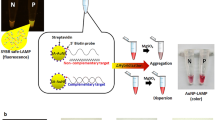
Abstract
Asymptomatic visceral leishmaniasis cases increase continuously, particularly among patients with HIV who are at risk to develop further symptoms of leishmaniasis. A simple, sensitive and reliable diagnosis is crucially needed due to risk populations mostly residing in rural communities with limited resources of laboratory equipment. In this study, a highly sensitive and selective determination of Leishmania among asymptomatic patients with Leishmania/HIV co-infection was achieved to simultaneously interpret and semi-quantify using colorimetric precipitates (gold-nanoparticle probe; AuNP-probe) and fluorescence (SYBR safe dye and distance-based paper device; dPAD) in one-step loop-mediated isothermal amplification (LAMP) assay. The sensitivities and specificities of 3 detection methods were equivalent and had reliable performances achieving as high as 95.5%. Detection limits were 102 parasites/mL (0.0147 ng/µL) which were 10 times more sensitive than other related studies. To empower leishmaniasis surveillance as well as prevention and control, this dPAD combined with SYBR safe and gold nanoparticle probe LAMP assay is reliably fast, simple, inexpensive and practical for field diagnostics to point-of-care settings in resource-limited areas which can be set up in all levels of healthcare facilities, especially in low to middle income countries.
Similar content being viewed by others
Introduction
Leishmaniasis is a vector-borne disease caused by flagellate protozoan parasites of the genus Leishmania remaining one of the global health challenges affecting millions of people worldwide. The parasite is transmitted by the bite of female blood feeding phlebotomine sandfly. More than 1 billion people live in leishmaniasis endemic areas where approximately 0.9 and 1.7 million people are newly infected. Only a small fraction will further develop the disease and 20,000 to 30,000 will eventually die1. The manifestations range from self-healing cutaneous leishmaniasis (CL) to potentially fatal outcome, visceral leishmaniasis (VL). In Thailand, the number of cases has continuously increased among patients with HIV/AIDS living in northern and southern regions due to the emergence of CL and VL caused by L. (Mundinia) martiniquensis and L. (Mundinia) orientalis, originally reported as L. (Mundinia) siamensis2,3,4. Remarkably, HIV probably influences the transmission of leishmaniasis in the affected areas due to a decline in host immune response.
The primary diagnostic test for HIV co-infection cases is currently polymerase chain reaction (PCR) because the method is trustworthy with high sensitivity and specificity3,5. However, PCR may be less practical and inappropriate in field diagnostics to point-of-care settings because it requires sophisticated and well-maintained laboratory equipment. Additionally, due to prolonged turnaround times of PCR operating procedures, the detection step may require 5 to 8 h or more6. On the contrary in terms of simplicity, the loop-mediated isothermal amplification (LAMP) assay is a simple approach to rapidly amplify large amounts of DNA within an hour by strand displacement activity of Bst polymerase based on isothermal conditions setting between 60 and 65 °C, and using a set of 4 to 6 primers without the use of complicated equipments7,8,9.
Several detection methods including turbidity, fluorescence and color have been developed to simply visualize and measure LAMP products using the naked eye10,11,12,13,14,15,16. To avoid the common contamination prone during the postamplification preparation step in the LAMP assay, a fluorescent detection reagent (FDR) is occasionally used in a prereaction17,18,19. A low cost and nonmutagenic fluorescent dye, SYBR safe, was used recently to reduce ambiguous evaluation in prereaction preparation; however, a fluorescent light source was needed to visualize the LAMP amplicons14.
Alternately, to avoid ambiguous reading and mutagenic effect, the LAMP products can be traced with functionalized gold nanoparticles (AuNPs) and analyzed using colorimetric detection15,20,21,22,23. Based on bio-nanoprobes, Thiol-linking of DNA and chemical functionalization of AuNPs for specific protein/antibody binding, simple and inexpensive methods were commonly applied to detect specific DNA sequences and are presently being expanded to diverse fields of pathogen diagnosis24,25,26,27,28. After hybridization with complementary DNA targets, the red solution indicating positive results remains because the complementary targets and AuNP probes form the polymeric network of cross-linked AuNPs preventing them from aggregation by salt induction. On the other hand, non-crosslinking hybridization induces aggregation of the AuNP probes by increasing salt concentration and results in a color change from red to blue (negative result)29,30 for which unambiguous interpretation of LAMP targets can be clearly observed by the naked eye13,15.
In 2017, a novel distance-based paper device (dPAD) using LAMP method was initially developed for detecting E. coli31. Less Mg2+ indirectly correlated to the distance on the paper device demonstrating a high concentration of LAMP products. Unreliable interpretation was observed when Mg2+ concentration was initially over or below narrow ranges of Mg2+ detection of the device. The dPAD could detect DNA lower than 103 copies/µL using several preparation steps; at least, and a 25 µL loading sample was required. According to indirect detection, visualization using direct DNA binding dyes, for example, SYBR Green, EvaGreen and SYBR safe indicators10,14,32,33, could be more reliable and preferable as an appropriate choice for dPAD. Recently, fluorescent dPAD using LAMP method has been reported to semi-quantify E. coli in urine and showed considerable promise for use as a fast and inexpensive UTI (urinary tract infection) diagnostics option due to fewer procedures to prepare using simple equipment12. Indeed, the approach could detect E. coli in less than 2 h without the postreaction staining process which was faster than conventional culture as well as the PCR method. Moreover, the length of fluorescent migratory distance directly correlated with the amount of LAMP products and reflected initial E. coli concentration. In addition to Bst polymerase, recent innovative works that combined the use of AuNPs with Recombinase Polymerase Amplification (RPA) isothermal amplification could also be used for Leishmania detection in animals using electrochemical and paper-based detection34,35.
In this present study, we reported a broad application of LAMP technique in a one-step assay that could be used to screen the infection as well as semi-quantify the Leishmania parasites in the buffy coat collected from patients with Leishmania/HIV co-infection. The development of a simplified and inexpensive dPAD combined with SYBR safe and gold‑nanoparticle probe LAMP assay was demonstrated that allowed rapid interpretation within a few minutes with high sensitivity and specificity.
