Abstract
Ralstonia pseudosolanacearum GMI1000 (Rpso GMI1000) is a soil-borne vascular phytopathogen that infects host plants through the root system causing wilting disease in a wide range of agro-economic interest crops, producing economical losses. Several features contribute to the full bacterial virulence. In this work we study the participation of light, an important environmental factor, in the regulation of the physiological attributes and infectivity of Rpso GMI1000. In silico analysis of the Rpso genome revealed the presence of a Rsp0254 gene, which encodes a putative blue light LOV-type photoreceptor. We constructed a mutant strain of Rpso lacking the LOV protein and found that the loss of this protein and light, influenced characteristics involved in the pathogenicity process such as motility, adhesion and the biofilms development, which allows the successful host plant colonization, rendering bacterial wilt. This protein could be involved in the adaptive responses to environmental changes. We demonstrated that light sensing and the LOV protein, would be used as a location signal in the host plant, to regulate the expression of several virulence factors, in a time and tissue dependent way. Consequently, bacteria could use an external signal and Rpsolov gene to know their location within plant tissue during the colonization process.
Similar content being viewed by others
Introduction
Light is an important environmental factor in all ecosystems because it is a source of energy and information. Almost all organisms can use light to sense their surroundings and thus be able to adapt to environmental changes, allowing them survival1.
Plant physiology is deeply regulated by environmental factors, being light probably one of the most relevant. As well as direct effects on plant metabolism, growth and development, light inevitably influences many other plant responses, including those induced by pathogen attack2. The role of light in host defense responses has been widely studied and it is known that an appropriate light environment is required for a full defense response3,4,5,6.
In phytopathogenic bacteria, light can define the result of plant-pathogen interactions, not only by affecting the plant's defense responses but also by modulating the virulence of the pathogens7. Recent reports revealed the light influence on bacterial lifestyle transitions, motility, and virulence8. Bacterial plant pathogens evolved to detect light conditions associated with different levels of plant resistance. Xanthomonas citri subsp. citri (Xcc) is a non-vascular hemibiotrophic phytopathogen responsible for citrus canker disease. Xcc physiology and its ability to colonize the host plant tissue are modulated by light perception9. In addition, Pseudomonas syringae pv. tomato DC3000 (Psto), another hemibiotrophic bacterium that causes bacterial speck in tomatoes, regulates its motility and virulence under different light conditions10,11. These bacteria, before the colonization of the host plant apoplast, grow epiphytically on the leaves surface having an important dose of solar radiation.
Light signals, their wavelengths, fluctuations in intensity and degree of polarization are perceived and transmitted by photoreceptor proteins. These proteins are classified into six different families: rhodopsins, phytochromes, photoactive proteins yellow (PYP, also called xanthopsins), LOV proteins (Light, Oxygen or Voltage), cryptochromes and BLUF (Blue-Light Sensing Using Flavin) proteins7,12. LOV proteins are a type of blue light photoreceptors, which are flavin binding proteins that use a flavin mononucleotide (FMN) as a chromophore13. The prominent role of LOV photoreceptor in the virulence processes of different pathogenic bacteria such as Brucella abortus14,15, Pseudomonas syringae pv. syringae16, Pseudomonas syringae pv. tomato10,11,17 and beneficial bacteria such as Rhizobium leguminosarum18, and Mesorhizobium loti19 was studied.
Ralstonia solanacearum (Rso) is a Gram negative β-proteobacteria responsible for multiple diseases related to the wilting of more than 200 plant species, causing huge economic losses worldwide, especially in developing tropical countries. This phytopathogen invades the vascular tissue in a systemic way20,21. Due to its wide range of hosts, large geographic distribution and diverse pathogenic behavior, this heterogeneous group is recognized today as a "species complex" (RSSC, Ralstonia solanacearum Species Complex)22. Within the RSSC, four subdivisions called phylotypes are recognized and each phylotype is divided into secuevars. Among the strains representing the phylotype I R. pseudosolanacearum GMI1000 (Rpso GMI1000) is found, a strain whose genome was completely sequenced23. Although R. solanacearum is considered a plant pathogen, it mainly behaves as a soil bacterium of saprophytic life with an extremely versatile lifestyle, which allows the bacteria to survive in the soil for long periods in the absence of its host plant. Rso moves toward the plant roots by different motilities such as swimming and twitching, searching for favorable growth conditions. After invading the host plant root system, the bacterium adheres to host cells and develops a biofilm to colonize the root cortex24. Then it reaches the vascular tissue spreading systemically to all plant tissues through the xylem. Finally, the exopolysaccharide (EPS) overproduction and bacterial active proliferation produce the obstruction of the xylem vessels, rendering the characteristic bacterial wilting phenotype, due to the lack of water and nutrients25.
In silico analysis of the R. pseudosolanacearum (Rpso) GMI1000 genome revealed the presence of a gene Rsp0254 encoding a putative LOV protein, the only photoreceptor protein detected, which led us to hypothesize that light could influence Rpso lifestyle and its interaction with the host plant23.
A complex regulatory network that responds to environmental conditions controls the expression of virulence factors in Rso. The global regulator PhcA presents the largest regulon described to date in the Rso species complex that directly or indirectly controls the expression of many genes26. Furthermore, the type III secretion system (T3SS), encoded by the hrp cluster, that allows effector proteins translocation into plant cells, is a key determinant of pathogenicity required for the disease development in host plants. The HrpG transcription factor controls the expression of many genes that promote the bacterial adaptation to the plant, including detoxifying enzymes, phytohormones, lectins, metabolic enzymes and transporters25. In addition, HrpG also functions as an activator of hrpB, which induces the expression of the structural units of T3SS and its associated effectors27,28. Rso is also capable of perceiving signals derived from the host cell wall during initial bacterial-plant cell contact, activating the expression of hrp genes29. The VsrAD two-component system controls the transcription of genes involved in EPS synthesis and other traits, some of which contribute strongly to Rso ability to colonize tomato stems and multiply in planta, regardless of the effect of the regulator on the EPS production30. EPS is required in the early and the late disease stages, during root colonization and later xylem physical obstruction, since it forms the necessary structural scaffold required for biofilm formation in both stages. Biochemical and genetic studies indicate that EPS and the enzymes that degrade the plant cell wall are necessary for the complete virulence. pehR gene controls early virulence factors and is also a positive regulator of the swimming motility cascade31. In this context, the physiological base of the bacterial wilt disease is multifactorial. Besides HrpG and PrhG transcriptional regulators, the Rso regulation network also includes numerous well-studied regulators such as PhcA, PrhN, PrhO, and XpsR cascades26.
In this work, the involvement of light and LOV protein in the regulation of Rpso physiological attributes and infectivity was elucidated. With this aim, we constructed a mutant strain lacking a functional Rpsolov gene (RpsoΔlov) and studied the effect of the absence of this gene on bacterial physiological characteristics. In addition, it was studied how certain environmental factors, such as light, affect the interaction between Rpso GMI1000 and its host plants. We demonstrated that light and the LOV protein control motility, adhesion and biofilm formation in Rpso allowing the successful colonization of the tomato plant rendering the bacterial wilt disease. This is the first report revealing the role of light of the vascular phytopathogen Rpso GMI1000.
Results
Rsp0254 is a LOV type photoreceptor putative fused to diguanylate cyclase-phosphodiesterase (DGC-PDE) response regulator
The 5.8 Mbp genome of the model strain R. pseudosolanacearum GMI1000 (Rpso GMI1000) is fully sequenced and organized into two circular replicons: a 3.7 Mbp chromosome and a 2.1 Mbp megaplasmid23. According to the in-silico analysis of the Rpso genome, in the megaplasmid there is an open reading frame coding for a putative LOV domain protein Rsp0254 (named Rpsolov gene for clarity purposes), a transmembrane predictive protein of 1178 amino acids. The Rpsolov gene presents different domains: a HAMP transmembrane signaling domain, a family of PAS domains that contain the LOV domain (635–738aa) (Supplementary Material 1 (S1)), and the domain responsible for regulating the response made up of a diguanylate cyclase (GGDEF) fused to a phosphodiesterase (EAL) domain32,33. In transmembrane proteins, the HAMP domains are found on the cytoplasmic side, where they convert intracellular transmembrane signals to response signals34. In the case of PAS domains, they can act as direct receptors or, as in the case of LOV domains, possess a cofactor responsible for the perception of light. LOV domains contain a molecule of flavin mononucleotide (FMN) as a non-covalently bound chromophore. The Rpso LOV protein presents a conserved key functional amino acid residue, the cysteine Cys 672, known to be important for photochemistry and signaling.
The Rpsolov gene distribution in the Ralstonia solanacearum species complex (RSSC)
Multiple alignments of the deduced amino acid sequences of LOV proteins from representative strains belonging to the four phylotypes including: Rpso GMI1000 (phylotype I)23, Rpso strain OE1-1 (phylotype I)35, Rpso FQY_4 (phylotype I)36, Rso K60 (phylotype IIA)37, Rso CFBP2957 (phylotype IIA)38, Rso IPO1609 (phylotype IIB)38, Rso UW551 (phylotype IIB)38, Rso Po82 (phylotype IIB)39, Rso UY331 (phylotype IIB)40, Rpso CMR15 (phylotype III)38, R. syzygii R24 (phylotype IV)41 and R. syzygii PSI07 (phylotype IV)38 revealed that the LOV protein is present in all Rso strains sequenced and possess highly conserved domains suggesting that light would play an important role in the Rso free lifestyle and during the plant-interaction (Supplementary material S1).
