Abstract
The studies recommended the relationship between lots of polymorphisms with the head and neck cancers (HNCs) risk. Herein, we reported the association between the CYP1A1 MspI polymorphism and the risk of HNC in an updated meta-analysis. The PubMed/MEDLINE, Web of Science, Cochrane Library, and Scopus databases were searched until March 31, 2021, without any restrictions. Odds ratios (ORs) and 95% confidence intervals (CIs) were applied to assess a relationship between CYP1A1 MspI polymorphism and the HNC risk based on five applied genetic models by RevMan 5.3 software. Other analyses (sensitivity analysis, meta-regression, and bias analysis) were performed by CMA 2.0 software. Trial sequential analysis (TSA) was done by TSA software (version 0.9.5.10 beta). Among the databases and other sources, 501 recorded were identified that at last, 29 studies were obtained for the analysis. The pooled ORs were 1.28 (95%CI 1.09, 1.51; P = 0.003), 1.68 (95%CI 1.16, 2.45; P = 0.007), 1.24 (95%CI 1.03, 1.50; P = 0.02), 1.26 (95%CI 1.07, 1.48; P = 0.005), and 1.66 (95%CI 1.27, 2.16; P = 0.0002) for allelic, homozygous, heterozygous, recessive, and dominant models, respectively. Therefore, the m2 allele and m1/m2 and m2/m2 genotypes had significantly increased risks in HNC patients. With regards to stable results and enough samples, the findings of the present meta-analysis recommended that there was an association between CYP1A1 MspI polymorphism and the HNC risk.
Similar content being viewed by others
Introduction
Head and neck cancer (HNC) affects more than 650,000 cases and 330,000 deaths each year1 and has remained a significant public health burden worldwide2. Men are significantly more affected by this type of cancer than women with a ratio of 2: 1 to 4: 1 and the prevalence of important anatomical sites of HNC (oral cavity, pharynx, and larynx) varies in different parts of the world3,4. HNC's current and future estimated load is shifting to less developed areas that may not have the equipment to cope with this increased load and this requires immediate attention by policymakers through the implementation of effective cancer control policies with population-based interventions2. There are many factors that can increase the incidence or prevalence of HNC, including the relative distribution of major risk factors such as alcohol consumption, tobacco, and smoking5. Genetic elements have also been implicated in the pathogenesis of this cancer. In support of this statement, several recent meta-analyses have confirmed the relationship of various polymorphisms with the risk of HNC6,7,8,9,10,11. Two reviews confirmed the relationship between several polymorphisms with the risk of HNC12,13. Therefore, HNCs are a complex multifactorial disorder that includes genetic, lifestyle, and environmental factors14,15. Cytochrome P450 (CYP) enzymes perform a major role in the metabolic activation of polycyclic aromatic hydrocarbons (PAHs) to epoxide intermediates, suggesting a link between PAHs, the CYP pathway, and cancer development that cytochrome P450 1A1 (CYP1A1) is believed to be the most important enzyme in this link16 and CYP1A1, as a drug-metabolizing enzyme, is among the main enzymes imported in the processing of tobacco-related carcinogens17. A studied polymorphism in the CYP1A1 gene (located on chromosome 15, including 9 exons or chromosome 15q22–24) has been shown to be related to the cancer risk, known as CYP1A1 MspI polymorphism (CYP1A1*2A)18 that CYP1A1 MspI is a T → C transition placed downstream of exon 7, in 3′ noncoding region19. This polymorphism may change the gene expression level or the messenger RNA stability due to highly induced enzymatic activity20. Seven meta-analyses checked the relationship between CYP1A1 MspI polymorphism and the risk of HNC including two case–control studies21, twelve in Asians with oral cancer22, seven23, thirty-two24, twelve including oral cancer25, twelve26, and twelve27. The meta-analysis He et al.24, although had more studies than other meta-analyses, focused on several types of cancer at the same time and didn’t provide information on sensitivity analysis, meta-regression, trial sequential analysis (TSA), and publication bias for HNC. In comparison with our study and other meta-analyses, this meta-analysis included thyroid cancer and different sites of head and neck as HNC, apart from that oral cavity, larynx, and pharynx. In comparison with the meta-analysis of He et al.24, we excluded studies that did not have a sufficient number of cases in their groups or their control groups had a deviation from Hardy–Weinberg equilibrium (HWE), because reducing the bias across the studies. Therefore, we aimed to evaluate the connection between the polymorphism of CYP1A1 MspI and the risk of HNC with twenty-nine studies in a meta-analysis, meta-regression, and TSA.
Materials and methods
Study design
This present study was designed by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) protocols28. The PICO (participants of interest, intervention, control, and outcome of interest) question was: Is CYP1A1 MspI polymorphism related to the HNC susceptibility comparing the prevalence of its alleles and genotypes in HNC patients in comparison with controls according to five genetic models?