Results
Semi-quantitative SYBR safe and AuNP-LAMP assay using dPAD
A schematic of the experimental principle and signal detection to visualize LAMP products in prereaction detection using fluorescent SYBR safe for rapid screening, and to visualize and quantify LAMP products in postamplification detection using LAMP probe-streptavidin conjugated AuNPs (SA-AuNP) for specific colorimetric precipitate induction as well as a distance-based paper device (dPAD) for semi-quantifying LAMP amplicons using DNA immobilized cellulose Whatman No.1, respectively, are shown in Fig. 1. Rapid qualitative screening could be done by closed tube SYBR safe LAMP method and directly visualized by the naked eye without a requirement of postamplification preparation step using blue light (BL) transilluminator (1). To re-evaluate and verify the first screening detection, 5 µL of LAMP reaction was transferred to a new tube for colorimetric precipitate detection by post hybridization of an AuNP-probe (2). Three processes were included, that is, complementary hybridization of the single strand probe-SA-AuNP to LAMP target, colorimetric and precipitate induction by MgSO4 salt and detection of colorimetric precipitate by the naked eye. The polymeric network of cross-linked AuNPs is formed according to the hybridization of the single strand probe-SA-AuNP that prevents the aggregation by salt induction, leading to unchanged red color of the reaction mixture. Conversely, the reaction mixture turns to pale purple with solid precipitates due to an absence of the LAMP targets that leads the aggregation of free probe-SA-AuNPs. The dPAD can be performed simultaneously to evaluate the degree of Leishmania infection in the samples by pipetting 2.5 µL of LAMP reaction of the closed tube SYBR safe LAMP method to the sample zone of the dPAD and waiting for 10 min (3). This simple process of detection includes LAMP reaction flowing into the channel of the detection zone, anionic DNA immobilized by cellulose Whatman No.1 filter paper, and distance measurement of the immobilized LAMP amplicons by the naked eye under BL transilluminator. The fluorescence of migratory distance correlates to the final amounts of LAMP products. DNA is a strongly anionic molecule and could be immobilized on the cellulose paper due to the physical adsorption and the charge interaction between DNA and the immobilized cationic polymers.
Optimization of dPAD for quantitative LAMP assay
The illustration of fabrication process is shown in Fig. 2a. After inactivating penetration of the hydrophobic wax ink and installing transparent adhesive tape to limit the boundary channel of sample and detection zones of the dPAD, the filter paper was dropped so that the LAMP reaction could flow into the detection channel zone without leakage of the flow samples. Five different patterns of dPAD were shown to immobilize the LAMP reaction at different lengths of migratory distance and could be directly measured by BL visualization. To acquire the suitable loading sample volume in the dPAD, the length difference of migratory distance between the negative and positive LAMP reactions at different parasites’ DNA concentration was evaluated. The optimization of each sample volume is shown in Fig. 2b. The samples were loaded at 0.5, 1.5 and 2.5 µL and the distance lengths of each load volume in millimeter scale (mm) were compared. The mean fluorescent distance of each concentration load volume was 1.78 ± 1.30 (mean ± SD) mm for negative control, 3.78 ± 2.11 mm for 105 parasites/mL and 5.11 ± 2.67 mm for 107 parasites/mL, respectively. The fluorescent distances were significantly differences in negative (N) and two positives (105 and 107) (F = 25.85, 79.15, 130.85; P-value = 0.001, < 0.001, < 0.001, respectively). The differences of the fluorescent migratory distance between positive and negative results at 0.5, 1.5 and 2.5 µL were 1.00, 2.00 and 3.00 mm with RSD values as 0.00 (n = 3), 0.00 (n = 3) and 0.00 (n = 3), respectively for 105 parasites/mL, and 1.67, 3.34 and 5.00 mm with RSD values as 0.35 (n = 3), 1.73 (n = 3) and 0.20 (n = 3), respectively for 107 parasites/mL. According to the difference of the flow results, the load volume at 2.5 µL demonstrated the longest immobilized length of fluorescent migratory distance on the filter paper as the best migratory distance. Of these, 2.5 µL which gained the highest fluorescent length of reaction flow distance were chosen as a standard load volume for all five patterns of the dPAD.
Fabrication process of a distance-based paper device (dPAD) and optimization of load sample volume, (a) schematic illustration of fabrication process, (b) fluorescent migratory distance of SYBR safe on the dPAD at different load volumes of LAMP reaction mixture (0.5, 1.5 and 2.5 µL, respectively): negative (N); no Leishmania DNA control, positives (105 and 107); 105 and 107 parasites/mL of Leishmania DNA.
All five patterns of the designed dPADs could differentiate between negative (N) and positive samples (105 and 107) (Fig. 3). The migratory distances were significantly differences in patterns 1 to 5 (F = 7.84, 5.69, 11.44, 9.60, 30.41; P-value = 0.026, 0.048, 0.012, 0.017, < 0.001, respectively). The distances of Leishmania’ DNA concentration at 105 and 107 parasites/mL were similar to patterns 1 to 4. The differences ranged between 1.67 to 2.00 mm depending on the patterns of the devices (Fig. 3a). However, the highest difference at 3.67 mm was observed in pattern 5 between 105 and 107 parasites/mL (Fig. 3b) that constituted a simple straight pattern in which the LAMP reaction was loaded in the sample channel zone at the bottom of the device and flowed into the detection channel directly straight forward. Thus, pattern 5 was chosen to gain the optimal migratory distance. These were further used to calibrate a device using a linearity between the fluorescent distance and the standard concentrations of Leishmania’ DNA, then for quantitative measurement of the LAMP products.
Effect of different patterns of sample and detection channel zones to immobilize migratory distance of LAMP reaction mixture on a distance-based paper device (dPAD), (a) mean and standard deviation of fluorescent migratory distance of five different patterns of dPAD, (b) fluorescent visualization of migratory distance of five patterns on dPAD. Abbreviations of variable names: no template control (N), positive Leishmania template control at 105 and 107 parasites/mL (P5, P7, respectively).
Semi-quantitative analysis and specificity
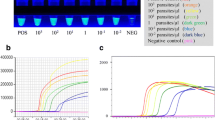
Detection of Leishmania in the dPAD and specific AuNP-probe, evaluated by tenfold serial dilution from 106 to 103 parasites/mL, were used to observe the calibration curve and linear range (Fig. 4). The results of serial dilution showed concordance of detection between the visual inspection of fluorescent dPAD (Fig. 4a) and colorimetric precipitate AuNPs probe, as well as gel electrophoresis (Fig. 4b). Gel electrophoresis exhibited various multiple bands of stem-loop DNA structures inferring successful amplification of LAMP products. The distance values of fluorescent migratory obtained from the dPAD at each concentration were plotted for observing the calibration curve and linear range. A calibration of the device showed a linearity between the fluorescent migration distance and Leishmania’s LAMP amplicons in the range of 103 to 106 parasites/mL with a linear equation as y = 1.132x + 4.439 and R2 as 0.997 (Fig. 4a).
Optimization of semi-quantitative analysis of a distance-based paper device (dPAD), (a) semi-quantitative measurement of fluorescent LAMP-dye-complex distance immobilized on dPAD (ten fold serial dilution; 106–103 parasites/mL), (b) detection of LAMP products at different concentration of Leishmania’ DNA using colorimetric precipitate of specific AuNP-LAMP probes and stem-loop DNA of LAMP amplicons in gel electrophoresis with 100 bp ladder: no template control (N).