Different light conditions and Rpsolov gene deletion do not affect the growth of Rpso
To determine whether light or the absence of the Rpsolov gene affect Rpso viability and growth kinetics, we analyzed the bacterial growth in white light and in darkness of Rpso GMI1000 and RpsoΔlov strains. The CFU/mL of culture were determined at different periods, but no significant change in the growth of Rpso GMI1000 or RpsoΔlov was found under the two lighting conditions tested (p = 0.5727) (Fig. 1).
Growth curves of Rpso GMI1000 and RpsoΔlov under different light conditions. Bacterial cells were cultured in BG medium at 28 °C under exposure to constant white light or darkness. Aliquots were taken at the indicated times and measured for colony-forming capacity by serial dilution and plating on BG-agar. Colonies were counted after 48 h incubation at 28 °C. No significant effect in the growth of Rpso GMI1000 or RpsoΔlov was found under the two lighting conditions (p = 0.5727).
Rpso swimming and twitching motilities are regulated by light and by Rpsolov gene
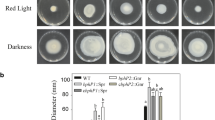
Rpso is a motile bacterium with one to four polar flagella able to slide on liquid medium by swimming. This motility contributes to the full virulence of this bacterium in the early stages of host invasion and colonization42. To determine the effect of light on Rpso wild type and RpsoΔlov swimming motility, bacteria were grown in white light and darkness (Fig. 2a), and the motility was quantified by measuring the diameter of the migration halo (Fig. 2b). Rpso GMI1000 produced smaller migration zones under white light compared to darkness (p = 0.0004). On the other hand, the mutant strain did not present swimming motility halos in the conditions assayed (p < 0.0001).
Effect of the Rpsolov gene mutation in swimming and twitching motility assays under different lighting conditions. (a) Representative images of both bacterial strains swimming agar plates incubated 48 h under white light or continuous darkness. (b) Measurement of the diameter of the bacterial migration halos (cm) from six independent experiments. Significant differences between conditions are represented by different asterisks (**-*p < 0.0001, ***-*p = 0.0002 and **-***p = 0.0004). (c) Images of Rspo GMI1000 and RpsoΔlov colony edges observed through an optical microscope at a 20x magnification. White arrows indicate the borders of the colonies, observing the typical raft of this motility. Images are representative of four independent experiments.
Twitching motility is a type IV pili-mediated translocation that allows bacterial adhesion to the plant roots43. The effect of light on Rpso twitching motility was evaluated in both bacterial strains under different lighting conditions. When the plates were incubated under darkness, colonies with layered edges and multiple irregular projections were observed which is typical of this type of bacterial motility. In contrast, under white light, Rpso produced colonies with smooth margins that are not characteristic of this motility (Fig. 2c). In the case of RpsoΔlov, the typical irregular projections of twitching motility were not observed in both lighting conditions.
White light affects in vitro adhesion
After invasion of the intercellular spaces, cells of R. solanacearum attach to the surfaces of plant cells as an initial step of host colonization and infection44. We study the binding capacity of Rpso GMI1000 and RpsoΔlov strain to an abiotic surface under white light and darkness (Fig. 3a). As shown Rpso exhibited increased adhesion ability when it was incubated in darkness compared to white light (p < 0.00001). However, the mutant strain showed decreased ability to adhere to the surface with respect to the wild type strain under both conditions tested (p < 0.0059 and p < 0.00001).
Evaluation of the adhesion of an abiotic surface and EPS production of Rpso strains. (a) Adhesion test of Rpso strains on abiotic surface. Rpso GMI1000 and RpsoΔlov bacterial cells were grown under different lighting conditions adhering to the surface of the polystyrene plates and were stained with 0.1% (w/v) Crystal violet. Quantification diagram of the bound dye solubilized with ethanol measuring the absorbance at 540 nm. Significant differences between conditions are represented by different asterisks (**-*p < 0.0059, ***-*p < 0.00001 and **-***p < 0.00001). (b) Quantification of the EPS production of the Rpso GMI1000 strain and the strain with the deleted Rpsolov gene. EPS was extracted from bacterial supernatants and quantified after 2 days of growth under different lighting conditions. The weight of EPS was normalized to the log (CFU/mL) of culture. The asterisks (*) indicate that there is a significant difference between the corresponding data (p = 0.0007).
In vitro production of Rpso extracellular polysaccharides depends on the Rpsolov gene
Ralstonia solanacearum generates an extracellular polysaccharide (EPS) composed of a complex polymer of N-acetylated sugars. EPS is an important virulence factor during bacterial wilt, being responsible for clogging the vessels of the xylem and triggering symptoms45. To determine the effect of light on EPS production in both Rpso strains, we quantified the precipitated EPS from the two-day Rpso GMI1000 and RpsoΔlov cultures grown under two different lighting conditions. No differences were observed in the production of EPS of the Rpso GMI1000 strain in white light and continuous darkness, but the results showed that in the absence of the Rpsolov gene there was a marked decrease in the generation of EPS in both lighting conditions (p = 0.0007) (Fig. 3b).
White light modifies the biofilm formation in Rpso
We analyzed the morphology of bacterial biofilms developed by a GFP-labeled Rpso strain GMI1000 and mCherry-labeled RpsoΔlov by confocal laser scanning microscopy (CLSM, Confocal Nikon C1SiR attached to a Nikon TE2000 inverted microscope)46. After three days of incubation under the different lighting treatments, Rpso GMI1000 generated a biofilm with a more structured, packed and organized topology, forming different layers in the dark than in white light. Bacteria appeared more dispersed in the last condition, similar to the mutant strain in both lighting conditions. In addition, a clear difference between the thickness of the biofilm of the wild strain with respect to the mutant strain was evidenced, the latter being thinner (Fig. 4).
Biofilm architecture of Rpso strains under different lighting conditions. Confocal laser scanning microscopy images showing orthogonal views of biofilm formed by GFP and mCherry labeled wild type Rpso and RpsoΔlov cultures after 3 days of static incubation in flat-bottomed microplates. Images are representative of results from three biological replicates (scale bar in inset, 100 µm).
Expression analysis of the Rpsolov gene
To analyze the Rpsolov gene expression, a reverse-transcription quantitative PCR (RT-qPCR) was performed with Rpso GMI1000 cultures grown under white light and darkness for 18 h in MM medium at 28 °C. Rpsolov gene was expressed in both lighting conditions, however a significantly higher level of expression was observed in darkness compared to white light (p = 4.114e−05) (Fig. 5). Experimental raw data in the software StepOne are shown in Supplementary material 3.
Gene expression analysis by quantitative real-time RT-PCR. The expression of the Rpsolov gene was assayed by RT-qPCR in Rpso GMI1000 under dark and light conditions using specific primers. Cultures grown for 18 h in MM medium were harvested to extract total RNA. The data shown report the relationship between the Rpsolov gene (Rsp0254) and the rplM reference gene in both lighting conditions tested. A significantly higher expression level was observed in the dark compared to white light. Asterisks (*) indicate significant differences between the corresponding data (p = 0.0004).
Different transcriptional regulators control Rpsolov gene expression in Rpso
To determine if the Rpsolov gene expression is modulated during the infection process, we analyzed the transcriptional regulation of this gene. For that purpose, transcriptional fusions were generated between the promoter region of the Rpsolov gene and lacZ, which encodes the β-galactosidase enzyme (lov::lacZ). Then, these constructions were introduced into mutant strains for different transcriptional regulators that are known to regulate virulence genes in Rpso. Figure 6 shows the level of β-galactosidase activity monitored for each reporter strain. According to these results Rpsolov expression is negatively modulated by HrpG as the β-galactosidase activity of the lov::lacZ fusion is increased by a ~ twofold factor in the hrpG mutant strain in comparison to its expression in the wild-type (p ≤ 0.001). On the other hand, β-galactosidase activity levels were comparable in the wild type strain and in the hrpB mutant background.
Expression of Rpsolov gene in different genetic backgrounds. (a) Schematic representation of the transcriptional fusion of the Rpsolov gene promoter with the lacZ gene in the different genetic backgrounds. (b) Rpso reporter strains were grown for 16 h in BG medium, β-galactosidase activity was measured and expressed in Miller units. The asterisks (*) in the dot plot indicate significant differences between the wild type strain and the ΔhprG (p < 0.001) and ΔvsrA (p = 0.0007) strains, respectively.
In addition, an effect on the lov::lacZ expression was observed for the vsrA mutant, which exhibited significantly reduced β-galactosidase activity compared to the wild type strain (p = 0.0007).
In the case of the pehR and hrpB mutant strains, there were no significant differences in the activity measures with respect to the wild type strain.
