phylogenetic characterization of bacterial endophytes from four Pinus species and their nematicidal activity against the pine wood nematode. Sci. Rep. 9, 1–11 (2019).
Ramalashmi, K. et al. A potential surface sterilization technique and culture media for the isolation of endophytic bacteria from Acalypha indica and its antibacterial activity. J. Med. Plant Stud. 6, 181–184 (2018).
Rasool, U. & Hemalatha, S. J. M. L. Marine endophytic actinomycetes assisted synthesis of copper nanoparticles (CUNPS): Characterization and antibacterial efficacy against human pathogens. Mater. Lett. 194, 176–180 (2017).
Zin, N., Loi, C., Sarmin, N. & Rosli, A. J. R. J. O. M. Cultivation-dependent characterization of endophytic actinomycetes. Res. J. Microbiol. 5, 717–724 (2010).
Coombs, J. T., Franco, C. M. J. A. & Microbiology, E. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 69, 5603–5608 (2003).
Van der Meij, A. et al. Inter-and intracellular colonization of Arabidopsis roots by endophytic actinobacteria and the impact of plant hormones on their antimicrobial activity. Antonie Van Leeuwenh. 111, 679–690 (2018).
Shan, W., Zhou, Y., Liu, H. & Yu, X. J. B. R. I. Endophytic actinomycetes from tea plants (Camellia sinensis): Isolation, abundance, antimicrobial, and plant-growth-promoting activities. BioMed. Res. Int. 2018, 5 (2018).
Thampi, A. & Bhai, R. S. J. B. C. Rhizosphere actinobacteria for combating Phytophthora capsici and Sclerotium rolfsii, the major soil borne pathogens of black pepper (Piper nigrum L.). Biol. Control 109, 1–13 (2017).
Kumar, P. S., Raj, J. P. P., Duraipandiyan, V. & Ignacimuthu, S. J. A. P. J. O. T. B. Antibacterial activity of some actinomycetes from Tamil Nadu, India. Asian Pac. J. Trop. Biomed. 2, 936–943 (2012).
Palla, M. S. et al. Isolation and molecular characterization of antifungal metabolite producing actinomycete from mangrove soil. Beni-Suef Univ. J. Basic Appl. Sci. 7, 250–256 (2018).
Li, Q., Chen, X., Jiang, Y., Jiang, C.J.A.-B. & Applications, B. Morphological identification of actinobacteria. Actinobacteria-basics Biotechnol. Appl. 2018, 59–86 (2016).
Nammali, A. et al. Streptomyces coffeae sp. nov., an endophytic actinomycete isolated from the root of Coffea arabica (L.). Int. J.f Syst. Evol. Microbiol. 71, 004834 (2021).
Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39, 783–791 (1985).
Passari, A. K. et al. Insights into the functionality of endophytic actinobacteria with a focus on their biosynthetic potential and secondary metabolites production. Sci. Rep. 7, 11809. https://doi.org/10.1038/s41598-017-12235-4 (2017).
Faja, O., Sharad, A., Younis, K. M., Usup, G. & Ahmad, A. J. I. J. C. R. Isolation, screening and antibiotic profiling of marine Actinomycetes extracts from the coastal of Peninsular Malaysia. Int. J. Chemtech. Res. 10, 212–224 (2017).
Acknowledgements
We would like to express our gratitude to the vice-chancellor for the research and technology of KUMS for financially supporting this project. The authors also would like to thank Medical Biotechnology lab at KUMS in particular Mr. Yasser Delbari and Miss. Hosna Hatami for their technical assistance.
Funding
Deputy for Research and Technology, Kermanshah University of Medical Sciences (Grant no. 4000793).
Author information
Authors and Affiliations
Contributions
The idea of this study was of Y.B. and Y.B. conceived and designed the research. R.A.M. collected the plants. R.A.M. and E.K. carried out the experiments with guidance of Y.B. Y.B. supervised the project. Y.B; and R.A.M. wrote the draft of manuscript. R. A.M. and E.K. collected and analyzed the data. Y.B. proofread, revised and approved the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mahdi, R.A., Bahrami, Y. & Kakaei, E. Identification and antibacterial evaluation of endophytic actinobacteria from Luffa cylindrica. Sci Rep 12, 18236 (2022). https://doi.org/10.1038/s41598-022-23073-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-23073-4
- Springer Nature Limited
This article is cited by
-
Diversity, molecular phylogenetics, and antibiotic biosynthetic potential of endophytic Actinobacteria isolated from medicinal plants in Nigeria
Brazilian Journal of Microbiology (2024)
-
The anticancer and antibacterial potential of bioactive secondary metabolites derived From bacterial endophytes in association with Artemisia absinthium
Scientific Reports (2023)
-
Identification and anti-bacterial property of endophytic actinobacteria from Thymes kotschyanus, Allium hooshidaryae, and Cerasus microcarpa
Scientific Reports (2023)