Using visual inspection of fluorescent dPAD and colorimetric precipitate AuNPs probe, cross amplifications were not detected except T. evansi when nonLeishmania pathogens’ DNA was tested against the LAMP primers and probe. Obviously, differences in the fluorescent migratory distance were found in the LAMP positive reaction (11.7–12.7 mm) as compared with those of the negative control or reaction mixtures of nonLeishmania pathogens’ DNA (10.0–11.0 mm) (Fig. 5a) in that the specificity could be distinguished and correlated with the results of specific AuNP-LAMP probes as shown in Fig. 5b. The original gel is presented in Supplementary Figure S1.
Specificity of semi-quantitative distance-based paper device (dPAD) combined with SYBR safe and AuNP-LAMP assay, (a) detection and migratory distance of LAMP amplicons of Leishmania’ and different pathogens’ DNA using dPAD, (b) LAMP detection of different pathogens’ DNA based on probe-SA-AuNP: tube 1; no template control (NTC), tube 2; human genomic DNA (GC), tubes 3–13; E. coli, G. intestinalis, L. donovani, L. martiniquensis, L. orientalis, T. evansi, T. vaginalis, P. falciparum, N. gonorrhea, S. pyogenes and S. flexneri, respectively.
Evaluation of sensitivity and specificity of semi-quantitative SYBR safe and AuNP-LAMP assay using dPAD
Fifty four DNA samples extracted from buffy coat including Leishmania/HIV cases (n = 22) and uninfected HIV cases (n = 32) were used to evaluate and validate the one-step semi-quantitative dPAD combined with SYBR safe screening and specific AuNP-LAMP assay. Using the nested PCR as a reference standard, the sensitivity and specificity of the dPAD were 95.5 (n = 21; 95% CI 77.2 to 99.9%) and 100.0% (n = 32; 95% CI 89.1 to 100.0%), respectively (Table 1). In addition, the sensitivity of fluorescent detection using SYBR safe screening and colorimetric precipitate detection using posthybridization of AuNP-LAMP probes were the same as the dPAD, 95.5% (nfluorescent = 21, ncolorimetric = 21; 95% CI, 77.2 to 99.9%, respectively) but the specificity was lower in only the fluorescent closed tube method, 98.5% (n = 31; 95% CI 83.8 to 99.9%). The three different methods of this one-step LAMP assay showed significant results that were in almost perfect agreement when compared with reference standard, nested PCR (P-value < 0.001, < 0.001, < 0.001; Kappa = 0.92, 0.96, 0.96, respectively). However, the dPAD was in almost perfect agreement when compared with fluorescent closed tube and colorimetric precipitate LAMPs (P-value < 0.001, < 0.001; Kappa = 0.96, 0.96, respectively). According to the migratory distance immobilized by the device, the length of the dPAD could be semi-quantified and classified in two levels: the uninfected (7.30 ± 0.64; n = 33) and infected groups (10.23 ± 0.70; n = 21), respectively (Fig. 6). A significant difference in the fluorescent migratory distance between uninfected and infected groups was observed at a 95% confidence level (uninfected vs infected, t-test, P-value < 0.001).
A boxplot representation of length variation of migratory distance (mm) immobilized on a distance-based paper device in an uninfected group (N; n = 33) and infected group (P; n = 21). Lower and upper boundaries of each box indicate the 25th and the 75th percentile, respectively. Ranges are represented as bars (whiskers) below and above the box, and indicate the 10th and 90th percentiles, respectively. Mean values are marked by the red dot marks on the boxplots. Migratory fluorescent distance length values of each sample (2.5 µL loading volume) were represented by white circle marks (nuninfected = 33 and ninfected = 21).
Discussion
The successful development of the dPAD combined with SYBR safe and gold‑nanoparticle probe LAMP assay was achieved to semi-quantify the amount of Leishmania’ DNA among patients with Leishmania/HIV co-infection. This highly sensitive LAMP determination could be selectively performed using dual indicators, fluorescent SYBR safe and colorimetric precipitate gold-nanoparticle probe, in the one-step LAMP method. The assay could be simultaneously visualized in prereaction detection using fluorescent SYBR safe for rapid screening, and in postamplification detection using AuNP-LAMP probes for specific colorimetric precipitate induction15 and the dPAD for semi-quantification of LAMP amplicons using DNA immobilized cellulose Whatman No. 112. For rapid qualitative screening, fluorescent SYBR safe closed-tube LAMP method was used in prereaction detection, whereas, simultaneously in postamplification detection, the specific verification and quantitative evaluation could be further performed to visualize colorimetric precipitate using AuNP-LAMP probes and to detect the degree of Leishmania infection using fluorescent measurement of the migratory distance on the dPAD. Obviously, the remaining SYBR safe dye in the postamplification mixture of the AuNP-LAMP method did not interfere with the hybridizing and aggregating of the AuNPs-probe15. Furthermore, the fluorescent emission of the dye could be observed in prereaction mixture and semi-quantitative postamplification mixture using the dPAD. For AuNP-probe hybridization to the target of LAMP amplicons, the polymeric network of cross-linked AuNPs was formed to interpret the unchanged red after salt induction29,30 but pale purple with precipitates was induced to the aggregate in the absence of LAMP targets regarding no polymeric network36,37,38.
The best interpretation of semi-quantitative analysis was demonstrated in pattern number five with the load sample volume of 2.5 µL that constituted a simple straight pattern of sample and detection channel zones. The LAMP assay normally generates a high yield mixture of various lengths of stem-loop DNA amplicons in the reaction12. Owing to physical adsorption and covalent bonding between the amine-terminated nucleic acid and the polysaccharide39, the immobilization of DNA on cellulose such as Whatman No. 1 filter paper is useful for the separation and purification of DNA-binding proteins as well as LAMP products12. DNA is a strongly anionic molecule at pH 4.0 or above, and the charge interaction between DNA and the immobilized cationic polymers on cellulose has been reported40. The cellulose Whatman No. 1 filter paper is generally available and commercially affordable in all laboratories. Therefore, the filter paper is comparatively suitable and cost effective for fabricating the device to detect and semi-quantify LAMP products12. Unlike the qPCR method that can directly measure the amounts of DNA at each amplification cycle, the device indirectly quantify LAMP amplicons after final amplification. Thus, the migratory distance is limited and depends on the final LAMP products at each reaction. The quantitative measurement using migratory distance depended on the final LAMP products at each reaction. When the DNA precursor such as dNTPs was exhausted, the amplification would stop producing loop DNA and enter the plateau phase of LAMP12. At this upper limit level, the length of fluorescent migratory distance would be equal and not reflect the true amount of initial parasites’ DNA concentration as well as infection density. Thus, the semi-quantitative measurement could be done by the dPAD to avoid unambiguous naked eye comparison between positive and negative samples using the millimeter scale of the device. The fluorescent distance of the no template control would represent the residual unbinding SYBR safe in the reaction that has comparatively lower anionic. Wax-printed pillars as hydrophobic barriers of microfluidics could delay the flow and increase the sensitivity of the device41.