Environmental light quality defines the successful colonization of the host plant
The virulence of wilt type and mutant Rpso strains grown in white light and darkness conditions was tested in susceptible tomato plants by inoculation with the Rpso GMI1000 Pps-GFP reporter strain and RpsoΔlov mCherry. The plants were kept in a normal photoperiod camera. 6 days after inoculation, before symptoms appeared, confocal laser scanning microscopy CLSM (Confocal Nikon C1SiR attached to a Nikon TE2000 inverted microscope) verified bacterial colonization in sections of root and plant stems.
It was observed that the analyzed strains were able to colonize the root system, noting an exacerbated invasion of the xylem vessels in tomato plants inoculated with Rpso GMI1000 cultures grown in darkness compared to plants that were inoculated with Rpso GMI1000 grown in light (Fig. 7a). On the other hand, the RpsoΔlov strain grown in both lighting conditions invades in less quantity the root system than the WT strain, being observed dispersed throughout the tissue and colonizing some xylem vessels (Fig. 7a). Stem colonization with RpsoΔlov is not observed (Fig. 7b). These representative images are supported by counting the CFU of bacteria obtained from root samples for the quantitative analysis of Rpso colonization. A greater number of bacteria were recovered from tomato roots inoculated with Rpso GMI1000 grown in white and dark light, the latter showing a greater difference (p = 0.0051536). The bacterial count of roots inoculated with the RpsoΔlov strain grown in both lighting conditions was lower than the growth of the WT strain under the same conditions (p < 0.00001). There was no statistical difference in root growth for the RpsoΔlov strain between both conditions (Fig. 7a).
Roots and stems tomato plants colonization by Rpso and RpsoΔlov. (a) Root cross sections observed by confocal laser microscopy indicating the presence of Rpso GMI1000 Pps-GFP and RpsoΔlov mCherry previously grown under different lighting conditions. (b) Stem cross sections observed by confocal laser microscopy indicating the presence of Rpso GMI1000 Pps-GFP and RpsoΔlov mCherry previously grown under different light conditions. The white arrows indicate the presence of pathogenic bacteria in the xylem vessels (xv) and other tissues present in the roots and stems of tomatoes. Each micrograph is a representative result of at least 10 sections of plant tissue from three biological replicates. Box plots of the bacterial population are shown under the different treatments in roots and stems of tomato plants at 6 dpi, respectively. Serial dilutions of the root and stem extracts were seeded on Rpso selective medium. The results were expressed as Log CFU/mL per gram of organ. The significant differences between the conditions are represented by different asterisks in the count of bacteria grown in both conditions in root and stem (root: **-*p < 0.00001, ***-*p < 0.00001 and **-***p = 0.0051536; stem: *p < 0.00001). xv, xylem vessels; vc, vascular cylinder; vb, vascular bundles.
On the other hand, it was observed that only the Rpso GMI1000 strain colonized the aerial part of the plant, showing a greater invasion tendency in the transversal sections of stems inoculated with the wild type strain grown in darkness. The RpsoΔlov strain lost the ability to ascend and colonize plant stems. These representative images are consistent with the CFU count of bacteria obtained from tomato stems supporting quantitatively the observations provided by microscopy analysis (p < 0.00001) (Fig. 7b).
Discussion
Environmental light is fundamental for the evolution and adaptation of all living organisms. Plants have developed abilities to maximize the capture of energy in their tissues and thus promote their development47. Until recently, light-induced signaling through photosensory proteins was considered an exclusive feature of photoautotrophic organisms. However, genome sequencing revealed the presence of photoreceptors in all life kingdoms48. The photoreceptor proteins identified in the genomes of several microorganisms, fungi, insects and plants suggest that the role of light goes far beyond the photosynthesis process. In the case of plants, light is not only essential for their survival but also to reinforce defense against pathogens3.
In phytopathogenic bacteria, a variety of photoreceptor proteins have been reported9,49. These proteins detect light to regulate various cellular processes such as motility, adhesion, morphology, multiplication, DNA repair, secondary metabolite production and bacterial colonization. Oberpichler et al. have provided evidence linking light perception and virulence through cell motility control in Agrobacterium tumefaciens50.
Previous studies have shown how light affects the plant-pathogen interaction, both regarding the host plant response and to the phytopathogen ability to infect the plant5,7,9,16. Several investigations have reported the influence of light on host and non-host plant interactions with Gram-negative bacteria: (1) biotrophic such as Agrobacterium tumefaciens50, (2) hemibiotrophic such as X. citri subsp. citri9, Xanthomonas campestris pv. campestris51 and Pseudomonas syringae pv. tomato10, (3) necrotrophic such as Botrytis cinerea52 and (4) endosymbionts such as Rhizobium leguminosarum18. In particular, the present work focuses on the study of the effect of light on interaction mechanisms between a vascular phytopathogen such as Rpso GMI1000 and tomato plants.
Rpso has a 5.8 Mbp genome formed by a chromosome and a megaplasmid. The megaplasmid genes analysis suggests that this replicon has a significant function in the bacterium adaptation to different environmental conditions23. A gene encoding a putative blue light photoreceptor (Rpsolov gene) was identified in this megaplasmid. Mandalari et al. studied in detail the organization of the LOV photoreceptor in the Ralstonia genus, mainly in Rpso GMI100033. Through the application of bioinformatics programs determined that it would be a transmembrane protein composed by 1178 amino acids, with a di-guanylate cyclase domain fused to a phosphodiesterase domain as response regulator domain, unlike of those present in Pseudomonas and Xanthomonas genus that are cytosolic and histidine kinase or hybrid histidine kinase9,10,53,54. Many in silico analysis of LOV photoreceptor, such as multiple sequence analysis, indicated that the LOV domain and the response regulation domain were found in several members of the Ralstonia solanacearum species complex (Supplementary material S1). Similar results were observed in Xanthomonas genus55. The organization of hybrid LOV-HK-RR proteins were conserved almost exclusively in bacterial plant pathogenic species and they are involved in the regulation of different virulence factors at some stage of the bacterial life cycle through blue light sensing9,56,57.
In this work, the role of light and LOV protein in Rpso physiology and in the pathogenesis process was studied. For this purpose, the wild type strain Rpso GMI1000 and RpsoΔlov, mutant strain due to complete deletion of the gene, were studied. In vitro growth curves of both strains were performed in white light and darkness (Fig. 1), observing that there are no differences in the bacterial numbers (CFU/mL) under the different light conditions, which indicates that the absence of Rpsolov gene and the light does not affect the bacterial viability and growth kinetics. These results agree with those described by Wu et al. and Kraiselburd et al., where the viability of Pseudomonas syringae pv. syringae and X. citri respectively was not affected by the different lighting conditions and Rpsolov gene deletion9,16.
Several reports show that light regulates the bacterial transition between a mobile and a sessile state7,8. Rpso moves towards a plant host when it perceives a stimulus or is attracted by the root exudates. We evaluated the effect of different lighting conditions on swimming and twitching motilities. Swimming motility is an individual translocation dependent on flagella that occurs in liquid media, water content being a critical factor for this displacement. As it is shown in Fig. 2a,b, the Rpso GMI1000 strain showed a greater displacement in darkness compared to white light, that is, white light inhibits swimming motility. In addition, it was observed that RpsoΔlov strain presented a lower displacement than wild type strain in both lighting conditions. In this context, the LOV protein would be involved in the regulation of motility. Similar results were obtained with P. syringae pv. tomato DC3000 where motility repression was observed, as with Rpso, under the same light condition, also a Psto DC3000 mutant in the Rpsolov gene showed decreased motility compared to the wild type strain in both light conditions, indicating that the LOV-HK photoreceptor positively regulates this type of motility10. Similar results were obtained by our group for the LOV protein of Xcc where motility was also modified in the mutant strain of the lov gene9. This behavior was also found in the phytopathogen A. tumefaciens, which has phytochrome type photoreceptors, observing that bacterial suspensions grown in white light showed less motility compared to dark-grown cultures50. In addition, in Xanthomonas oryzae pv. oryzae (Xoo) was observed that the complete deletion of the bacteriophytochrome gene (BphyP Knockout) rendered a similar behavior than RpsoΔlov, the mutant strain produced reduced swimming motility in all lighting conditions58.
Twitching is a type of translocation present in a wide variety of bacteria, including the genus Pseudomonas and Ralstonia. This type of motility depends on the type IV pili extension and active retraction and the moisture availability in the culture medium. The type IV pili are formed by polymerization of pilin monomers. This structure is involved in different biological processes, including adhesion, biofilm formation and horizontal gene transfer59. In Rpso it was demonstrated that these appendices are essential for pathogenicity60. Pilin is post-translational modified by glycosylation in Gram-negative bacteria and it has been reported that defective mutant strains in the pili production or glycosylation did not show contraction motility and caused reduced symptoms and slower disease progression. For example, in Rpso GMI1000, a mutant strain deficient in the gene that codes for an enzyme involved in the O-glycosylation of type IV pili, did not generate bacterial wilt symptoms when it was inoculated in tomato plants61. In our study when Rpso was grown under white light this motility was reduced, while, under dark conditions, the bacterium migrated via twitching. In absence of light, as can be seen in Fig. 2c, it was observed that colonies present irregular aspects and long bacterial extensions (raft) irradiated from the migration zones. Furthermore, in the presence of light, bacteria developed colonies with smooth edges, without visible bacterial extensions, suggesting that light inhibits Rpso capability to migrate via twitching motility. These results agree with those published by Bitrian62 and Hoff63 where light implication in the bacterial physiology of the environmental pathogen Acinetobacter baylyi was found. This bacterium has a blue light photoreceptor, BLUF type, responsible for this behavior. The evaluation of twitching motility in RpsoΔlov allowed us to observe a lower displacement both in white light and in darkness, compared to the wild type strain (Fig. 2c). These results suggest that Rpso LOV protein is an activator of twitching motility in the assay conditions. This behavior was also observed in the Rpsolov mutant of X. citri subsp. citri which presented colonies with smooth borders different from the starry edges of the wild type strain9.
