Abstract
Background
Despite evident success of malaria control in many sites in the Arabian Peninsula, malaria remains endemic in a few spots, in Yemen and south-west of Saudi Arabia. In addition to local transmission, imported malaria sustains an extra source of parasites that can challenge the strengths of local control strategies. This study examined the genetic diversity of Plasmodium falciparum in Yemen and mutations of drug resistant genes, to elucidate parasite structure and distribution of drug resistance genotypes in the region.
Methods
Five polymorphic loci (MSP-2, Pfg377 and three microsatellites on chromosome 8) not involved in anti-malarial drug resistance, and four drug resistant genes (pfcrt, pfmdr1, dhfr and dhps) were genotyped in 108 P. falciparum isolates collected in three sites in Yemen: Dhamar, Hodeidah and Taiz.
Results
High diversity was seen in non-drug genes, pfg377 (He = 0.66), msp-2 (He = 0.80) and three microsatellites on chr 8, 7.7 kb (He = 0.88), 4.3 kb (He = 0.77) and 0.8 kb (He = 0.71). There was a high level of mixed-genotype infections (57%), with an average 1.8 genotypes per patient. No linkage disequilibrium was seen between drug resistant genes and the non-drug markers (p < 0.05). Genetic differentiation between populations was low (most pair-wise FST values <0.03), indicating extensive gene flow between the parasites in the three sites.
There was a high prevalence of mutations in pfmdr1, pfcrt and dhfr; with four mutant pfmdr1 genotypes (NFCDD[57%], NFSND[21%], YFCDD[13%] and YFSND[8% ]), two mutant pfcrt genotypes (CVIET[89%] and SVMNT[4%]) and one mutant dhfr genotype (ICNI[53.7%]). However, no dhps mutations were detected.
Conclusion
The high diversity of P. falciparum in Yemen is indicative of a large parasite reservoir, which represents a challenge to control efforts. The presence of two distinct pfcrt genotype, CVIET and SVMNT, suggests that chloroquine resistance can possibly be related to a migratory path from Africa and Asia. The absence of the triple mutant dhfr genotype (IRN) and dhps mutations supports the use of artesunate + sulphadoxine-pyrimethamine as first-line therapy. However, the prevalent pfmdr1 genotype NFSND [21%] has previously been associated with tolerance/resistance response to artemisinin combination therapy (ACT). Regular surveys are, therefore, important to monitor spread of pfmdr1 and dhfr mutations and response to ACT.
Similar content being viewed by others
Background
The Arabian Peninsula lies at the fringes of malaria endemicity where successful control measures have brought local transmission to halt in many countries in the region, e.g. Bahrain, Kuwait and United Arab Emirates (UAE)[1]. However, some sites in Yemen and southern Saudi Arabia remain malarious, with a high prevalence of drug-resistant Plasmodium falciparum parasites[2, 3]. In addition, imported malaria cases, via asymptomatic travellers from malaria-endemic areas, sustain a major challenge for possible rebound of local transmission[4].
The region attracts numerous migrations from Africa and Asia. Over the past two decades political instability has resulted in mass movement of people from the Horne of Africa (Somalia, Eritrea and Ethiopia) into Yemen and Saudi Arabia[5]. These countries are known for high prevalence of drug-resistant P. falciparum malaria. In addition, the region attracts a lot of skilled workers from Asia. However, the impact of these migrations on the gene pool of local malaria parasites in the Arabian Peninsula remains unclear as information on malaria parasites genetics is scarce.
In Yemen, over 60% of the population live in areas with stable malaria transmission[6]. Plasmodium falciparum is responsible for about 90% of malaria cases. Indigenous chloroquine resistance (CQR) was reported in 1989[7, 8], and since then it spread to different regions in the highland and lowland areas in Taiz[7] and Hodeidah[9]. Cross- sectional surveys in 2008 and 2009 in Taiz, Hodeidah, Dhamar and Rymah showed high prevalence of the CQR marker pfcrt 76 T[2]. However, resistance to sulphadoxine-pyrimethamine (SP) is rare in Yemen[2, 10]. Currently SP is used in combination with artesunate (AS) as first-line treatment of uncomplicated falciparum malaria. In addition, artemether-lumefantrine (AL) is used as a second-line therapy for treatment failure[11].