In spite of the unambiguous interpretation, the results of serial dilution showed concordance of the LAMP detection between visual inspection of fluorescence using the device for measuring Leishmania density and colorimetric precipitate AuNPs probe as a second clear-cut indicator for LAMP amplicon control15, as well as gel electrophoresis. The limitation of sensitivity of the SYBR safe closed tube and colorimetric precipitate AuNP-LAMP probes methods were previously reported to 102 parasites/mL or 0.0147 ng/µL15, exhibiting ten times more sensitivity than those of MG and SYBR safe assays11,14, and was equal to a simplified closed tube assay, SYBR green I16. To gain linearity between the fluorescent migration distance and Leishmania’s LAMP amplicons, the range 103 to 106 parasites/mL was used except without template control (N) because the value of the mean migratory distance was underestimated probably due to no LAMP amplicons in the reaction mixture that could demote physical adsorption and covalent bonding on the dPAD. The pan-leishmania LAMP primers could detect L. orientalis, L. martiniquensis and L. donovani based on high-copy-number 18S rRNA gene. The pan-leishmania LAMP assay is suitable and cost-effective for Leishmania screening because, at present, four Leishmania species coinfected with HIV patients in Thailand3. Similar to related studies, the amplification and cross reactivity with DNA from Trypanosoma sp., a closely related blood protozoan, were observed using pan-leishmania primers11,15,16. According to different clinical presentations exhibited by trypanosomiasis, the cross-amplification would not significantly lead to misdiagnosis11. In co-infected endemic areas, additional LAMP primers specific to Trypanosoma spp. could be additionally used to selectively identify the co-existence of these two blood protozoa42. Multiplex gene detection in the LAMP assay maybe be possible when the fluorescent dye labeling LAMP primers are specifically synthesized10. The sensitivity (n = 21, 95.5%) and specificity (n = 32, 100.0%) of semi-quantitative dPAD using immobilized filter paper were almost equivalent to those of LAMP methods using fluorescent SYBR safe closed tube (n = 21 and 31, 94.1 and 97.1%, respectively) and colorimetric precipitates using posthybridization of the AuNP-probe (n = 21 and 32, 95.5 and 100.0%, respectively) for detecting and semi-quantifying Leishmania’ DNA among patients with Leishmania/HIV co-infection when nested PCR was used as the reference standard. In the reaction mixture with a number of LAMP amplicons, the ambiguous evaluation could be interpreted regarding lower fluorescent intensity emissions. Thus, the SYBR safe closed tube is suitable for rapid screening in the prereaction mixture, whereas specific colorimetric precipitate AuNP-LAMP probes and the dPAD could be used as a second clear-cut indicator and semi-quantitative detection of the parasite density, respectively. Furthermore, the kappa statistical test indicated almost perfect agreement between these LAMP detecting methods. Other samples, e.g., saliva, should be examined to further validate feasibility and simplified use of this one-step semi-quantitative LAMP assay.
The rapid diagnostic test kit (RDT) is simpler and faster than the nucleic acid amplification test (NAAT), however, the sensitivity and specificity are lower. Thus, the NAATs are preferred to detect causative agents. Due to the requirement of costly instruments and well-trained lab technicians, NAATs are inconvenient and may not be affordable to local health service centers. Alternatively, basic equipments are required for LAMP assay, whereas an inexperienced worker could simply achieve the assay using step-by-step operating procedures15. Unlike RDTs such as malaria and VL strip tests, LAMP reaction required initial DNA for detection; therefore, DNA extraction is inevitable in the LAMP as well as all NAAT techniques. Various simplified methods have been successfully modified and adapted to the assays to overcome the limitations of the LAMP assay in field sites, for example, selective colorimetric and fluorescent dyes11,14,16,43, colorimetric precipitate AuNP-probes13,15, lyophilized forms44, paper-based devices12 and dye capsules45,46. Recently, the semi-quantitative LAMP assay was successfully developed to detect E. coli in urine using inexpensive filter paper fabrication of dPAD12. In this present work, we have demonstrated the applied spectrum of the LAMP technique that could be integrated in a one-step LAMP assay to screen, specifically verify and semi-quantify the Leishmania parasites among asymptomatic patients with Leishmania/HIV co-infection who usually harbor low parasite levels. A selective detection step using DNA-binding fluorescent dye for closed tube and semi-quantitative dPAD, and colorimetric precipitate of specific AuNP-LAMP probe can be interpreted simultaneously, taking approximately 85 to 95 min after adding a DNA sample that is faster than the standard nested PCR method, 8 to 10 h3. The cost of the reagents was not much different, approximately US $5 per reaction, based on the cost in Thailand. However, LAMP assay needs a simple heating device or noninstrument nucleic acid amplification (NINA) that is comparatively cheaper than PCR; especially, qPCR47. Using a simplified boiling extraction method11, the cost per reaction could be reduced about 70% and DNA preparation could be done within 10 min if DNA samples require short term preservation. By introducing Loop primers (LF, loop-forward and LB, loop-backward), the reaction time could be shorten due to rapid synthesis of LAMP products48. During post-reaction preparation step, the risk of contamination could be minimized using a simple closed system cabinet. The study supported that LAMP assays could be modified and adapted to broad spectrum of detection ranging from screening with the closed tube, and second clear-cut specific AuNP-probes to the semi-quantitative dPAD in which transilluminator was required to excite fluorescence as a trade-off for unambiguous interpretation. Thus, the LAMP assay is a very promising approach to facilitate early diagnosis and promote suitable treatment that can reduce severity and prevent progression of the disease. It is comparatively simple to proceed selectively with a simultaneous qualitative and semi-quantitative examination in a single assay without the requirement of sophisticated device, however, the assay is not suitable and sensitive for detecting the very low density of Leishmania infection that could be achieved by detecting devices (Table 2) such as strip reader35 or electrochemical detection34.