Several factors contribute to bacterial adhesion on the host tissue, including fimbrial and non-fimbrial adhesins, extracellular polysaccharides (EPS) and flagella64,65. In this work, Rpso GMI1000 ability to adhere to abiotic surfaces under different lighting conditions was evaluated. A drastic decrease in adhesion under white light (Fig. 3a) was observed, indicating that it is a light-dependent process. This result differs from the obtained by Río Alvarez et al. where differences in the adhesion capacity of the wild Psto DC3000 strain on A. thaliana leaves were observed. Moreover, in dark conditions or under red light the wild type strain did not adhere to the leaf surface after 6 h of incubation. However, when bacteria were pre-treated for 10 min with red light and then incubated for 6 h in the dark, they recovered the ability to adhere to A. thaliana leaves. These differences can be attributed to other photoreceptors present in Psto beside LOV type photoreceptor10. On the other hand, Rpso has only one putative photoreceptor that would sense the light absence in the soil depth allowing the capture of a host plant signal, root adhesion and then to initiate plant colonization. In the evaluation of RpsoΔlov adhesion to abiotic surfaces, this strain lost the ability to adhere in all light conditions, this type of mutation generated by complete deletion of the gene and the observed phenotype allowed us to conclude that the LOV protein has a role as a positive regulator of adhesion in Rpso independently of light. These results agree with Kraiselburd et al. where the X. citri subsp. citri mutant in the lov gene presented in vitro and in vivo adhesion significantly diminished compared to the wild type, showing a strong dependence on light during bacterial growth9. Caulobacter crescentus is a Gram-negative bacterium widely distributed in soils, lakes and water of sea which plays a very important role in the carbon cycle. The genome of C. crescentus contains an operon that codes for a LOV-histidine-kinase protein (LovK) and a single domain response regulator (LovR) which interacts with LovK66. Studies by Purcell et al. revealed that a mutant in LovR of C. crescentus presented a severe loss in adhesion capacity compared to the wild type strain, indicating that this protein is also an adhesion positive regulator as LOV protein of Rpso66.
EPS is the main Rpso virulence factor that causes wilting by restricting the flow of water through the xylem vessels67 and also notably improves the speed and extent of stem colonization68. We analyzed the EPS content in minimal and rich media. In minimal medium the EPS production was similar in both bacterial strains and in all conditions tested (Supplementary material S2). In CPG rich medium no significant differences in EPS production was observed under the different lighting conditions in the Rpso GMI1000 strain. On the contrary, a marked decrease in EPS synthesis was observed for the RpsoΔlov strain with respect to the wild strain (Fig. 3a). These results suggest that EPS production in Rpso GMI1000 could be regulated by the LOV protein acting as a light-independent positive regulator of exopolysaccharide synthesis. Similar results were observed for X. citri subsp. citri, where light does not affect xanthan production under the lighting conditions tested9. This apparent absence of light regulation in the case of the wild strain is contrary to the expected results considering that the Rpsolov gene encodes a photoreceptor, but it has been shown that the activity of some LOV-type bacterial photoreceptors is modulated by other stimuli such as for example, the cytosolic redox state in conjunction with light and that they would also perceive not only blue light, but also red light. Bonomi et al. determined that one of the virulence factors regulated by the LOV-HK photoreceptor of Rhizobium leguminosarum is the production of EPS. The mutant in the lov gene showed, as in Rpso GMI1000, a lower capacity for EPS synthesis compared to the wild strain, however, the regulation of polysaccharide production in R. leguminosarum occurs through light , LOV-HK being the sensor involved in this process18.
Bacteria develop dense communities associated with a surface known as biofilms, which are essential for their persistence69 and play an important role in the virulence of many pathogenic bacteria70. The morphological form in multicellular aggregates arises from the interaction of bacterial genetic makeup and environmental cues71.
Initially Rpso invades the intercellular spaces of the roots, attaches itself to plant cells and then spreads within them. Quorum sensing is activated at this stage, leading to the formation of fungus-like biofilms72, which are necessary for the pathogenicity of Rpso. The planktonic bacterial cells released from the biofilms can invade the xylem vessels, ascend through it and secrete virulence factors such as EPS in the stem, again forming a thick biofilm as a structural scaffold in the vascular bundles to cause water obstruction. and thus induce wilt symptoms72,73.
When biofilms formation and architecture were analyzed using CLSM, we found that the macrocolony biofilm generated by Rpso GMI1000 in dark was structured with several layers leading to folds formation, rendering a more compact and organized biofilm compared to white light, where a macrocolony biofilm covers the surface more loosely. This last characteristic is also presented in the mutant strain in the Rpsolov gene, which shows the same phenotype (Fig. 4).
Our results agree with those of Mussi et al., where the opportunistic pathogen Acinetobacter baumanni develops a differential production of biofilm with a greater capacity to form biofilms in dark conditions74. In conclusion, the absence of light regulates the formation of biofilms in Rpso GMI1000 and A. baumanni.
On the other hand, we discovered a marked variability in the thickness of the biofilm structures between the two strains studied. The wild type strain was characterized by developing a thick biofilm with appreciable density, while RpsoΔlov was thin and dispersed, concluding that the LOV protein is involved in the biofilm formation.
In view of the results observed in different types of motilities, biofilm formation and abiotic adhesion, we infer that light would be behaving as an inhibitor of the different virulence factors mentioned above, but when deleting the Rpsolov gene it was observed that this protein would act as a positive regulator of virulence features. Rpso GMI1000 has a single encoded photoreceptor protein in its genome that exhibits multiple domains as described above. In this work, a mutant strain was constructed in the complete gene (Knockout gene), without observing a phenotype that validates this hypothesis its role as a photoreceptor but that corroborates its participation in the regulation of the modified attributes in Rpso. Site directed mutants of Rpsolov gene site in other domains could provide a clearer role for this gene, since the phenotypes obtained could be associated not only with the LOV domain but also with other domains of this gene, such as the response regulatory domain58.
Rpso virulence was examined in tomato host plants 6 days after inoculation with wild-type Rpso and RpsoΔlov grown under white or dark light conditions.
The wild type strain showed greater colonization of tomato roots and xylem vessels of stems inoculated with Rpso GMI1000 grown in the dark compared to those plants inoculated with bacteria grown in white light (Fig. 7a,b). Therefore, Rpso GMI1000 shows higher virulence in the dark condition. These results are consistent with the phenotype obtained with Psto, in a similar light treatment10,11. On the other hand, it was observed that RpsoΔlov colonizes and disperses through the root system but loses the ability to ascend and multiply in the stem, showing that the deletion of the Rpsolov gene causes a decrease in virulence in the host plant (Fig. 7a,b).
Therefore, the bacterial physiological alterations caused by the light environment and the contribution of the Rpsolov gene in the motility, adhesion and biofilm of Rpso, contribute to the successful propagation and colonization of roots and stems of the host plant.
In the case of the evaluation of Rpsolov gene expression in the two light conditions, real time quantitative analysis showed that in all conditions, the Rpsolov gene was expressed. The Rpsolov/rplm gene expression ratio in darkness was significantly greater than the ratio in white light (Fig. 5). This result shows an induction of the expression of Rpsolov gene in the dark. The same result was observed in Acinetobacter baumannii ATCC 17978. When this strain was incubated at 24 °C in light and darkness, the expression of blsA gene encoding a BLUF photoreceptor was higher in dark condition, but at 37 °C no differences in the blsA gene expression level was observed. These results indicate that temperature could play a role in the expression of blsA74. In this context, the induction of the Rpsolov gene in the dark could also be influenced by other environmental factors such as temperature, pH or redox state, as has been seen in other cases75,76,77. Further investigation of the Rpsolov gene will be essential to shed clarity on this issue.