Data sources and literature search
A systematic search was comprehensively used in PubMed/MEDLINE, Web of Science, Cochrane Library, and Scopus databases until March 31, 2021, without any restrictions. The used search terms were: (“cytochrome P4501A1” or “CYP1A1” or “AHH” or “aryl hydrocarbon hydroxylase”) and (“oral cancer” or “oral carcinoma” or “oral cavity cancer” or “OSCC” or “oral squamous cell carcinoma” or “oral SCC” or “tongue cancer” or “tongue carcinoma” or “mouth neoplasm” or “head and neck cancer” or “head and neck carcinoma” or “HNSCC” or “salivary gland cancer” or “salivary gland tumor” or “laryngeal cancer” or “larynx Cancer” or “nasopharyngeal cancer” or “nasopharynx cancer” or “Nasopharyngeal carcinoma” or “oropharyngeal cancer” or “oropharyngeal carcinoma” or “hypopharyngeal cancer” or “pharyngeal cancer” or “pharynx cancer” or “hypopharynx squamous cell carcinoma” or “hypopharynx SCC” or “larynx squamous cell carcinoma” or “larynx SCC”) and (“variant” or “polymorphism” or “genotype” or “gene” or “allele”). An independent review of titles and abstracts was conducted by two authors (H.M. and M.S.). A lack of consensus was resolved by a conversation with a third author (M.M.R). We manually checked other electronic sources for relevant studies and also the references of all subject-related studies that met the criteria so that no study was missed.
Criteria
Inclusion criteria were: (1) studies with a case–control design and reporting the association between CYP1A1 MspI polymorphism and the HNC susceptibility; (2) HNC was diagnosed by pathological or histological examinations; (3) sufficient data calculating the allele or genotype frequencies of CYP1A1 MspI polymorphism; (4) studies without a deviation from HWE in the control group or studies that HWE could not be computed (because there was no the prevalence of all genotypes separately); (5) Studies having 100 or more than 100 cases in both groups (case and control groups). Exclusion criteria were: (1) duplicate publications; (2) meta-analyses, reviews, letters to the editor, book chapters, conference papers, book chapters; (3) studies in the absence of control group; (4) studies reporting other polymorphisms of CYP1A1; and (5) studies reporting the CYP1A1 expression; (5) Studies with less than 100 cases in one or two groups; and (6) family-based studies. Among duplicate publications, we selected one with the newest date. An independent review of full-texts was conducted by two reviewers (H.R.M. and M.S.) and the disagreement was resolved by discussion between both reviewers.
Data extraction
The data of the involved studies were extracted independently by two reviewers (H.M. and M.S.) to retrieve the necessary information. In case of discrepancy between the data of the two reviewers, a new review was performed by other reviewers (M.M.R and F.R).
Quality assessment
The quality evaluation was performed according to a questionnaire from the Newcastle–Ottawa scale (NOS)29. The NOS included a maximum of nine scores for the least risk of bias in three domains: I) selection of study groups (four scores); II) comparability of groups (two scores); and III) ascertainment of exposure (three scores) for case–control studies30. Two reviewers (H.M. and M.S.) independently evaluated the quality of the included studies by scoring them according to a set of pre-established criteria and discrepancies were resolved by a short discussion.
Statistical analysis
Both odds ratio (OR) and 95% confidence interval (CI) were used to evaluate an association between the polymorphism of CYP1A1 MspI and the cancer risk. Five applied genetic models for CYP1A1 MspI polymorphism were (allelic (m2 vs. m1), homozygous (m2/m2 vs. m1/m1), heterozygous (m1/m2 vs. m1/m1), recessive (m2/m2 + m1/m2 vs. m1/m1), and dominant (m2/m2 vs. m1/m1 + m1/m2) models). To assess heterogeneity, a Chi-square-based Q test and inconsistency index I2 were applied31,32 that a P-value > 0.10 (I2 < 50%) presented a lack of heterogeneity and so we used fixed-effects model33 and if there was heterogeneity, the pooled results estimated by the random-effects model34.
Subgroup analysis is a method of analysis that involves dividing all participating data into smaller subsets based on a common feature and is often used to compare them and to examine the effects of different factors on the results. We divided the initial results based on ethnicity, control source, and tumor type.
Meta-regression is a quantitative method performed in meta-analysis to estimate the effect of moderators on the effect size of the study applying regression-based techniques35. We assessed the effect of publication year and sample size on the effect size.
There were two sensitivity analyses containing “one-study-removed” and cumulative analysis” to evaluate the stability/consistency of pooled results.
Funnel plots are visual tools for evaluating the types of biases in meta-analyses and are designed to examine whether publication bias can affect the reliability of estimates36. Both Begg’s37 and Egger’s38 tests were used for the diagnosis of asymmetry of these plots. Asymmetry can be a reason for bias in studies that in the state, P-values (two-sided) < 0.05 for the tests.
The P-values (two-sided) < 0.05 was as a significant index. The results of forest plot analyses were extracted by Review Manager 5.3 (RevMan 5.3) software and other analyses by Comprehensive Meta-Analysis version 2.0 (CMA 2.0) software.
We used TSA due to false-positive or negative conclusion39 in the meta-analysis using TSA software (version 0.9.5.10 beta) (Copenhagen Trial Unit, Centre for Clinical Intervention Research, Rigshospitalet, Copenhagen, Denmark) to reduce these statistical errors40. The required information size (RIS) was calculated when an alpha risk of 5%, a beta risk of 20%, and a two-sided boundary type were used. While the Z-curve reached the RIS line or monitoring the boundary line or futility area, it illustrated that enough samples are involved in the studies, and therefore their results were valid. Otherwise, the value of information was not great enough, and additional studies were needed.
Ethics approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations.