Conclusion
In this study, we developed a simplified and inexpensive dPAD combined with SYBR safe and gold‑nanoparticle probe LAMP assay to detect and semi-quantify Leishmania parasites in buffy coat of asymptomatic patients with Leishmania/HIV co-infection using colorimetric precipitate and fluorescence, respectively. The two additional steps of AuNPs-probes hybridization and semi-quantitative immobilized dPAD in postamplification preparation of closed tube SYBR safe LAMP assay required 5 and 10 min to clearly interpret the specific results and indirectly measure the number of LAMP amplicons. The sensitivities and specificities of closed tube SYBR safe, colorimetric precipitate AuNP-probes and the dPAD were equivalent, which were as high as 95.5%. The one-step semi-quantitative dPAD combined with SYBR safe screening and specific AuNP-LAMP assay is selective and suitable for simultaneous multipurpose Leishmania detection, for example, rapid direct screening in field environments using the closed tube SYBR safe method to reduce contamination prone14,17 or, re-evaluation and verification using posthybridization of the AuNPs-probe method to control and assure the first detection22,25 as well as semi-quantitative measurement using the dPAD to evaluate the level of infection for appropriate treatment12. These applications could empower either asymptomatic leishmaniasis surveillance among patients with HIV or prevention and control in the future. Hence, the assay could be used as an alternative method in addition to traditional PCR detection, especially in areas with limited resources that is reliably fast and practical for field diagnostics to healthcare services in low resource settings that is easily delivered to end-users, especially in low to middle income countries.
Materials and methods
Ethics statement
All participants aged > 18 years were informed and enrolled in the study. Written informed consent was received from all participants before collecting samples and analyzing anonymously. All methods were carried out according to relevant guidelines and regulations. This study was approved by the Ethics Committee of the Royal Thai Army Medical Department (IRBRTA 952/2562).
DNA extraction and PCR amplification for clinical samples and Leishmania parasites culture
Sample preparation
Eight milliliters of EDTA anti-coagulated blood samples were collected from eligible participants visiting the HIV Clinic at Satun Hospital, Satun Province during a 6-month follow-up period to receive antiretroviral therapy (ART)15,16. The whole blood specimen was centrifuged at 900×g for 10 min to separate the plasma and buffy coat, and was then kept at −20 °C until further used in DNA extraction15,16.
Owing to rapid cell growth and differentiation of Leishmania promastigotes in nutritious Schneider’s Drosophila medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 20% heat inactivated fetal bovine serum (GE Healthcare, Chicago, IL, USA) at 26 °C, a high yield of parasites could be obtained for DNA extraction15,16. A total of 107 promastigotes of L. orientalis (MON-324; MHOM/TH/2010/ TR), L. martiniquensis (MON-229; WHOM/TH/2011/PG) and L. donovani (MHOM/ET/67/ HU3) were harvested and washed three times with phosphate buffered saline (PBS) before DNA extraction15.
DNA extraction
DNA of buffy coat samples was extracted using the Geneaid™ DNA Isolation Kit (blood) (New Taipei, Taiwan), whereas genomic DNA of each species of Leishmania parasites was extracted using the DNeasy Extraction Kit (tissue) (Qiagen, Hilden, Germany) according to manufacturer protocols15,16. The concentration and quality of the extracted DNA were measured using a Nanodrop spectrophotometer (Denovix, Wilmington, DE, USA) at 260/280 and 260/230 ratios15,16. The DNA samples were kept at −20 °C until further use (Fig. S1).
PCR amplification and DNA detection
The internal transcribed spacer (ITS1) region of the ribosomal RNA (rRNA) gene of Leishmania was amplified based on nested PCR described by Manomat et al.3 using a FlexCycler2 Thermocycler (Analytik Jena, Jena, Germany). Promastigotes’ DNA of L. martiniquensis (WHOM/TH/2011/PG) was used as a positive control. The PCR products were electrophoresis on 1.5% agarose gel and visualized using a Molecular Imager® Gel Doc™ XR + System with Imager LabTM3.0 (Bio-Rad). Purified PCR products of positive samples were sent to Bionics Co. Ltd. (Seoul, South Korea) for sequencing. The sequencing chromatograms were validated and multiple-aligned with reference Leishmania strains retrieved from GenBank using BioEdit, Version 7.0.1.
Loop-mediated isothermal amplification (LAMP) assay
LAMP amplification
The conserved 18S rRNA gene of Leishmania was amplified based on LAMP assay as described by Ruang-areerate et al.15. A total volume of 25 µL LAMP reaction was prepared. Briefly, each reaction of LAMP mixture consisted of 40 pmol of FIP and BIP primers, 10 pmol of F3 and B3 primers, 1× Thermopol buffer, 8 mM MgSO4, 1.4 mM of each dNTP (Biotechrabbit, Hennigsdorf, Germany), 0.8 M betaine (Sigma-Aldrich), 8 U of Bst DNA Polymerase Large Fragment (New England Biolabs, Ipswich, MA, USA) and 2 µL of DNA template15,16. Additionally, 1× SYBR™ Safe DNA Gel Stain (Thermo Fisher Scientific, Waltham, MA, USA) was added to each reaction mixture14 as the nonspecific fluorescent dye indicator of LAMP products. The reaction was incubated at 65 °C for 75 min and then heated at 80 °C for 10 min to inactivate the reaction.
LAMP visualization
After incubating, the LAMP amplicons of target DNA were analyzed and confirmed based on direct visual inspection of the reaction tubes using either a blue light (BL) or an ultraviolet light UV transilluminator14,15. A positive amplification showed vivid fluorescent emission, whereas no fluorescent emission was observed in the absence of amplification. To determine a mixture of various lengths of the stem-loop DNA of LAMP products, 5 µL of reaction mixture was electrophoresed on 2% agarose gel stained with SYBR™ Safe (Thermo Fisher Scientific) and visualized using an UV transilluminator15,16.
Synthesis of LAMP probe conjugated AuNPs
Biotin LAMP probe labeling
The biotin labeling LAMP probes designed by Ruang-areerate et al.15 were used to hybridize the Leishmania LAMP targets. In brief, the loop forward (LF) region of 18S rRNA LAMP amplicons located between F2 and F1C regions was chosen and designed for specific probes that were complementary to the loop region. Finally, the LF probe was modified by functionalizing with biotin at 5′ region and custom synthesized by Bionics Co. Ltd. (Seoul, South Korea).
LAMP probe conjugated AuNPs
As described by Ruang-areerate et al.15, a 40 nm particle diameter of colloidal AuNPs stabilized in phosphate buffer that had been conjugated with streptavidin; OD = 10, were commercially synthesized by Kestrel Bioscience Thailand Co. Ltd., Thailand. Previously, streptavidin-AuNPs were diluted at 1:1 with phosphate buffer and subsequently mixed with 100 µM 5′-biotin labeling LF probe at 1:10 ratio by vigorous vortex for 15 to 30 s at room temperature in the dark to obtain 1 nmole streptavidin-AuNP-5′-biotin-probes structure (OD = 0.5). The LAMP probe conjugated AuNPs were then stored in the dark at 4 °C until used15.
To detect Leishmania LAMP amplicons, the ratio of the hybridization was conducted in a total volume of 15 µL at 65 °C for 5 min15. Briefly, the LAMP probe conjugated AuNPs and LAMP reaction mixture were mixed at 1:1 ratio. The concentration of MgSO4 was used at 100 mM in a 5 µL fix volume of postamplification mixture (15 µL) to induce aggregation and to visually detect color and colloidal change. A positive result indicated that free AuNP-LAMP probe-amplicon complexes were formed in the reaction remained deep red after incubating, whereas pale purple with dark insoluble precipitate was observed in the absence of LAMP products.