Considering the wide range of biological functions affected by various environmental conditions, many of which are perceived by photoreceptor proteins, and according to the results described above where a light regulation of the Rso pathogenicity was observed, we decided to examine and provide an overview of the implication of the Rpsolov gene in the Rpso GMI1000 virulence factor regulation cascade which is sensitive to internal metabolism and environment. For this purpose, a transcriptional fusion was generated between the Rpsolov gene promoter and lacZ gene in the wild type Rpso strain and in different transcriptional regulators mutant strains. β galactosidase activity measurements then performed indicated that the Rpsolov gene is part of this network. These results showed that HrpG negatively regulates the Rpsolov gene expression under in vitro culture conditions (Fig. 6). HrpG, a response regulator belonging to the OmpR family, was originally discovered by positively regulating the HrpB expression, which controls the T3SS and activates the synthesis of 3-hydroxy-oxindole, a compound related to quorum sensing in early stages of Rpso infection29,78. Transcriptomic studies with Rpso revealed that the complete HrpG regulon controls several genes in addition to those regulated by HrpB59. HrpG controls functions that promote the bacteria adaptation to life within the host, as well as some virulence factors79. Our results suggest in this case that Rpsolov gene expression is controlled by HrpG in a HrpB-independent manner. On the other hand, the VsrA transcriptional regulator positively controls the Rpsolov gene expression. All these assays were performed in in vitro conditions. Despite extensive knowledge about how these networks work in culture, there are very few reports of the processes that occur in vivo during pathogenesis22. Recently it was shown that the expression of some of these transcriptional regulators depend on the conditions where the bacteria were grown. Perrier et al. studied the expression of these Rpso regulators in a complete medium and in planta conditions26. They showed that virulence functions corresponding to the HrpB and HrpG regulons are repressed by PhcA in complete medium but are specifically activated in planta. These regulons represent a set of key genes required for Rpso pathogenesis. Furthermore, it was reported that the expression of Rso T3SS genes are still effective in the xylem. Taking into account that the experiments to define the regulation cascade in relation to the Rpsolov gene were carried out under in vitro conditions and that the regulation of HrpG presents a contrasting regulation in vitro and in planta conditions26, it is necessary to carry out more investigation to clarify the role of light and HrpG in vivo. Furthermore, we observed in the case of the pehR strain that there were no significant differences with respect to the wild type strain under the conditions tested. The pehR gene is strongly expressed at low cell densities because it controls early virulence factors and is also a positive regulator of the swimming-type motility cascade31. PehR regulates both in minimal medium and in the plant, the expression of flhDC, an open reading frame that encodes the main regulator of flagellar biosynthesis and bacterial motility80. Probably, the expression of these regulators and the participation of the Rpsolov gene dependent on environmental factors in the Rpso regulatory cascade will ensure the expression of genes related to virulence at the appropriate time.
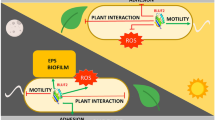
Finally, we have proposed a model to integrate the results obtained in Rpso physiological characterization and in the pathogenicity under the different lighting conditions (Fig. 8). Briefly, when bacteria are in the soil, in darkness, the so-called “very early and early virulence factors'' are activated, rendering a higher motility both swimming and twitching, greater adhesion and biofilm in the intercellular spaces of roots. Once Rpso enters the host plant, it invades the xylem vessels where it is capable to perceive the daylight in the aerial parts of the plant repressing the virulence factors, allowing the bacterium to raise the aerial host tissues and thus avoiding plant defense mechanisms. During the night, again in the darkness, the plant became more susceptible to biotic stress3. Under this situation, the bacterium activates the late virulence factors, producing more biofilm and allowing greater plant colonization (Fig. 8). These modifications in the bacterial behavior agree with the variation in the gene profile expression of R. solanacearum detected by RNAseq analysis using bacteria isolated from different regions of the plant tissues27,81,82.
Integrative model illustrating Rpso GMI1000 light detection on virulence factors during interaction with tomato host plants. When the bacterium is in the ground (in the dark) and receives a specific stimulus from the host, the "very early virulence factors" are activated to reach, enter and colonize the roots through the "early virulence factors". Once inside the plant, propagation by xylem beams begins, detection of daylight begins and repression of "early virulence factors" occurs. During the night, again in the darkness, the bacterium detects the absence of light and takes advantage of the fact that the plant is more susceptible to attack by pathogens and, therefore, activates the "late virulence factors" that trigger bacterial wilt.
It can be concluded that light act regulating several Rpso features directly involved in the pathogenicity process allowing a successful host colonization and infection. Furthermore, the Rpsolov gene and light would act as an essential bacterial factor that indicates position in the host plant, to regulate expression of virulence genes. Consequently, bacteria use an external signal and the LOV protein to know their location within plant tissue during the colonization process. Since Rpsolov gene presents a diguanylate cyclase and a phosphodiesterase C-terminal domain as a response regulator, the phenotypes observed for mutant bacteria could be associated with the pleiotropic effect modulated by a second messenger c-di-GMP. Further investigation of the putative blue light photoreceptor encoded by Rpsolov gene, will be essential to shed light into this question.
In summary, in this work is presented for the first time the role of light in the lifestyle of R. pseudosolanacearum, a vascular phytopathogen, demonstrating that the quality of this factor enables successful interaction with the host plant.
Materials and methods
Plasmids, bacterial strains and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Rpso cells were cultured in different media, mainly in Bacto-glucose (BG) medium or BG-1.5% (w/v) agar supplemented with 0.005% (w/v) tetrazolium chloride and 0.5% (w/v) glucose83. Alternatively, Casaminoacids-Peptone-Glucose (CPG) medium84, Boucher’s minimal medium (MM) supplemented with 20 mM l-glutamate as a carbon source85 or semi-selective SMSA medium (mSMSA) supplemented with 25 mg/L Bacitracin, 100 mg/L Polymyxin B sulphate, 5 mg/L Chloramphenicol, 0.5 mg/L Penicillin-G, 5 mg/L Crystal violet, 1 mg/L Cicloheximide and 50 mg/L 2,3,5-triphenyl tetrazolium chloride were used for Rpso growth86. Escherichia coli JM109 used for genetic constructions was cultured at 37 °C in Luria–Bertani medium87,88. For selection of the reporter strains gentamicin, 5 and 10 µg/mL was used in liquid and solid media, respectively.
Physiological assays were performed under different lighting conditions. For light condition, bacteria were grown in a chamber with continuous white light (130 μmol/m2s) provided by LEDs. For dark conditions, flasks or plates were covered with aluminum foil.
Construction of the RpsoΔlov mutant strain
To study the possible participation of the Rpso LOV protein in bacterial physiology, a mutant strain in the Rsp0254 gene was constructed. For this, the gene was replaced by a Gm resistance cassette present in the suicide vector pCM35195. The upstream and downstream regions to the Rsp0254 gene were amplified by PCR using the primers RpsoDOWNLOV-Fw (5′-GTTAACGCGCGCTTCACGGTGTAG-3′), RpsoDOWNLOV-Rv (5′-GAGCTCGACTGGCTGTGGCTCACC-3′), RpsoUPLOV-Fw (5′-GGAATTCCTGGCCCGACGATATAG-3′) and RpsoUPLOV-Rv (5′-GGGGTACCTTGGATGACCGGTAGAGCC-3′). The fragments were cloned on pCM35194 using the corresponding restriction sites. Rpso cells were transformed with the recombinant plasmid by natural transformation. The mutant strain was obtained by integration of the cloned fragment into the megaplasmid through a double homologous recombination event and selected by gentamicin resistance.
Growth curves in different lighting conditions
Saturated cultures of Rpso GMI1000 and RpsoΔlov grown in the darkness were sub-cultivated at 1% inoculum in BG fresh medium and incubated under white light or darkness conditions at 28 °C with shaking at 200 rpm. In order to determine the colony forming units (CFU)/mL, aliquots of cell suspensions were taken at different times. Three biological replicates in each lighting condition were used for the wild strain, while 2 were used for the mutant strain.
Swimming assay
Overnight cultures of Rpso GMI1000 and RpsoΔlov strains grown in darkness were washed with distilled water and adjusted to 107 CFU/mL. Aliquots of 3 µL of these suspensions were inoculated on the center of BG-0.3% (w/v) agar plates and incubated at 28 °C under white light or darkness. The diameters of the swimming areas were measured at 48 h post-inoculation97. Six biological replicates were used in each condition tested.
Twitching assay
Twitching motility tests were carried out following the protocol described by Siri et al.98. Petri dishes were prepared with CPG-1.6% (w/v) agar. Bacteria were grown overnight in darkness at 28° C in liquid CPG medium with shaking. The wild-type and mutant Rpso cultures were diluted to obtain a final concentration of 109 CFU/mL, and 10 μL of the bacterial suspensions were then inoculated on the surface of the CPG plates. The plates were incubated in different lighting conditions at 28° C in a humid chamber for 24 h. Motility was examined by optical microscopy (Carl Zeiss, Axiostar, Germany), using a 20× objective.
In vitro adhesion assay
In vitro adhesion of the studied strains was determined using polyvinyl chloride microtiter plates (Nunc MicroWell plate; Thermo Fisher Scientific Inc., Waltham, MA, USA). Rpso GMI1000 and RpsoΔlov saturated cultures grown in MM medium were adjusted to 106 CFU/mL and 100 µL of cell suspension were placed on said plates. Plates were incubated statically in different lighting conditions at 28 °C for 6 h. To quantify cell aggregation, 25 µL of 1% (w/v) Crystal violet solution was added to the wells. After 15 min incubation, unbound Crystal violet was gently removed with a pipette and the wells were washed with distilled water. Subsequently, 200 μL of 95% (v/v) ethanol were added and carefully resuspended the Crystal violet adhered to the cells. Bacterial adhesion was quantified by measuring the absorbance at 540 nm of the obtained solution44.