The present study examined genetic diversity of P. falciparum in Yemen, and distribution of genes implicated in resistance to widely used anti-malarial drugs, including, chloroquine, SP, mefloquine and artemisinin. Such analysis will provide desirable information on the genetics structure of P. falciparum in the Arabian Peninsula, and the prospect of evolution of drug resistance following recent shift of malaria therapy to artemisinin combination therapy (ACT).
Methods
Study sites and sampling
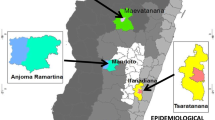
One hundred and fifteen finger-prick blood samples were collected, from microscopy-confirmed malaria cases, in three localities in Yemen (Dhamar, Hodeidah and Taiz) (Figure 1) Dhamar (Latitude: 14°32′33″ N, Longitude: 44°24′18″ E ) is a highland (2407 m above sea) where transmission occurs throughout the year. Hodeidah (Latitude: 14°48′08″N, Longitude: 42°57′04″E) is a coastal area where malaria transmission occurs in winter and Taiz (Latitude: 13°34′44″N, Longitude: 44°01′19″E) is a mountainous area (1311 m above sea) and transmission occurs in summer[2]. Ethical clearance was obtained from Faculty of Medicine, Sana’a University, Yemen. Informed consent was obtained from each participant before enrolment in the study.
Non drug-resistance genes
pfg377 and msp-2
The pfg377 gene primers amplify a polymorphic region, encoding a seven degenerate amino acid repeats; alleles of this gene, vary by multiples of 21 base pairs[12]. Polymerase chain reaction (PCR) primers and conditions for amplification were as described previously[12], with minor modification where the nested primer 377R3D1 was labelled.
msp-2 alleles fall into two sequence types, FC27 and 3D7, which differ in the sequence of the dimorphic central region, block 3[13]. PCR primers and conditions for amplification of each type were described by Aubouy et al.[14].
The fluorescent PCR products of pfg377 and msp-2 were analysed in an ABI 3130XL Genetic Analyzer, and alleles were visualized and sized on Genesmapper v. 4 (Applied Biosystems).
Microsatellites
Three single copy microsatellites on chr 8 flanking the dhps (0.8, 4.3 and 7.7 kb from the 3′ end of the gene) were types. All isolates examined in this study harbour the wild-type dhps, therefore, the above microsatellites are not influenced by drug pressure and were used to examine parasite diversity and population structure.
The microsatellites were examined as described by Roper et al.[15]. Fluorescent PCR products were analysed in an ABI 3130XL Genetic Analyzer, and alleles were visualized and sized on Genemapper v.4 (Applied Biosystems).
Drug resistance genes
pfcrt, pfmdr1, dhfr and dhps
Alleles of the pfcrt, pfmdr1, dhfr and dhps genes were amplified using two rounds of PCR[1, 16–19]. The amplified fragments of each gene encompasses mutations associated with CQR (pfcrt-72,-74,-75, and −76), and multidrug resistant gene-1 (pfmdr1-86, -184, 1034,1042 and 1246), pyrimethamine resistance (dhfr-51, -59, -108, and −164), sulphadoxine resistance (dhps-436, -437, -540, and −613).