Device fabrication and quantitative LAMP assay
Fabrication of distance-based paper device
The dPADs were fabricated from Whatman filter paper No. 1 (Cole-Parmer, Vernon Hills, IL, USA). Five different patterns of dPADs were designed to semi-quantify LAMP amplicons that were divided in sample and detection zones with channel length 20 mm and width 2 mm using Adobe Illustrator, Version 25.1 (Adobe Inc., San Jose, CA, USA). All designs were printed by laser printer (Xerox ColorQube 8570, Japan) on the filter paper that the hydrophobic wax ink with a width 1 mm that controlled and limited the territory flow of the LAMP reaction sample. Then dPAD was baked at 120 °C for 3 min to induce the wax to penetrate into the filter paper and then cooled at room temperature to limit the boundary of the sample and detection zones of the dPAD. The back side of the device was sealed with a stacked transparent adhesive tape to prevent any leakage of the flow samples12.
Optimization of sample volume and pattern selection
To optimize the sample volume for the dPAD, loading volume of LAMP reaction was evaluated at 0.5, 1.5, and 2.5 µL. The sample volume that provided the highest fluorescent length of reaction flow distance regarding SYBR Safe indicator was chosen. The fluorescent distance of each loading volume was undertaken in triplicate. The migratory distances of five patterns of dPADs in triplicates were compared to select the dPAD that gained the clearest fluorescent distances at all concentrations. Linear regression was performed to assess the independent association of fluorescent distance and loading volumes at different concentrations (105 and 107 parasites/mL) as well as fluorescent distance and concentrations of five patterns.
Measurement of LAMP amplicons
To semi-quantify Leishmania parasites, the LAMP reaction mixture was dropped on the sample zone of the dPAD in which the sample flowed into the detection zone and dried within 10 min at room temperature. The device was evaluated by visual inspection under the blue light (BL) so that the migratory distance of fluorescence was measured in millimeter scale.
Semi-quantitative analysis and specificity
At the mid log phase, 20 µL of Leishmania parasites were collected and resuspended with 20 µL of phosphate buffered saline (PBS) containing 0.2% glutaraldehyde (GE, Healthcare, USA). The total parasite densities (parasites/mL) were counted at 400× magnification under light microscope using a Neubauer Chamber. To calibrate semi-quantitative analysis of the dPAD, purified genomic DNA of L. orientalis were tenfold serially diluted from 106 to 103 parasites/mL using a Nanodrop spectrophotometer, and were equivalent to 1.147 µg/µL to 0.147 ng/µL, respectively. The experiments were performed in triplicate, and nuclease-free water was used as negative control.
The positive detection using the paper device was confirmed by colorimetric precipitation of specific AuNP-LAMP probes and gel electrophoresis. The optimal volume of LAMP reaction mixture was pipetted to the sample zone of the dPAD as described previously. To achieve a ready use of the readout measurement, the millimeter scale was printed on the right side of the device. Then the mean distance of all LAMP reaction mixtures including negative and tenfold serial dilution of Leishmania’ DNA samples were substituted into a linear equation of calibration curve to calculate correlation coefficient (R2) of the visual readout obtained from the semi-quantitative fluorescent distance-based paper device.
To ensure that AuNP-LAMP probes were specific to Leishmania and cross-amplification with other pathogens was unlikely, the LAMP reactions were examined against human genomic DNA extracted from buffy coat and nonLeishmania’ DNA including genomic DNA extracted from Escherichia coli, Shigella flexneri, Streptococcus pyogenes, Neisseria gonorrhea, Plasmodium falciparum, Trichomonas vaginalis, Trypanosoma evansi and Giardia intestinalis as described by Ruang-areerate et al.15. Extracted DNA of three different Leishmania species (L. orientalis, L. martiniquensis and L. donovani) were used as positive controls, and nuclease-free water was used for no template control (NTC)15.
Evaluation of sensitivity and specificity of semi-quantitative SYBR safe and AuNP-LAMP assay using dPAD
Sensitivity and specificity of SYBR safe screening, specific AuNP-LAMP probes and semi-quantitative dPAD were determined using a total of 54 genomic DNA samples from buffy coat, consisting of 22 confirmed asymptomatic VL and 32 uninfected cases. Diagnosis of VL was confirmed when nested PCR targeting the ITS1 region of rRNA gene was positive and DNA sequence of the PCR amplicons was identical to Leishmania’ DNA. Nested PCR results are considered a reference standard method due to the unavailability of any gold standard15,16,49. These data were used to validate and evaluate the sensitivity and specificity of SYBR safe, specific AuNP-LAMP probes and dPAD in one-step LAMP assay. The strength of the agreement was determined between prereaction detection, including fluorescent closed tube (SYBR safe), and postamplification detection using AuNP-LAMP probes and the dPAD. LAMP methods were assessed using the kappa statistical test at 95% confidence intervals (CI) and P-value < 0.05 was considered statistically significant. The analysis was performed using STATA, Version SE14 (Stata Corporation, College Station, TX, USA).
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request.
References
Steverding, D. The history of leishmaniasis. Parasit. Vectors 10, 82. https://doi.org/10.1186/s13071-017-2028-5 (2017).
Leelayoova, S. et al. Leishmaniasis in Thailand: A review of causative agents and situations. Am. J. Trop. Med. Hyg. 96, 534–542. https://doi.org/10.4269/ajtmh.16-0604 (2017).
Manomat, J. et al. Prevalence and risk factors associated with Leishmania infection in Trang Province, southern Thailand. PLoS Negl. Trop. Dis. 11, e0006095. https://doi.org/10.1371/journal.pntd.0006095 (2017).
Jariyapan, N. et al. Leishmania (Mundinia) orientalis n. sp. (Trypanosomatidae), a parasite from Thailand responsible for localised cutaneous leishmaniasis. Parasit. Vectors 11, 351. https://doi.org/10.1186/s13071-018-2908-3 (2018).
Georgiadou, S. P., Makaritsis, K. P. & Dalekos, G. N. Leishmaniasis revisited: Current aspects on epidemiology, diagnosis and treatment. J. Transl. Med. 3, 43–50. https://doi.org/10.1515/jtim-2015-0002 (2015).
Rajapaksha, P. et al. A review of methods for the detection of pathogenic microorganisms. Analyst 144, 396–411. https://doi.org/10.1039/c8an01488d (2019).
Fallahi, S. et al. Diagnosis of Candida albicans: Conventional diagnostic methods compared to the loop-mediated isothermal amplification (LAMP) assay. Arch. Microbiol. 202, 275–282. https://doi.org/10.1007/s00203-019-01736-7 (2020).