Biofilm formation assay
Biofilm formation analyses were performed with a modified Rpso strain that constitutively expresses the green fluorescence protein (GFP)46 and a LOV protein mutant strain transformed with a plasmid overexpressing mCherry96. Saturated cultures Rpso grown in CPG medium in darkness were adjusted to 107 CFU/mL, diluted 1∶20 in fresh medium and then 300 µL of the bacterial suspensions were placed into chamber covered glass slides (N°155411, Lab-Tek, NUNC, Naperville. IL, U.S.A.). Chambers were statically incubated in a humidified polyvinyl chloride (PVC)-box at 28 °C under the different light conditions. Biofilm formation was visualized by confocal laser scanning microscopy (CLSM,Confocal Nikon C1SiR attached to a Nikon TE2000 inverted microscope)99. Images obtained were analyzed with ImageJ software.
EPS production
Quantification of EPS production by Rpso was performed following the protocol described by Peyraud et al. with some modifications45. Rpso GMI1000 and RpsoΔlov saturated cultures grown in darkness were subcultured in 100 mL of minimal medium (MM) supplemented with 20 mM l-glutamate as a carbon source. Subsequently, they were incubated for 48 h at 28 °C in the two different lighting conditions. Aliquots of 5 mL of cell suspensions were filtered with 0.22 µm pore filters and the supernatants were collected. In order to precipitate the EPS, 20 mL of isopropanol and 0.36 mL of 0.3 M NaCl were added to the supernatants followed by the incubation at 4 °C for 72 h. Then, the mixtures were centrifuged at 4 °C for 10 min at 16,000g and the supernatants discarded. Pellets were dried for 15 min at room temperature and the dry weights determined. Subsequently, we made a modification in the way of obtaining bacterial suspensions where the saturated cultures of Rpso GMI1000 and RpsoΔlov that grew in the dark were subcultured in 10 mL of medium rich in CPG. Then continued with protocol described by Peyraud et al45.
RNA extraction, reverse transcription (RT), and quantitative real-time PCR (qPCR)
Rspo GMI1000 was cultured 18 h in MM medium under white light and darkness. Total RNA was isolated using TRIzol reagent (Invitrogen), according to the manufacturer's instructions. The extracted RNA was treated with RNase-free DNase (Promega) and its integrity was checked by agarose gel electrophoresis. For cDNA synthesis, total RNA (1 µg) was added to a 20 µl reverse transcription reaction medium containing 4 µl 5 × M-MLV buffer (Promega), 0.5 mM dNTP mixture, 0.5 µg random hexamer primer (Invitrogen), 200 U M-MLV reverse transcriptase (Promega) and incubated for 60 min at 42 °C. Reverse transcription was terminated by incubating for 5 min at 94 °C. qPCR was carried out using HOT FIREPol EvaGreen qPCR Mix Plus (Solis Biodyne), following the manufacturer's instructions. Primers RTlov-Fw (5′-TCAACATCGACCGCTTCAAG-3′) y RT2lov-RV (5′-AGCGCGAAGACGTCGCC-3′) were used for the Rpsolov gene and primers RTrplM-Fw (5′-GCGCAATTGGTTCGTGATTG-3′) y RT2-rplM-RV (5′-GGCTGCGTTGATCACGATG-3′) were used for constitutive control gene rplM. The StepOne Real-Time PCR system (Applied Biosystems) was used. qPCR reactions were carried out under the following conditions: initial denaturalization at 95 °C for 12 s, and 40 cycles of amplification at 95 °C for 15 s, annealing 60 °C for 25 s and extension at 72 °C for 20 s. Three biological replicates were analyzed three times. The amount of transcripts was presented as the ratio between the gene of interest and the reference gene (applying 2−ΔCt where ΔCt refers to the difference in the threshold cycles between the genes of interest and reference).
Generation of Rpsolov reporter strains
A transcriptional fusion of the Rpsolov gene promoter with the lacZ gene was generated by using integration plasmid pCZ36794. Briefly, a 1000 bp fragment containing the promoter region and the beginning of the coding sequence of Rpsolov gene was PCR amplified with primers LOVFT-Fw (5′ AAGCTTTCTCGTACGAAACCCAGAGC 3′) and LOVFT-Rv (5′ TCTAGAGTCAGGTGGTGGACGGTCT 3′) and cloned into the HindIII and XbaI sites of pCZ367. The resulting plasmid was then introduced into the different genetic backgrounds (GMI1000, ΔhrpG, ΔpehR, ΔvsrA, ΔhrpB) by electroporation (2.5 kV, 200 W, 25 µF, 0.2-cm cuvette gap) and the recombinant clones were selected by pCZ367 Gentamicin resistance. Integration of the vector in the correct site of the bacterial genome by a simple recombination event was checked by PCR using the primers UPLOVFT-Fw (5′CATGCTTTCTTTCCCACCAC3′) and Lacseq-Rv (5′TGTAAAACGACGGGATCCAT 3′), which hybridize upstream of the Rspolov fragment used for the recombination and in the lacZ gene, respectively. Measurements of β-galactosidase activity were performed as described by Brito et al.100. All these assays were realized without light treatment.
Virulence assay
For pathogenicity tests, night cultures of the reporter strain Rpso GMI1000 Pps-GFP and RpsoΔlov mCherry grown in dark and white light at 28 °C were adjusted to a concentration of 107 CFU/mL. Tomato plants (Solanum lycopersicum var. Minitomato) were inoculated with 20 mL of the bacterial suspensions to achieve a final concentration of 106 CFU/g101. The roots were injured before inoculation. Plants inoculated with sterile water were used as negative controls. To determine the amount of bacteria (CFU) at 6 days post-inoculation, the plants were disinfected with 70% ethanol (v/v) for 3 min, immersed in sterilized water for 3 min and dried with sterile absorbent paper. The roots and 1 cm sections of the stems were cut and weighed. Subsequently, both tissues were ground in sterile water and serial dilutions of the bacterial suspensions were streaked onto mSMSA plates and incubated 7 days at 28 °C. In addition, 10 cross-sections of the main root and stem of the control and inoculated plants were cut with a disinfected scalpel by hand and visualized by CLSM (Confocal Nikon C1SiR attached to a Nikon TE2000 inverted microscope) to analyze the bacterial colonization in stem and root xylem vessels101. Images obtained were analyzed with ImageJ software.
Statistical analyses
Statistical analysis was performed with R statistical software (R Foundation for Statistical Computing, Vienna, Austria). To compare the growth curves, the data were analyzed using a mixed model of repeated measures (longitudinal data), considering three fixed factors, condition (at two levels: growth in white light and darkness), strain (Rpso GMI1000 and RpsoΔlov) and time (at 10 levels) and culture as a random factor. It was considered significant when p < 0.05.
To fulfill the objective of the work, a non-parametric bifactorial ANOVA (two-way ANOVA) test was applied in the motility, adhesion, EPS production, and bacterial count in roots and stem tests. A p < 0.05 was considered statistically significant.
The comparison of continuous variables (ß-galactosidase activity and Rpsolov gene expression) in different subgroups was performed using the Mann–Whitney U test. The analysis was performed at a significance level of 5% and a p value less than 0.05 was considered statistically significant.
References
Petroutsos, D. et al. A blue-light photoreceptor mediates the feedback regulation of photosynthesis. Nature 537, 563–566 (2016).
Kangasjärvi, S., Neukermans, J., Li, S., Aro, E. M. & Noctor, G. Photosynthesis, photorespiration, and light signalling in defence responses. J. Exp. Bot. 63, 1619–1636 (2012).
Roden, L. C. & Ingle, R. A. Lights, rhythms, infection: The role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell 21, 2546–2552 (2009).
Mühlenbock, P. et al. Chloroplast signaling and lesion simulating disease1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20, 2339–2356 (2008).
Yang, Y. X. et al. RNA-seq analysis reveals the role of red light in resistance against Pseudomonas syringae pv. tomato DC3000 in tomato plants. BMC Genomics 16, 1–16 (2015).
Svyatyna, K. & Riemann, M. Light-dependent regulation of the jasmonate pathway. Protoplasma 249, 137–145 (2012).
Kraiselburd, I., Moyano, L., Carrau, A., Tano, J. & Orellano, E. G. Bacterial photosensory proteins and their role in plant–pathogen interactions. Photochem. Photobiol. 93, 666–674 (2017).
Gomelsky, M. & Hoff, W. D. Light helps bacteria make important lifestyle decisions. Trends Microbiol. 19, 441–448 (2011).
Kraiselburd, I. et al. A LOV protein modulates the physiological attributes of Xanthomonas axonopodis pv. citri relevant for host plant colonization. PLoS ONE 7, e38226 (2012).
Río-Álvarez, I. et al. Light regulates motility, attachment and virulence in the plant pathogen Pseudomonas syringae pv. tomato DC3000. Environ. Microbiol. 16, 2072–2085 (2014).
Santamaría-Hernando, S. et al. Pseudomonas syringae pv. tomato exploits light signals to optimize virulence and colonization of leaves. Environ. Microbiol. 20, 4261–4280 (2018).
Kottke, T., Xie, A., Larsen, D. S. & Hoff, W. D. Photoreceptors take charge: Emerging principles for light sensing. Annu. Rev. Biophys. 47, 291–313 (2018).