Results
Study selection
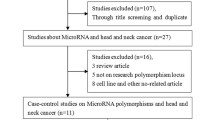
Among the databases and other sources, 501 recorded were identified (Fig. 1). After omitting duplicates and unrelated records, 91 full-text articles were evaluated for eligibility. Then, 57 articles excluded with reasons (one mixed oral precancerous and cancer cases, one had no control group, two reviews, four reported CYP1A1 expression, two didn’t report the prevalence of alleles and genotypes, one book chapter, twenty-two reported other polymorphisms of CYP1A1, two reported duplicate publications, one family-based study, one had no sufficient data, one reported oral precancerous cases, twelve studies reported less than 100 cases in one or two groups (case and control groups), and seven meta-analyses). After that, 34 studies17,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73 were included systematic review and we deleted 5 studies41,53,66,71,72 with a deviation from HWE in their control groups. Finally, 29 studies were entered into the analysis.
Basic characteristics
Table 1 is shown the characteristics of the studies17,42,43,44,45,46,47,48,49,50,51,52,54,55,56,57,58,59,60,61,62,63,64,65,67,68,69,70,73 involved in the meta-analysis. The studies were published from 1996 to 2019 including 8392 HNC cases and 8646 controls. Eighteen studies43,44,45,46,48,52,55,59,61,62,63,64,65,66,68,69,70,73 were reported in Asians, seven17,42,49,51,54,57,58 in Caucasians, and four47,50,56,60 in mixed ethnicity. The control source in eighteen studies42,43,44,47,49,50,51,52,54,58,60,61,62,63,66,69,70,73 was hospital-based and in eleven17,45,46,48,55,56,57,59,64,65,68 was population-based. The type of tumor and the genotyping method were other variables for the studies.
Quality assessment
Ten criteria were identified to evaluate the quality of the studies contained in the meta-analysis (Table 2). Twenty-five studies had a high quality (score ≥ 7).
Genotype prevalence
Table 3 is shown the genotype prevalence of CYP1A1 MspI polymorphism in the HNC patients and the controls. Seven studies17,44,46,48,50,52,62 had not reported any data about HWE.
Pooled analyses
Figures 2, 3, 4, 5 and 6 are shown the random-effects analyses of allelic, homozygous, heterozygous, recessive, and dominant models of the association between CYP1A1 MspI polymorphism and the risk of HNC, respectively. The pooled ORs were 1.28 (95%CI 1.09, 1.51; P = 0.003; I2 = 75%) for allelic, 1.68 (95%CI 1.16, 2.45; P = 0.007; I2 = 68%) for homozygous, 1.24 (95%CI 1.03, 1.50; P = 0.02; I2 = 66%) for heterozygous, 1.26 (95%CI 1.07, 1.48; P = 0.005; I2 = 75%) for recessive, and 1.66 (95%CI 1.27, 2.16; P = 0.0002; I2 = 64%) for dominant models. The m2 allele and m1/m2 and m2/m2 genotypes had significantly an elevated risk in HNC patients.
Subgroup analyses
The subgroup analysis was performed on the ethnicity, the control source, and the tumor type (Table 4). The results showed that ethnicity, control source, and tumor type could be effective factors on the pooled ORs. With regard to the ethnicity, the association of CYP1A1 MspI polymorphism and HNC risk based on five models (allelic, homozygous, heterozygous, recessive, and dominant), two models (allelic and heterozygous)), and two models (allelic and heterozygous) were statistically significant for Asian, Caucasian, and mixed ethnicities, respectively, that in contrast with Asian and Caucasian ethnicities, there was a decreased risk of m2 allele and m1/m2 genotype in mixed ethnicity. For the control source, the association was statistically significant in four models (allelic, homozygous, and dominant) for hospital-based controls and three models (heterozygous and recessive) for population-based controls. For tumor type, the association in four models (allelic, homozygous, recessive, and dominant) for oral cancer, three models (allelic, homozygous, and dominant) for laryngeal cancer, and three models (allelic, heterozygous, and recessive) for pharyngeal cancer was statistically significant.
Publication bias
Figure 7 is shown the funnel plots of the relationship between CYP1A1 MspI polymorphism and the risk of HNC based on the genetic models. Both Egger’s and Begg’s tests were: (allelic model: 0.322 and 0.151; homozygous model: 0.340 and 0.471; heterozygous model: 0.570 and 0.421; recessive model: 0.030 and 0.050; and dominant model: 0.064 and 0.243). The P-values > 0.05 were for both tests that determined lack of any publication bias across the studies, exception for Egger’s test in dominant model (P < 0.05) that showed the publication bias across the studies in this model.
Trial sequential analysis
The Z-curve (blue line) of the allelic, homozygous, heterozygous, recessive, and dominant models reached the RIS line (vertical red line), revealing that the CYP1A1 MspI polymorphism was related to the HNC risk with enough samples and reliable results that we selected the graphs for four models because of the better quality of the graphs (Fig. 8).
Trial sequential analyses for CYP1A1 MspI polymorphism and the head and neck risk. (A,B,C,D,E) show allelic, homozygous, heterozygous, and recessive models, respectively. Abbreviation: D2, diversity; RRR, relative risk reduction; IIA, incidence in intervention arm; ICA, incidence in control arm. IIA and ICA were calculated from the average incidence in case and control groups. Error α and 1 − β were defined as 5% and 80%, respectively in each model.
Sensitivity analysis
The sensitivity analyses including “one-study-removed” (Fig. 9) and “cumulative analysis” (Fig. 10) showed the stability of the initial pooled ORs. We included the results of the sensitivity analyses for the recessive model.