Ghodsian, S., Rouhani, S., Fallahi, S., Seyyedtabaei, S. J. & Taghipour, N. Detection of spiked Fasciola hepatica eggs in stool specimens using LAMP technique. Iran. J. Parasitol. 14, 387–393 (2019).
Notomi, T., Mori, Y., Tomita, N. & Kanda, H. Loop-mediated isothermal amplification (LAMP): Principle, features, and future prospects. J. Microbiol. 53, 1–5. https://doi.org/10.1007/s12275-015-4656-9 (2015).
Gadkar, V. J., Goldfarb, D. M., Gantt, S. & Tilley, P. A. G. Real-time detection and monitoring of loop mediated amplification (LAMP) reaction using self-quenching and de-quenching fluorogenic probes. Sci. Rep. 8, 5548. https://doi.org/10.1038/s41598-018-23930-1 (2018).
Sriworarat, C., Phumee, A., Mungthin, M., Leelayoova, S. & Siriyasatien, P. Development of loop-mediated isothermal amplification (LAMP) for simple detection of Leishmania infection. Parasit. Vectors 8, 591. https://doi.org/10.1186/s13071-015-1202-x (2015).
Saengsawang, N. et al. Development of a fluorescent distance-based paper device using loop-mediated isothermal amplification to detect Escherichia coli in urine. Analyst. 145, 8077–8086. https://doi.org/10.1039/D0AN01306D (2020).
Suwannin, P. et al. Heat-enhancing aggregation of gold nanoparticles combined with loop-mediated isothermal amplification (HAG-LAMP) for Plasmodium falciparum detection. J. Pharm. Biomed. Anal. 203, 114178. https://doi.org/10.1016/j.jpba.2021.114178 (2021).
Thita, T., Manomat, J., Leelayoova, S., Mungthin, M. & Ruang-areerate, T. Reliable interpretation and long-term stability using SYBR™ safe fluorescent assay for loop-mediated isothermal amplification (LAMP) detection of Leishmania spp. Trop. Biomed. 36, 495–504 (2019).
Ruang-areerate, T. et al. Development of loop-mediated isothermal amplification (LAMP) assay using SYBR safe and gold-nanoparticle probe for detection of Leishmania in HIV patients. Sci. Rep. 11, 12152. https://doi.org/10.1038/s41598-021-91540-5 (2021).
Sukphattanaudomchoke, C. et al. Simplified closed tube loop mediated isothermal amplification (LAMP) assay for visual diagnosis of Leishmania infection. Acta Trop. 212, 105651. https://doi.org/10.1016/j.actatropica.2020.105651 (2020).
Adams, E. R., Schoone, G. J., Ageed, A. F., Safi, S. E. & Schallig, H. D. Development of a reverse transcriptase loop-mediated isothermal amplification (LAMP) assay for the sensitive detection of Leishmania parasites in clinical samples. Am. J. Trop. Med. Hyg. 82, 591–596. https://doi.org/10.4269/ajtmh.2010.09-0369 (2010).
Adams, E. R. et al. Development and evaluation of a novel loop-mediated isothermal amplification assay for diagnosis of cutaneous and visceral leishmaniasis. J. Clin. Microbiol. https://doi.org/10.1128/JCM.00386-18 (2018).
Nzelu, C. O., Kato, H. & Peters, N. C. Loop-mediated isothermal amplification (LAMP): An advanced molecular point-of-care technique for the detection of Leishmania infection. PLoS Negl. Trop. Dis. 13, e0007698. https://doi.org/10.1371/journal.pntd.0007698 (2019).
Muangchuen, A., Chaumpluk, P., Suriyasomboon, A. & Ekgasit, S. Colorimetric detection of Ehrlichia canis via nucleic acid hybridization in gold nano-colloids. Sensors. https://doi.org/10.3390/s140814472 (2014).
Garrido-Maestu, A. et al. Combination of microfluidic loop-mediated isothermal amplification with gold nanoparticles for rapid detection of Salmonella spp. in food samples. Front. Microbiol. 8, 2159. https://doi.org/10.3389/fmicb.2017.02159 (2017).
Arunrut, N. et al. Sensitive visual detection of AHPND bacteria using loop-mediated isothermal amplification combined with DNA-functionalized gold nanoparticles as probes. PLoS ONE 11, e0151769. https://doi.org/10.1371/journal.pone.0151769 (2016).
Wong, J. K. F., Yip, S. P. & Lee, T. M. H. Ultrasensitive and closed-tube colorimetric loop-mediated isothermal amplification assay using carboxyl-modified gold nanoparticles. Small 10, 1495–1499. https://doi.org/10.1002/smll.201302348 (2014).
Baptista, P. et al. Gold nanoparticles for the development of clinical diagnosis methods. Anal. Bioanal. Chem. 391, 943–950. https://doi.org/10.1007/s00216-007-1768-z (2008).
Wachiralurpan, S. et al. Rapid colorimetric assay for detection of Listeria monocytogenes in food samples using LAMP formation of DNA concatemers and gold nanoparticle-DNA probe complex. Front. Chem. 6, 90. https://doi.org/10.3389/fchem.2018.00090 (2018).
Pal, D. et al. Visual detection of Brucella in bovine biological samples using DNA-activated gold nanoparticles. PLoS ONE 12, e0180919. https://doi.org/10.1371/journal.pone.0180919 (2017).
Suebsing, R., Prombun, P. & Kiatpathomchai, W. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) combined with colorimetric gold nanoparticle (AuNP) probe assay for visual detection of Penaeus vannamei nodavirus (PvNV). Lett. Appl. Microbiol. 56, 428–435. https://doi.org/10.1111/lam.12065 (2013).
Kong, C. et al. Loop-mediated isothermal amplification for visual detection of Vibrio parahaemolyticus using gold nanoparticles. Microchim. Acta 185, 35. https://doi.org/10.1007/s00604-017-2594-4 (2018).
Baptista, P., Doria, G., Henriques, D., Pereira, E. & Franco, R. Colorimetric detection of eukaryotic gene expression with DNA-derivatized gold nanoparticles. J. Biotechnol. 119, 111–117. https://doi.org/10.1016/j.jbiotec.2005.02.019 (2005).
Baptista, P. V., Koziol-Montewka, M., Paluch-Oles, J., Doria, G. & Franco, R. Gold-nanoparticle-probe-based assay for rapid and direct detection of Mycobacterium tuberculosis DNA in clinical samples. Clin. Chem. 52, 1433–1434. https://doi.org/10.1373/clinchem.2005.065391 (2006).
Hongwarittorrn, I., Chaichanawongsaroj, N. & Laiwattanapaisal, W. Semi-quantitative visual detection of loop mediated isothermal amplification (LAMP)-generated DNA by distance-based measurement on a paper device. Talanta 175, 135–142. https://doi.org/10.1016/j.talanta.2017.07.019 (2017).