PCR fragments of the isolates, together with control clones (3D7, Hb3 and Dd2) of known sequence and drug response, were then sequenced. The parasites were obtained from The European Malaria Reference Reagent Repository. 3D7 and Dd2, are chloroquine sensitive and resistant clones, with pfcrt76K and pfcrt76T codons, respectively[20], and codons pfmdr1 86 N and 86Y associated with susceptible and resistance response to choloroquine, respectively[21]. The 3D7 clone is known to be pyrimethamine and sulphadoxine sensitive carrying wild-type dhfr and dhps gene, implicated in resistance to the two drug respectively[22], while Dd2 is a pyrimethamine and sulphadoxine resistance, carrying mutations in dhfr and dhps associated with in vitro resistance[23]. The PCR products were first purified using ExoSAP-IT (USB) as described by the manufacturer. Sequencing was carried out using BigDye Terminator v3.1 cycle (ABI, UK) and run on a thermocycler. Initial sequences were compared with reference sequences in the PlasmoDB database of P. falciparum genomic using BLASTN, BioEdit and Lasergene software.
Statistical analysis
The prevalence of an allele and genotype were calculated as percentage of all the alleles and genotypes detected at a given locus among the examined isolates. The expected heterozygosity index (H e), which measures the diversity at a locus examined, was calculated for non-drug resistant genes using microsatellite Toolkit software as described elsewhere[24].
The multiplicity of infection was defined as the proportion of people who carry more than one allele (genotype) for any of the examined genes, and the minimum number of clones per infection was estimated as the largest number of alleles at any of the examined loci[25].
Gamete linkage disequilibrium (LD) values between all pairs of loci were calculated for all samples GENEPOP v.4.1[26]. For these analyses, only the predominant alleles at each locus, indicating clonal infections, were used.
To examine whether allele frequencies differ between parasite populations in Dhamar, Hodeidah and Taiz, F ST indices were calculated using Weir and Cockerham's method[27] estimator of Wright's F-statistics using the computer package GENEPOP v.4.1[26]. A permutation test (n = 10,000) was applied (permuting alleles over populations) to test whether F ST indices were significantly different from zero.
Principal coordinate analysis (PCoA) was carried out to examine the genetic similarities (of alleles of non-drug resistant genes) between P. falciparum isolates in the three coding sites using Genalex Version 6.5[28]. The two-dimensional PCoA plot shows the relationships between the haplotypic variants found in the three populations.
Results
Out of the 115 samples with microscopy-confirmed malaria parasites, 108 proved to be P. falciparum and produced the expected PCR amplicons for the examined genes (pfcrt, pfmdr1, dhfr, dhps, pfg377 and msp-2). The other seven (6.4%) isolates were most likely to have been Plasmodium vivax, which is found in Yemen.
Diversity and structure of non drug-resistant genes
The five non drug-resistant loci (pfg377, msp-2 and three microsatellites on chr 8) were highly polymorphic, with a number of alleles per locus ranging from five (pfg377) to 45 (msp-2). Diversity, expressed as expected hetrozygosity (He), was high for all loci, ranging from 0.51 to 0.89, with average of 0.66 (for pfg377) and 0.80 (for msp-2) (Table 1), reflecting high diversity among the examined isolates.
No LD was seen between genes involved in drug resistance (pfcrt, pfmdr1 and dhfr) and non-drug loci (pfg377 and msp-2) (P < 0.001). However, loci involving resistance to one drug (pfcrt and pfmdr1) had a greater average LD than those not involved in drug resistance loci (P < 0.01).
Alleles at non drug-resistance loci were distributed widely across parasites in different sites in Yemen (Table 1), and any ‘private’ alleles (detected in only one population) existed at very low frequencies (Additional file1). Comparison of parasites in the three sites showed low F st values, ranged from 0.141 to 0.003, suggesting frequent gene flow (Table 1). Furthermore, PCoA analysis showed no pattern of clustering of parasites in any of the three geographical sites (Figure 2).
Graphical plotting of the PCoA based on genetic variation of non drug-resistant genes ( Pfg377 , MSP-2 and three microsatellites in chr 8) among Plasmodium falciparum parasites in three sites in Yemen, and variation representation by each axis. Blue icon = Taiz, red icon = Dhamar and green icon = Hodeidah.