Meta-regression
A meta-regression analysis based on the publication year and the sample size were carried out on the relationship between the HNC risk and CYP1A1 MspI polymorphism (Table 5). The analysis showed the sample size in recessive and dominant models, the tumor type in allelic, homozygous, and heterozygous models, and the ethnicity in allelic, homozygous, recessive, and dominant models could be important confounding factors for the association between the HNC risk and CYP1A1 MspI polymorphism (P < 0.05). Increasing the sample size, the risk of HNC significantly increased (a direct correlation).
Discussion
A recent systematic review reported that 242 genes have associated with the risk of HNC74. Our meta-analysis reported the association of one of the polymorphisms (CYP1A1 MspI) in these genes with the HNC susceptibility. The results were stable and showed elevated risks of m2 allele and m2/m2 and m1/m2 genotypes in HNC patients with enough samples that the results were under the influence of the ethnicity, the tumor type, and the control source. In addition, the sample size, the tumor type, and the ethnicity could be confounding factors on the results.
A 5.71-fold risk of nasopharyngeal cancer has been reported in cases carrying glutathione-S-transferases (GSTs) such as GSTT1, GSTM1, and CYP1A1 MspI genotypes, suggesting that cross-linking between these genes may modulate nasopharyngeal cancer susceptibility, with similar results reported in HNCs17,46,62,71,75. As the results of one study showed, CYP1A1 polymorphisms alone were not related to an increased risk of oral cancer and the moderate risk for oral cancer was combining this polymorphism with GST polymorphisms69. Cha et al.76 showed the role of combined genotypes of CYP1A1 m2/m2 and GSTM1 null in the oral cancer risk. Cyp1A1 MspI polymorphism in lung cancer was associated with PAH-DNA adduct levels77 and the frequency of p53 gene mutations78. Smokers had the significant elevated risk (OR 7.13, P < 0.0001) of nasopharyngeal cancer among individuals carrying CYP1A1 MspI m2/m2 + m1/m2 genotype71.
One study in Northeast India found that the susceptibility to HNC related to tobacco and alcohol consumption is modulated by CYP1A1 MspI polymorphism, showing the interaction of gene-environment in prediction the HNC susceptibility and therefore this polymorphism is a predisposing risk factor for HNC70. In addition, another study reported tobacco use (particularly tobacco chewing) appeared as a significant moderator in cases with variant genotypes of CYP1A1 in India69. Sharma et al.65 expressed that CYP1A1 gene haplotype (C2453A, A2455G, and T3801C) frequency distribution in HNC patients was significantly higher than controls. Therefore, it is important to consider the haplotype and the combined impacts of genetic and environmental factors when examining the genetic risk to complex illnesses such as HNC62,65.
The discrepancies between results of the association between CYP1A1 polymorphisms and HNCs in Indians may be due to ethnic differences in culture, linguistics, and diets in this population, or they may be because of a difference in the sample size of the studies63. As our meta-analysis confirmed that the sample size was a confounding factor on the association and increasing the sample size, OR increased.
Wang et al.79 reported that the association between CYP1A1 polymorphisms and the risk of HNC could be affected by tumor type as that they observe an elevated risk of laryngeal and pharyngeal cancers, but no for oral cancer. Another study70 showed that the m2/m2 genotype of CYP1A1 MspI polymorphism had a significantly elevated risk in oral cancer patients, but there was no significant relationship between this polymorphism and pharyngeal and laryngeal cancers that one review80 confirmed it. Also, Sam et al.63 showed the association between m1/m2 genotype had just significant risk in laryngeal and pharyngeal cancers, not oral cancer. In our meta-analysis, the m1/m2 genotype had just a significant association with pharyngeal cancer and m2/m2 just in oral and laryngeal cancers.
Seven meta-analyses21,22,23,24,25,26,27 reported the association between CYP1A1 MspI polymorphism and the risk of HNCs. Two meta-analyses22,25 illustrated that the CYP1A1 MspI polymorphism may be associated with oral cancer susceptibility in Asians as well as Xie et al.81 in a stratified analysis by ethnicity, showed significant evidence of the association of CYP1A1 MspI polymorphism with the HNC risk in Asian ethnicity, but not mixed and Caucasian ethnicities. These results showed the impact of ethnicity on the relationship between CYP1A1 MspI polymorphism and the HNC risk as the meta-analysis of He et al.24 and our meta-analysis reported. In addition, our meta-analysis showed an association between other cancers (laryngeal and pharyngeal cancers), both in Asians and in other ethnicities (Caucasian and mixed ethnicities). Some studies82,83,84 and our meta-analysis to follow them, classified Indians in Caucasian ethnicity and some other studies85,86 as Asians, but based other studies87,88, Indians include several ethnicities (mixed). One possible difference between the results of studies can be due to the different classification of the ethnicity for each region. Therefore, it should be noted that there is a need for a comprehensive classification to select the type of ethnicity of each country or region in the future so that the results of meta-analyzes based on the ethnicity be more homogeneous. In addition, in the meta-analyzes mentioned21,22,23,24,25,26,27, the number of studies was different and this could be another reason for the difference between their results. So, more studies are needed in different areas in the world to reduce this difference in results between studies. As the results of different studies and their contradictions showed, the relationship between this polymorphism and HNC risk is influenced by various factors, and paying attention to the effective factors in future studies can provide a way to find more dominant factors. As a result, treatment of these patients and as a result of increasing their survival can be done more easily and under more effective and better conditions.