Parida, M. et al. Rapid detection and differentiation of dengue virus serotypes by a real-time reverse transcription-loop-mediated isothermal amplification assay. J. Clin. Microbiol. 43, 2895–2903. https://doi.org/10.1128/JCM.43.6.2895-2903.2005 (2005).
Sun, Y. et al. A lab-on-a-chip system with integrated sample preparation and loop-mediated isothermal amplification for rapid and quantitative detection of Salmonella spp. in food samples. Lab Chip 15, 1898–1904. https://doi.org/10.1039/C4LC01459F (2015).
de la Escosura-Muñiz, A. et al. Magnetic bead/gold nanoparticle double-labeled primers for electrochemical detection of isothermal amplified Leishmania DNA. Small 12, 205–213. https://doi.org/10.1002/smll.201502350 (2016).
Rivas, L. et al. Triple lines gold nanoparticle-based lateral flow assay for enhanced and simultaneous detection of Leishmania DNA and endogenous control. Nano Res. 8, 3704–3714. https://doi.org/10.1007/s12274-015-0870-3 (2015).
Han, X., Liu, Y. & Yin, Y. Colorimetric stress memory sensor based on disassembly of gold nanoparticle chains. Nano Lett. 14, 2466–2470. https://doi.org/10.1021/nl500144k (2014).
Shi, H.-Y. et al. A gold nanoparticle-based colorimetric strategy coupled to duplex-specific nuclease signal amplification for the determination of microRNA. Microchim. Acta 184, 525–531. https://doi.org/10.1007/s00604-016-2030-1 (2017).
Ge, Y. et al. Detection of influenza viruses by coupling multiplex reverse-transcription loop-mediated isothermal amplification with cascade invasive reaction using nanoparticles as a sensor. Int. J. Nanomed. 12, 2645–2656. https://doi.org/10.2147/IJN.S132670 (2017).
Su, S., Nutiu, R., Filipe, C. D. M., Li, Y. & Pelton, R. Adsorption and covalent coupling of ATP-binding DNA aptamers onto cellulose. Langmuir 23, 1300–1302. https://doi.org/10.1021/la060961c (2007).
Aied, A., Zheng, Y., Pandit, A. & Wang, W. DNA immobilization and detection on cellulose paper using a surface grown cationic polymer via ATRP. ACS Appl. Mater. Interfaces 4, 826–831. https://doi.org/10.1021/am201483h (2012).
Rivas, L., Medina-Sánchez, M., de la Escosura-Muñiz, A. & Merkoçi, A. Improving sensitivity of gold nanoparticle-based lateral flow assays by using wax-printed pillars as delay barriers of microfluidics. Lab Chip 14, 4406–4414. https://doi.org/10.1039/C4LC00972J (2014).
Njiru, Z. K. Rapid and sensitive detection of human African trypanosomiasis by loop-mediated isothermal amplification combined with a lateral-flow dipstick. Diagn. Microbiol. Infect. Dis. 69, 205–209. https://doi.org/10.1016/j.diagmicrobio.2010.08.026 (2011).
García-Bernalt Diego, J., Fernández-Soto, P. & Muro, A. LAMP in neglected tropical diseases: A focus on parasites. Diagnostics. 11(3), 521. https://doi.org/10.3390/diagnostics11030521 (2021).
García-Bernalt Diego, J. et al. Progress in loop-mediated isothermal amplification assay for detection of Schistosoma mansoni DNA: Towards a ready-to-use test. Sci. Rep. 9, 14744. https://doi.org/10.1038/s41598-019-51342-2 (2019).
Karthik, K. et al. New closed tube loop mediated isothermal amplification assay for prevention of product cross-contamination. MethodsX 1, 137–143. https://doi.org/10.1016/j.mex.2014.08.009 (2014).
Tao, Z. Y. et al. Adaptation of a visualized loop-mediated isothermal amplification technique for field detection of Plasmodium vivax infection. Parasit. Vectors 4, 115. https://doi.org/10.1186/1756-3305-4-115 (2011).
Curtis, K. A. et al. Isothermal amplification using a chemical heating device for point-of-care detection of HIV-1. PLoS ONE 7, e31432. https://doi.org/10.1371/journal.pone.0031432 (2012).
Nagamine, K., Hase, T. & Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16, 223–229. https://doi.org/10.1006/mcpr.2002.0415 (2002).
Rodríguez-Cortés, A. et al. Leishmania infection: Laboratory diagnosing in the absence of a “gold standard”. Am. J. Trop. Med. Hyg. 82, 251–256. https://doi.org/10.4269/ajtmh.2010.09-0366 (2010).
Acknowledgements
The authors are thankful to Worarachanee Imjaijitt, Office of Research and Development, Phramongkutklao Hospital & Phramongkutklao College of Medicine (ORD, PMK & PCM), for data analysis. We also acknowledge Asst. Prof. Tawin Inpankaew for sharing Trypanosoma evansi DNA. This research was supported by the Phramongkutklao College of Medicine, Kasetsart University Research and Development Institute; KURDI (FF (KU) 4.65), and Mahidol University (Basic Research Fund: fiscal year 2021 and 2022). Toon Ruang-areerate (T.R.) received supplementary financial support from the Anandamahidol Foundation.
Author information
Authors and Affiliations
Contributions
T.R. and W.D. conceived and designed the experiments. T.R., N.S., N.R. and T.T. conducted the experiments. T.R., K.C. and S.S. supported funding acquisition. T.R. and P.R. prepared figures. T.R. N.S., P.R. and N.R. analyzed the results. T.R., P.R., S.L., K.C. and W.D. contributed to the supervision and method. T.R., P.R., S.L. and W.D. wrote and reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ruang-areerate, T., Saengsawang, N., Ruang-areerate, P. et al. Distance-based paper device using combined SYBR safe and gold nanoparticle probe LAMP assay to detect Leishmania among patients with HIV. Sci Rep 12, 14558 (2022). https://doi.org/10.1038/s41598-022-18765-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-18765-w
- Springer Nature Limited
This article is cited by
-
Ultra-sensitive colorimetric detection of SARS-CoV-2 by novel gold nanoparticle (AuNP)-assisted loop-mediated isothermal amplification (LAMP) and freezing methods
Microchimica Acta (2024)
-
Validation of quantitative loop-mediated isothermal amplification assay using a fluorescent distance-based paper device for detection of Escherichia coli in urine
Scientific Reports (2023)
-
Genetic variation and geographic distribution of Leishmania orientalis and Leishmania martiniquensis among Leishmania/HIV co-infection in Thailand
Scientific Reports (2023)
-
The efficacy of AuNP-probe conjugate nanobiosensor in non-amplification and amplification forms for the diagnosis of leishmaniasis
BMC Infectious Diseases (2022)