Crosson, S., Rajagopal, S. & Moffat, K. The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry 42, 2–10 (2003).
Swartz, T. E. et al. Blue-light-activated histidine kinases: Two-component sensors in bacteria. Science 317, 1090–1093 (2007).
Rinaldi, J. et al. Structural insights into the HWE histidine kinase family: The Brucella blue light-activated histidine kinase domain. J. Mol. Biol. 428, 1165–1179 (2016).
Wu, L., McGrane, R. S. & Beattie, G. A. Light regulation of swarming motility in Pseudomonas syringae integrates signaling pathways mediated by a bacteriophytochrome and a LOV protein. MBio 4, 1–9 (2013).
Moriconi, V. et al. LOV-domain photoreceptor, encoded in a genomic island, attenuates the virulence of Pseudomonas syringae in light-exposed Arabidopsis leaves. Plant J. 76, 322–331 (2013).
Bonomi, H. R. et al. Light regulates attachment, exopolysaccharide production, and nodulation in Rhizobium leguminosarum through a LOV-histidine kinase photoreceptor. Proc. Natl. Acad. Sci. USA. 109, 12135–12140 (2012).
Shimomura, A. et al. Blue light perception by both roots and rhizobia inhibits nodule formation in Lotus japonicus. Mol. Plant-Microbe Interact. 29, 786–796 (2016).
Allen C., Prior P. & Hayward A. C. The current bacterial wilt situation: a global overview. In Bacterial Wilt Disease and the Ralstonia solanacearum Species Complex. Am. Phytopathol. Soc. 10, 9–28 (2005).
Hayward, A. C. Bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 29, 65–87 (1991).
Genin, S. & Denny, T. P. Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89 (2012).
Salanoubat, M. et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502 (2002).
Hikichi, Y. et al. Regulation involved in colonization of intercellular spaces of host plants in Ralstonia solanacearum. Front. Plant Sci. 8, 1–6 (2017).
Genin, S. Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 187, 920–928 (2010).
Perrier, A. et al. Comparative transcriptomic studies identify specific expression patterns of virulence factors under the control of the master regulator PhcA in the Ralstonia solanacearum species complex. Microb. Pathog. 116, 273–278 (2018).
Genin, S., Brito, B., Denny, T. P. & Boucher, C. Control of the Ralstonia solanacearum Type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 579, 2077–2081 (2005).
Puigvert, M. et al. Type III secretion inhibitors for the management of bacterial plant diseases. Mol. Plant Pathol. 20, 20–32 (2019).
Aldon, D., Brito, B., Boucher, C. & Genin, S. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 19, 2304–2314 (2000).
Yao, J. & Allen, C. Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 188, 3697–3708 (2006).
Allen, C., Gay, J. & Simon-Buela, L. A regulatory locus, pehSR, controls polygalacturonase production and other virulence functions in Ralstonia solanacearum. Mol. Plant-Microbe Interact. 10, 1054–1064 (1997).
Losi, A. The bacterial counterparts of plant phototropins. Photochem. Photobiol. Sci. 3, 566–574 (2004).
Mandalari, C., Losi, A. & Gärtner, W. Distance-tree analysis, distribution and co-presence of bilin- and flavin-binding prokaryotic photoreceptors for visible light. Photochem. Photobiol. Sci. 12, 1144–1157 (2013).
Sahoo, B. R. & Fujiwara, T. Conformational states of HAMP domains interacting with sensory rhodopsin membrane systems: An integrated all-atom and coarse-grained molecular dynamics simulation approach. Mol. Biosyst. 13, 193–207 (2017).
Kanda, A. et al. An amino acid substitution at position 740 in σ70 of Ralstonia solanacearum strain OE1-1 affects its in planta growth. Appl. Environ. Microbiol. 74, 5841–5844 (2008).
Cao, Y. et al. Genome sequencing of Ralstonia solanacearum FQY_4, isolated from a bacterial wilt nursery used for breeding crop resistance. Genome Announc. 1, 6088–6089 (2013).
Hayes, M. M., MacIntyre, A. M. & Allen, C. Complete genome sequences of the plant pathogens Ralstonia solanacearum type Strain K60 and R. solanacearum Race 3 Biovar 2 Strain UW551. Am. Soc. Microbiol. 5, 1–2 (2017).
Remenant, et al. Genomes of three tomato pathogens within the Ralstonia solanacearum species complex reveal significant evolutionary divergence. BMC Genomics 11, no pagination (2010).
Xu, J. et al. Complete genome sequence of the plant pathogen Ralstonia solanacearum strain Po82. J. Bacteriol. 193, 4261–4262 (2011).
Guarischi-Sousa, R. et al. Complete genome sequence of the potato pathogen Ralstonia solanacearum UY031. Stand. Genomic Sci. 11, 1–8 (2016).
Remenant, B. et al. Ralstonia syzygii, the blood disease bacterium and some asian R. solanacearum strains form a single genomic species despite divergent lifestyles. PLoS ONE 6, 1–10 (2011).
Meng, F., Yao, J. & Allen, C. A MotN mutant of Ralstonia solanacearum is hypermotile and has reduced virulence. J. Bacteriol. 193, 2477–2486 (2011).
Liu, H., Kang, Y., Genin, S., Schell, M. A. & Denny, T. P. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147, 3215–3229 (2001).
Hayashi, K. et al. Contribution of a lectin, LecM, to the quorum sensing signalling pathway of Ralstonia solanacearum strain OE1-1. Mol. Microbiol. 20, 334–345 (2019).
Peyraud, R., Denny, T. & Genin, S. Exopolysaccharide quantification for the plant pathogen Ralstonia solanacearum. Bio-Protoc. 7, 2–9 (2017).
Monteiro, F., Genin, S., van Dijk, I. & Valls, M. A luminescent reporter evidences active expression of Ralstonia solanacearum type III secretion system genes throughout plant infection. Microbiology 158, 2107–2116 (2012).
Beattie, G. A., Hatfield, B. M., Dong, H. & McGrane, R. S. Seeing the light: The roles of red- and blue-light sensing in plant microbes. Annu. Rev. Phytopathol. 56, 41–66 (2018).
Van der Horst, M. A., Key, J. & Hellingwerf, K. J. Photosensing in chemotrophic, non-phototrophic bacteria: Let there be light sensing too. Trends Microbiol. 15, 554–562 (2007).
Moyano, L. et al. Red light delays programmed cell death in non-host interaction between Pseudomonas syringae pv tomato DC3000 and tobacco plants. Plant Sci. 291, 110361 (2020).
Oberpichler, I. et al. Light affects motility and infectivity of Agrobacterium tumefaciens. Environ. Microbiol. 10, 2020–2029 (2008).
Villeth, G. R. et al. Comparative proteome analysis of Xanthomonas campestris pv. campestris in the interaction with the susceptible and the resistant cultivars of Brassica oleracea. FEMS Microbiol. Lett. 298, 260–266 (2009).
Schumacher, J. How light affects the life of Botrytis. Fungal Genet. Biol. 106, 26–41 (2017).
Kraiselburd, I. et al. The LOV protein of Xanthomonas citri subsp. citri plays a significant role in the counteraction of plant immune responses during citrus canker. PLoS ONE 8, 1–16 (2013).
Glantz, S. T. et al. Functional and topological diversity of LOV domain photoreceptors. Proc. Natl. Acad. Sci. USA. 113, E1442–E1451 (2016).
Qian, W., Han, Z. J. & He, C. Two-component signal transduction systems of Xanthomonas spp.: A lesson from genomics. Mol. Plant-Microbe Interact. 21, 151–161 (2008).
Losi, A. & Gärtner, W. Old chromophores, new photoactivation paradigms, trendy applications: Flavins in blue light-sensing photoreceptors. Photochem. Photobiol. 87, 491–510 (2011).
Losi, A. & Gärtner, W. Bacterial bilin- and flavin-binding photoreceptors. Photochem. Photobiol. Sci. 7, 1168–1178 (2008).
Verma, R. K. et al. A bacteriophytochrome mediates Interplay between light sensing and the second messenger cyclic Di-GMP to control social behavior and virulence. Cell Rep. 32, 108202 (2020).
Craig, L., Forest, K. T. & Maier, B. Type IV pili: Dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 17, 429–440 (2019).
Kang, Y., Liu, H., Genin, S., Schell, M. A. & Denny, T. P. Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46, 427–437 (2002).
Elhenawy, W. et al. Protein O-linked glycosylation in the plant pathogen Ralstonia solanacearum. Glycobiology 26, 301–311 (2015).
Bitrian, M., González, R. H., Paris, G., Hellingwerf, K. J. & Nudel, C. B. Blue-light-dependent inhibition of twitching motility in Acinetobacter baylyi ADP1: Additive involvement of three BLUF-domain-containing proteins. Microbiology 159, 1828–1841 (2013).
Hoff, W. D. et al. Prokaryotic phototaxis. In Chemotaxis, Methods in Molecular Biology, Vol 571, 25–49 (2009).
Branda, S. S., Vik, Å., Friedman, L. & Kolter, R. Biofilms: The matrix revisited. Trends Microbiol. 13, 20–26 (2005).
Rigano, L. A. et al. Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri. Mol. Plant-Microbe Interact. 20, 1222–1230 (2007).