Apart from several strengths (enough samples, stability of the results, and lack of publication bias across the studies), the present meta-analysis also had some limitations as (1) High heterogeneity between the studies. (2) Few numbers of studies in Asian and mixed ethnicities. (3) The impact of risk factors on the results with different distributions in included studies. (4) We just included published studies.
Conclusions
The findings of the present meta-analysis recommended the association between CYP1A1 MspI polymorphism and the HNC susceptibility with enough samples and stable results. The ethnicity, the tumor type, the control source, and the sample size were significant risk factors for the results. Therefore, pay attention to these factors can be important in relation to the association of CYP1A1 MspI polymorphism and the HNC risk in future studies. In addition, well-designed studies with large samples in various areas of the world with precise matching criteria are required to reveal the present meta-analysis conclusions.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- HNC:
-
Head and neck cancer
- CYP:
-
Cytochrome P450
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- PAH:
-
Polycyclic aromatic hydrocarbon
- TSA:
-
Trial sequential analysis
- GST:
-
Glutathione-S-transferase
References
Bray, F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018).
Gupta, B., Johnson, N. W. & Kumar, N. Global epidemiology of head and neck cancers: A continuing challenge. Oncology 91, 13–23 (2016).
Bray, F., Ren, J. S., Masuyer, E. & Ferlay, J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int. J. Cancer 132, 1133–1145 (2013).
Lambert, R., Sauvaget, C., de Camargo Cancela, M. & Sankaranarayanan, R. Epidemiology of cancer from the oral cavity and oropharynx. Eur. J. Gastroenterol. Hepatol. 23, 633–641 (2011).
Sankaranarayanan, R., Masuyer, E., Swaminathan, R., Ferlay, J. & Whelan, S. Head and neck cancer: A global perspective on epidemiology and prognosis. Anticancer Res. 18, 4779–4786 (1998).
Ge, W., Jiao, Y. & Chang, L. The association between MTHFR gene polymorphisms (C677T, A1298C) and oral squamous cell carcinoma: A systematic review and meta-analysis. PLoS ONE 13, e0202959 (2018).
Li, L. & Zhang, Z.-T. Genetic association between NFKBIA and NFKB1 gene polymorphisms and the susceptibility to head and neck cancer: A meta-analysis. Dis. Mark. 2019, 6523837 (2019).
Rezaei, F. et al. Evaluation of serum and salivary interleukin-6 and interleukin-8 levels in oral squamous cell carcinoma patients: Systematic review and meta-analysis. J. Interferon Cytokine Res. 39, 727–739 (2019).
Rezaei, F., Doulatparast, D. & Sadeghi, M. Common polymorphisms of interleukin-10 (-1082A/G,-592A/C, and-819C/T) in oral cancers: An updated meta-analysis. J. Interferon Cytokine Res. 40, 357–369 (2020).
Du, X. Y. et al. Association between the aldehyde dehydrogenase-2 rs671 G> a polymorphism and head and neck cancer susceptibility: A meta-analysis in east asians. Alcoholism Clin. Exp. Res. 45, 307–317 (2021).
Mozaffari, H. R. et al. A PRISMA-compliant meta-analysis on association between X-ray repair cross complementing (XRCC1, XRCC2, and XRCC3) polymorphisms and oral cancer susceptibility. Gene 781, 145524 (2021).
Hiyama, T., Yoshihara, M., Tanaka, S. & Chayama, K. Genetic polymorphisms and head and neck cancer risk. Int. J. Oncol. 32, 945–973 (2008).
Okoturo, E., Osasuyi, A. & Opaleye, T. Genetic polymorphism of head and neck cancers in African populations: A systematic review. OTO Open 4, 24739742X0942202 (2020).
Abusail, M. S., Dirweesh, A. M. A., Salih, R. A. A. & Gadelkarim, A. H. Expression of EGFR and p53 in head and neck tumors among Sudanese patients. Asian Pac. J. Cancer Prev. 14, 6415–6418 (2013).
Maasland, D. H., van den Brandt, P. A., Kremer, B., Goldbohm, R. A. S. & Schouten, L. J. Alcohol consumption, cigarette smoking and the risk of subtypes of head-neck cancer: Results from the Netherlands Cohort Study. BMC Cancer 14, 1–14 (2014).
Quiñones, L. & Gil, L. Induction of rat hepatic cytochrome P450 1A1 isozyme by organic extracts from airborne particulate matter. Xenobiotica 25, 571–579 (1995).
Zakiullah, Z. et al. Genetic susceptibility to oral cancer due to combined effects of GSTT1, GSTM1 and CYP1A1 gene variants in tobacco addicted patients of Pashtun ethnicity of Khyber Pakhtunkhwa province of Pakistan. Asian Pac. J. Cancer Prev. 16, 1145–1150 (2015).
Crofts, F. et al. A novel CYP1A1 gene polymorphism in African-Americans. Carcinogenesis 14, 1729–1731 (1993).
Cascorbi, I., Brockmöller, J. & Roots, I. A C4887A polymorphism in exon 7 of human CYP1A1: Population frequency, mutation linkages, and impact on lung cancer susceptibility. Can. Res. 56, 4965–4969 (1996).
Shah, P. P., Saurabh, K., Pant, M. C., Mathur, N. & Parmar, D. Evidence for increased cytochrome P450 1A1 expression in blood lymphocytes of lung cancer patients. Mutat. Res./Fundam. Mol. Mech. Mutagen. 670, 74–78 (2009).
Brunotto, M., Zarate, A., Bono, A., Barra, J. L. & Berra, S. Risk genes in head and neck cancer: A systematic review and meta-analysis of last 5 years. Oral Oncol. 50, 178–188 (2014).
Xu, J. et al. Association of CYP1A1 MspI polymorphism with oral cancer risk in Asian populations: A meta-analysis. Genet Mol. Res. 15, gmr7688 (2016).
Porchia, L. M., Meneses-Sanchez, P., Ruiz-Vivanco, G., Perez-Fuentes, R. & Gonzalez-Mejia, M. E. CYP1A1 MspI polymorphism and cancer susceptibility among Latinos: A meta-analysis. Meta Gene 11, 197–204 (2017).
He, X.-F. et al. Association between the CYP1A1 T3801C polymorphism and risk of cancer: Evidence from 268 case–control studies. Gene 534, 324–344 (2014).
Yu, K.-T. et al. CYP1A1 polymorphism interactions with smoking status in oral cancer risk: Evidence from epidemiological studies. Tumor Biol. 35, 11183–11191 (2014).
Han, J., Wang, L., Yang, Y. & Zhang, J. Meta-analyses of the effect of CYP1A1 and CYP2D6 polymorphisms on the risk of head and neck squamous cell carcinoma. Oncol. Res. Treatment 37, 406–411 (2014).
Zhuo, X. et al. Quantitative assessment of CYP1A1* 2A variations with oral carcinoma susceptibility: Evidence from 1,438 cases and 2,086 controls. Cancer Invest. 30, 552–559 (2012).
Page, M. J. et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 372, n71 (2021).
Wells, G. A. et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses (accessed 19 October 2009); http://www.ohri.ca/programs/clinical_epidemiology/oxford.htm (2009).
Lo, C.K.-L., Mertz, D. & Loeb, M. Newcastle-Ottawa Scale: Comparing reviewers’ to authors’ assessments. BMC Med. Res. Methodol. 14, 1–5 (2014).
Higgins, J. P. & Thompson, S. G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 21, 1539–1558 (2002).
Higgins, J. P., Thompson, S. G., Deeks, J. J. & Altman, D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003).
Mantel, N. & Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl Cancer Inst. 22, 719–748 (1959).
DerSimonian, R. & Laird, N. Meta-analysis in clinical trials. Control. Clin. Trials 7, 177–188 (1986).
López-López, J. A., Van den Noortgate, W., Tanner-Smith, E. E., Wilson, S. J. & Lipsey, M. W. Assessing meta-regression methods for examining moderator relationships with dependent effect sizes: AM onte C arlo simulation. Res. Synth. Methods 8, 435–450 (2017).
Sterne, J. A. & Egger, M. Funnel plots for detecting bias in meta-analysis: Guidelines on choice of axis. J. Clin. Epidemiol. 54, 1046–1055 (2001).
Begg, C. B. & Mazumdar, M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–1101 (1994).
Egger, M., Smith, G. D., Schneider, M. & Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–634 (1997).
Imberger, G., Thorlund, K., Gluud, C. & Wetterslev, J. False-positive findings in Cochrane meta-analyses with and without application of trial sequential analysis: An empirical review. BMJ Open 6, e011890 (2016).
Wetterslev, J., Jakobsen, J. C. & Gluud, C. Trial sequential analysis in systematic reviews with meta-analysis. BMC Med. Res. Methodol. 17, 1–18 (2017).
Anantharaman, D., Chaubal, P. M., Kannan, S., Bhisey, R. A. & Mahimkar, M. B. Susceptibility to oral cancer by genetic polymorphisms at CYP1A1, GSTM1 and GSTT1 loci among Indians: Tobacco exposure as a risk modulator. Carcinogenesis 28, 1455–1462 (2007).
Boccia, S. et al. CYP1A1, CYP2E1, GSTM1, GSTT1, EPHX1 exons 3 and 4, and NAT2 polymorphisms, smoking, consumption of alcohol and fruit and vegetables and risk of head and neck cancer. J. Cancer Res. Clin. Oncol. 134, 93–100 (2008).
Chatterjee, S. et al. Polymorphisms of CYP1A1, GSTM1 and GSTT1 loci as the genetic predispositions of oral cancers and other oral pathologies: Tobacco and alcohol as risk modifiers. Indian J. Clin. Biochem. 25, 260–272 (2010).
Chen, W., Mu, J. & Guo, M. Association of EC-SOD, CYP1A1 gene polymorphisms, and smoking with oral cancer. Int. J. Clin. Exp. Med. 10, 12412–12418 (2017).
Cheng, Y.-J. et al. No association between genetic polymorphisms of CYP1A1, GSTM1, GSTT1, GSTP1, NAT2, and nasopharyngeal carcinoma in Taiwan. Cancer Epidemiol. Prevent. Biomark. 12, 179–180 (2003).
Choudhury, J. H. et al. Tobacco carcinogen-metabolizing genes CYP1A1, GSTM1, and GSTT1 polymorphisms and their interaction with tobacco exposure influence the risk of head and neck cancer in Northeast Indian population. Tumor Biol. 36, 5773–5783 (2015).
Cury, N. M. et al. Polymorphisms of the CYP1A1 and CYP2E1 genes in head and neck squamous cell carcinoma risk. Mol. Biol. Rep. 39, 1055–1063 (2012).