Purcell, E. B., Boutte, C. C. & Crosson, S. Two-component signaling systems and cell cycle control in Caulobacter crescentus. Adv Exp Med Biol 631, 30 (2008).
Denny, T. P., Carney, B. F. & Schell, M. A. Inactivation of multiple virulence genes reduces the ability of Pseudomonas solanacearum to cause wilt symptoms. Mol. Plant Microbe Interact. 3, 293–300 (1990).
Saile, E., McGarvey, J. A., Schell, M. A. & Denny, T. P. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 87, 1264–1271 (1997).
Oliveira, N. M. et al. Biofilm formation as a response to ecological competition. PLoS Biol. 13, 1–23 (2015).
Karatan, E. & Watnick, P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347 (2009).
Serra, D. O., Richter, A. M. & Hengge, R. Cellulose as an architectural element in spatially structured Escherichia coli biofilms. J. Bacteriol. 195, 5540–5554 (2013).
Mori, Y. et al. The vascular plant-pathogenic bacterium Ralstonia solanacearum produces biofilms required for its virulence on the surfaces of tomato cells adjacent to intercellular spaces. Mol. Plant Pathol. 17, 890–902 (2016).
Lowe-Power, T. M., Khokhani, D. & Allen, C. How Ralstonia solanacearum exploits and thrives in the flowing plant xylem environment. Trends Microbiol. 26, 929–942 (2018).
Mussi, M. A. et al. The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J. Bacteriol. 192, 6336–6345 (2010).
Njimona, I. & Lamparter, T. Temperature effects on Agrobacterium phytochrome agp1. PLoS ONE 6, e25977 (2011).
Golic, A. et al. Staring at the cold sun: Blue light regulation Is distributed within the genus Acinetobacter. PLoS ONE 8, e55059 (2013).
Braatsch, S., Gomelsky, M., Kuphal, S. & Klug, G. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 45, 827–836 (2002).
Delaspre, F. et al. The Ralstonia solanacearum pathogenicity regulator HrpB induces 3-hydroxy-oxindole synthesis. Proc. Natl. Acad. Sci. USA. 104, 15870–15875 (2007).
Vasse, J., Genin, S., Frey, P., Boucher, C. & Brito, B. The hrpB and hrpG regulatory genes of Ralstonia solanacearum are required for different stages of the tomato root infection process. Mol. Plant-Microbe Interact. 13, 259–267 (2000).
Tans-Kersten, J., Brown, D. & Allen, C. Swimming motility, a virulence trait of Ralstonia solanacearum, is regulated by FlhDC and the plant host environment. Mol. Plant-Microbe Interact. 17, 686–695 (2004).
Puigvert, M. et al. Transcriptomes of Ralstonia solanacearum during root colonization of Solanum commersonii. Front. Plant Sci. 8, 370 (2017).
Jacobs, J. M. et al. The in planta transcriptome of Ralstonia solanacearum: Conserved physiological and virulence strategies during bacterial wilt of tomato. MBio 3, 1–11 (2012).
Clough, S., Schell, M. A. & Denny, T. Evidence for involvement of a volatile extracellular factor in Pseudomonas solanacearum virulence gene expression. Mol. Plant Microbe Interact. 7, 621–630 (1994).
Hendrickt, C. A. & Sequeira, L. Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl. Environ. Microbiol. 48, 94–101 (1984).
Bertolla, F., Van Gijsegem, F., Nesme, X. & Simonet, P. Conditions for natural transformation of Ralstonia solanacearum. Appl. Environ. Microbiol. 63, 4965–4968 (1997).
Elphinstone, J., Hennessy, J., Wilson, J. & Stead, D. E. Sensitivity of different methods for the detection of Ralstonia solanacearum in potato tuber extracts. Bull. OEPP/EPPO 26, 663–678 (1996).
Sambrook, J., Fitsch, E. F. & Maniatis, T. Molecular Cloning: A Laboratory Manual 2nd edn. (Cold Spring Harbor Laboratory Press, 1989).
Safni, I. et al. Polyphasic taxonomic revision of the Ralstonia solanacearum species complex: Proposal to emend the descriptions of Ralstonia solanacearum and Ralstonia syzygii and reclassify current R. syzygii strains as Ralstonia syzygii subsp. syzygii subsp. nov., R. s.. Int. J. Syst. Evol. Microbiol. 64, 3087–3103 (2014).
Prior, P. et al. Genomic and proteomic evidence supporting the division of the plant pathogen Ralstonia solanacearum into three species. BMC Genomics 17, 1–11 (2016).
Valls, M., Genin, S. & Boucher, C. Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLoS Pathog. 2, 0798–0807 (2006).
Ray, S. K., Kumar, R., Peeters, N., Boucher, C. & Genin, S. rpoN1, but not rpoN2, is required for twitching motility, natural competence, growth on nitrate, and virulence of Ralstonia solanacearum. Front. Microbiol. 6, 1–11 (2015).
Boucher, C. A., Barberis, P. A., Trigalet, A. P. & Demery, D. A. Transposon mutagenesis of Pseudomonas solanacearum: Isolation of Tn5-induced avirulent mutants. J. Gen. Microbiol. 131, 2449–2457 (1985).
Genin, S., Gough, C. L., Zischek, C. & Boucher, C. A. Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum. Mol. Microbiol. 6, 3065–3076 (1992).
Cunnac, S., Occhialini, A., Barberis, P., Boucher, C. & Genin, S. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: Identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol. Microbiol. 53, 115–128 (2004).
Marx, C. J. & Lidstrom, M. E. Broad-host-range cre-lox system for antibiotic marker recycling in Gram-negative bacteria. Biotechniques 33, 1062–1067 (2002).
Huet, S. et al. Nuclear import and assembly of Influenza a virus RNA polymerase studied in live cells by fluorescence cross-correlation spectroscopy. J. Virol. 84, 1254–1264 (2010).
Mori, Y. et al. Involvement of ralfuranones in the quorum sensing signaling pathway and virulence of Ralstonia solanacearum strain OE1-1 Yuka. Mol. Plant Pathol. 19, 454–463 (2018).
Siri, M. I., Sanabria, A., Boucher, C. & Pianzzola, M. J. New type IV pili-related genes involved in early stages of Ralstonia solanacearum potato infection. Mol. Plant-Microbe Interact. 27, 712–724 (2014).
Petrocelli, S., Tondo, M. L., Daurelio, L. D. & Orellano, E. G. Modifications of Xanthomonas axonopodis pv. citri lipopolysaccharide affect the basal response and the virulence process during citrus canker. PLoS ONE 7, e40051 (2012).
Brito, B., Marenda, M. & Barberis, P. prhJ and hrpG, two new components of the plant signal-dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum. Mol. Microbiol. 31, 237–251 (1999).
Cruz, A. P. Z. et al. A novel, sensitive method to evaluate potato germplasm for bacterialwilt resistance ysing a luminescent Ralstonia solanacearum Reporter Strain. Mol. Plant-Microbe Interact. 27, 277–285 (2014).
Acknowledgements
Thanks to Stephane Genin (LIPME, Université de Toulouse, INRAE, CNRS, Castanet-Tolosan, France) for providing mutant Ralstonia solanacearum strains in transcriptional regulators for this study and Marc Valls (Centre for Research in Agricultural Genomics (CRAG), CSIC-IRTA-UAB-UB, Campus UAB, Bellaterra, Barcelona, Spain and Department of Genetics, University of Barcelona, Barcelona, Spain) for providing R. solanacearum GMI1000 Pps-GFP. We also thank Rodrigo Vena for assistance with the microscopy facility. This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT) (PICT2017-2242 to E.G.O.) and Universidad Nacional de Rosario (UNR), (Grant N° 1BIO432) to E.G.O. E.G.O., M.V.R. and M.L.T. are staff members; A.C. and J.T. are fellows of Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina), M.B.R. is fellow of ANPCyT; S.P. and E.G.O. are staff member of CIUNR-UNR.
Funding
Funding was funded by Agencia Nacional de Promoción Científica y Tecnológica (grant no. PICT 2017-2242) and Universidad Nacional de Rosario (UNR), Argentina (Grant no. 1BIO432).
Author information
Authors and Affiliations
Contributions
J.T., M.B.R., M.L.T., and E.G.O.: conceived and designed all experiments. J.T., M.B.R., M.L.T., A.C., M.V.R., S.P., V.F.: collection and analysis of biological data. E.G.O.: contributed reagents, materials and analysis tools. M.B.R. and A.C.: prepared the figures and tables. L.P.: statistical analysis of data. J.T., M.B.R. and E.G.O.: wrote the main text. J.T., M.B.R., M.L.T., A.C., S.P., M.V.R., V.F., M.I.S., L.P. and E.G.O.: they reviewed and approved the final version of the document.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tano, J., Ripa, M.B., Tondo, M.L. et al. Light modulates important physiological features of Ralstonia pseudosolanacearum during the colonization of tomato plants. Sci Rep 11, 14531 (2021). https://doi.org/10.1038/s41598-021-93871-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-93871-9
- Springer Nature Limited
This article is cited by
-
Bacteria phototaxis optimizer
Neural Computing and Applications (2023)