Dong, T., Wang, L., Liu, L. & Ma, S. Susceptibility to oral squamous cell carcinoma: Correlation with variants of CYP1A1-MspI, GSTT1, GSTM1, ALDH2, EC-SOD and Lifestyle factors. Balkan J. Med. Genet. 19, 61–70 (2016).
Gajecka, M. et al. CYP1A1, CYP2D6, CYP2E1, NAT2, GSTM1 and GSTT1 polymorphisms or their combinations are associated with the increased risk of the laryngeal squamous cell carcinoma. Mutat. Res./Fundam. Mol. Mech. Mutagen. 574, 112–123 (2005).
Gattás, G. J. F. et al. Genetic polymorphisms of CYP1A1, CYP2E1, GSTM1, and GSTT1 associated with head and neck cancer. Head Neck J. Sci. Spec. Head Neck 28, 819–826 (2006).
Gronau, S., Koenig-Greger, D., Jerg, M. & Riechelmann, H. Gene polymorphisms in detoxification enzymes as susceptibility factor for head and neck cancer?. Otolaryngol. Head Neck Surg. 128, 674–680 (2003).
Guo, L., Zhang, C., Shi, S. & Guo, X. Correlation between smoking and the polymorphisms of cytochrome P450 1A1-Msp I and glutathione S-transferase T1 genes and oral cancer. Hua xi kou qiang yi xue za zhi = Huaxi kouqiang yixue zazhi = West China J. Stomatol. 30, 187–191 (2012).
Kao, S. Y. et al. Genetic polymorphism of cytochrome P4501A1 and susceptibility to oral squamous cell carcinoma and oral precancer lesions associated with smoking/betel use. J. Oral Pathol. Med. 31, 505–511 (2002).
Ko, Y. et al. Association of CYP1B1 codon 432 mutant allele in head and neck squamous cell cancer is reflected by somatic mutations of p53 in tumor tissue. Can. Res. 61, 4398–4404 (2001).
Lourembam, D. S. et al. Evaluation of risk factors for nasopharyngeal carcinoma in a high-risk area of India, the Northeastern Region. Asian Pac. J. Cancer Prev. 16, 4927–4935 (2015).
Lourenço, G. J., Silva, E. F. S. N., Rinck-Junior, J. A., Chone, C. T. & Lima, C. S. P. CYP1A1, GSTM1 and GSTT1 polymorphisms, tobacco and alcohol status and risk of head and neck squamous cell carcinoma. Tumor Biol. 32, 1209–1215 (2011).
Lucas, D. et al. Cytochromes P4502E1 and P4501A1 genotypes and susceptibility to cirrhosis or upper aerodigestive tract cancer in alcoholic caucasians. Alcoholism Clin. Exp. Res. 20, 1033–1037 (1996).
Matthias, C., Jahnke, V., Fryer, A. & Strange, R. First results on the influence of polymorphisms at glutathione S-transferase, cytochrome P450, and tumor necrosis factor gene loci on the development of multiple head and neck cancer. Laryngorhinootologie 82, 25–30 (2003).
Maurya, S. S. et al. Gene-environment interactions in determining differences in genetic susceptibility to cancer in subsites of the head and neck. Environ. Mol. Mutagen. 56, 313–321 (2015).
Olivieri, E. H. R. et al. CYP1A2* 1C, CYP2E1* 5B, and GSTM1 polymorphisms are predictors of risk and poor outcome in head and neck squamous cell carcinoma patients. Oral Oncol. 45, e73–e79 (2009).
Sabitha, K., Reddy, M. V. & Jamil, K. Smoking related risk involved in individuals carrying genetic variants of CYP1A1 gene in head and neck cancer. Cancer Epidemiol. 34, 587–592 (2010).
Sam, S. S., Thomas, V., Reddy, K., Surianarayanan, G. & Chandrasekaran, A. Gene–environment interactions associated with CYP1A1 MspI and GST polymorphisms and the risk of upper aerodigestive tract cancers in an Indian population. J. Cancer Res. Clin. Oncol. 136, 945–951 (2010).
Sam, S. S., Thomas, V., Reddy, S. K., Surianarayanan, G. & Chandrasekaran, A. CYP1A1 polymorphisms and the risk of upper aerodigestive tract cancers in an Indian population. Head Neck J. Sci. Spec. Head Neck 30, 1566–1574 (2008).
Sato, M., Sato, T., Izumo, T. & Amagasa, T. Genetic polymorphism of drug-metabolizing enzymes and susceptibility to oral cancer. Carcinogenesis 20, 1927–1931 (1999).
Sharma, R., Ahuja, M., Panda, N. K. & Khullar, M. Combined effect of smoking and polymorphisms in tobacco carcinogen-metabolizing enzymes CYP1A1 and GSTM1 on the head and neck cancer risk in North Indians. DNA Cell Biol. 29, 441–448 (2010).
Shukla, D., Dinesh Kale, A., Hallikerimath, S., Vivekanandhan, S. & Venkatakanthaiah, Y. Genetic polymorphism of drug metabolizing enzymes (GSTM1 and CYP1A1) as risk factors for oral premalignant lesions and oral cancer. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 156, 253–259 (2012).
Shukla, D. et al. Association between GSTM1 and CYP1A1 polymorphisms and survival in oral cancer patients. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 157, 304–310 (2013).
Singh, A. P. et al. Polymorphism in cytochrome P4501A1 is significantly associated with head and neck cancer risk. Cancer Invest. 27, 869–876 (2009).
Singh, R. et al. Gene polymorphisms, tobacco exposure and oral cancer susceptibility: A study from G ujarat, West India. Oral Dis. 20, 84–93 (2014).
Singh, S. A. et al. Influence of the CYP1A1 T3801C polymorphism on tobacco and alcohol-associated head and neck cancer susceptibility in Northeast India. Asian Pac. J. Cancer Prev. 16, 6953–6961 (2015).
Singh, S. A. & Ghosh, S. K. Metabolic phase I (CYPs) and phase II (GSTs) gene polymorphisms and their interaction with environmental factors in nasopharyngeal cancer from the ethnic population of northeast India. Pathol. Oncol. Res. 25, 33–44 (2019).
Tai, J. et al. Genetic polymorphisms in cytochrome P450 genes are associated with an increased risk of squamous cell carcinoma of the larynx and hypopharynx in a Chinese population. Cancer Genet. Cytogenet. 196, 76–82 (2010).
Tanimoto, K., Hayashi, S.-I., Yoshiga, K. & Ichikawa, T. Polymorphisms of the CYP1A1 and GSTM1 gene involved in oral squamous cell carcinoma in association with a cigarette dose. Oral Oncol. 35, 191–196 (1999).
Kasradze, D., Juodzbalys, G., Guobis, Z., Gervickas, A. & Cicciù, M. Genetic and proteomic biomarkers of head-and-neck cancer: A systematic review. J. Cancer Res. Ther. 16, 410 (2020).
Sánchez-Siles, M. et al. Genotype of null polymorphisms in genes GSTM1, GSTT1, CYP1A1, and CYP1A1* 2A (rs4646903 T> C)/CYP1A1* 2C (rs1048943 A> G) in patients with larynx cancer in southeast spain. Cancers 12, 2478 (2020).
Cha, I.-H. et al. Polymorphisms of CYP1A1 and GSTM1 genes and susceptibility to oral cancer. Yonsei Med. J. 48, 233–239 (2007).
Hemminki, K. et al. Aromatic DNA adducts in foundry workers in relation to exposure, life style and CYP1A1 and glutathione transferase M1 genotype. Carcinogenesis 18, 345–350 (1997).
Kawajiri, K., Eguchi, H., Nakachi, K., Sekiya, T. & Yamamoto, M. Association of CYP1A1 germ line polymorphisms with mutations of the p53 gene in lung cancer. Can. Res. 56, 72–76 (1996).
Wang, Y., Yang, H., Duan, G. & Wang, H. The association of the CYP1A1 Ile462Val polymorphism with head and neck cancer risk: Evidence based on a cumulative meta-analysis. Onco. Targets. Ther. 9, 2927 (2016).
Liu, L. et al. Functional CYP1A1 genetic variants, alone and in combination with smoking, contribute to development of head and neck cancers. Eur. J. Cancer 49, 2143–2151 (2013).
Xie, S. et al. CYP1A1 MspI polymorphism and the risk of oral squamous cell carcinoma: Evidence from a meta-analysis. Mol. Clin. Oncol. 4, 660–666 (2016).
Luo, H., Li, H., Yao, N., Hu, L. & He, T. Association between 3801T> C polymorphism of CYP1A1 and idiopathic male infertility risk: A systematic review and meta-analysis. PLoS ONE 9, e86649 (2014).
Dong, L.-P. & Da Huang, W.-J.C. Meta-analysis of the association between Apo-A1 rs670, rs5069 polymorphisms and coronary artery diseases. Int. J. Clin. Exp. Med. 11, 6445–6453 (2018).
Han, M. et al. Associations of MMP-2− 1306 C/T and MMP-9− 1562 C/T polymorphisms with breast cancer risk among different populations: A meta-analysis. Genes Genom. 39, 331–340 (2017).
He, F. et al. Association between the Glutathione-S-transferase T1 null genotype and esophageal cancer susceptibility: A meta-analysis involving 11,163 subjects. Oncotarget 9, 15111 (2018).
Dar, S. A. et al. Interleukin-6-174G> C (rs1800795) polymorphism distribution and its association with rheumatoid arthritis: A case-control study and meta-analysis. Autoimmunity 50, 158–169 (2017).
Kashyap, V. et al. Genetic structure of Indian populations based on fifteen autosomal microsatellite loci. BMC Genet. 7, 1–9 (2006).
Wang, Y., Yang, J., Huang, J. & Tian, Z. Tumor necrosis factor-α polymorphisms and cervical cancer: Evidence from a meta-analysis. Gynecol. Obstet. Invest. 85, 153–158 (2020).
Author information
Authors and Affiliations
Contributions
Conceptualization: H.M. Methodology: M.S. Validation: M.M.R. Formal analysis: M.S. Resources: M.S. Writing- original draft preparation: M.S. Writing-review and editing: M.M.R., F.R., A.G., H.H., B.G. Visualization, supervision: H.M. Project administration: H.M. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mohammadi, H., Momeni Roochi, M., Rezaei, F. et al. Association between the CYP1A1 MspI polymorphism and risk of head and neck cancer: a meta-analysis. Sci Rep 12, 1527 (2022). https://doi.org/10.1038/s41598-022-05274-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-05274-z
- Springer Nature Limited