Multiplicity of Plasmodium falciparum infection
There was a high prevalence of mixed genotype infection, 57% of the isolates carried more than one genotype. The number of genotypes per infected person (the maximum number of alleles observed at any locus) was estimated as 1.8, assuming that each clone is readily transmissible to mosquito, this reflects a high rate of cross-mating[25].
Distribution of drug resistance markers
pfcrt
Out of the 108 P. falciparum isolates successfully examined, four (4%), 96 (89%), 96 (89%) and 100 (93%) harboured mutations at codons 72, 74, 75 and 76 respectively (Table 2). Ninety-six (89%), four (4%) and eight (7%) isolates carried the pfcrt genotypes CVIET, S VMNT and the wild-type (CVMNK), respectively (Table 2).
All isolates carrying the wild-type pfcrt genotype (CVMNK) were detected in Taiz, except one seen in Hodeidah. Similarly, isolates with the mutant pfcrt genotype S VMNT were only seen in Hodeidah.
pfmdr1
There was a, high prevalence of pfmdr1 mutation at codon N86Y (20%), Y184F (99%), S1034C (70%) and N1042D (70%), while no mutation was seen at codon 1246 (Table 2). The majority of isolates (57%) carried the mutant genotype (NFCD D), while 21%, 13%, and 8% of them harboured the mutant genotype (NF SND), (YFCD D), and (YF SND) respectively. However, only one isolate carried the wild-type (1%) (Table 2).
dhfr
No mutations were seen at codons 59 and 164. However, 58 (54%) isolates showed mutations at codons N51I and S108N (Table 2). A large proportion of the isolates (54%) carried the mutant genotype (I CN I), while 50 (46%) isolates harboured the wild-type (NCSI) (Table 2).
dhps
Unlike the other drug-resistant genes, no mutation was detected at all codons 436, 437, 540 and 613, which are implicated in sulphadoxine resistance.
Discussion
Many sites in the Arabian Peninsula turned into “malaria-free” following intensive control efforts mounted over a relatively short period of time[1]. In contrast, in Yemen and southwest Saudi Arabia malaria remains resilient to control efforts. Some of the challenges facing the prospect of elimination, in these sites, is the introduction of drug resistance parasites via asymptomatic carriers. The present study examined the extent of genetic diversity and distribution of drug-resistant genes among of P. falciparum in some major foci in Yemen.
A high degree of diversity of non-drug-resistant genes (msp-2 and pfg377 and microsatellites in Chr 8) was seen among P. falciparum in Yemen. The He index ranged between 0.51 and 0.89, suggesting a high rate of transmission and a bigger parasite population size than anticipated in this region. Both allelic diversity and prevalence of mixed-genotype infection were high, 57% of the isolates harboured more than one allele at any of the examined loci, with a mean of 1.8 genotypes per infected individual. Assuming that each clone is readily transmissible to mosquito, the rate of cross-mating and subsequent recombination is expected to be high[25]. This agrees with the observed random association between loci and absence of geographical differentiation among P. falciparum in the three sites (Dhamar, Hodeidah and Taiz) (F st < 0.05). Such a pattern of parasite structure is similar to that seen among P. falciparum populations in Africa[29, 30]. These findings imply presence of a large effective population size (Ne) of P. falciparum in Yemen, as there is a direct relationship between the level of diversity and Ne[31–33]. This represents a challenge to control efforts in the whole region, as parasites can readily migrate from Yemen into other sites in the Arabian Peninsula, such as Oman[11], UAE[34] and Saudi Arabia[35], where transmission has been interrupted. In addition to local foci, imported malaria, via numerous migrations from Africa/Asia, can enrich diversity of parasites in the Arabian Peninsula. Over the past two decades political instability has resulted in mass movement of people from the Horn of Africa (Somalia, Eritrea and Ethiopia) into Yemen and Saudi Arabia[5]. Travellers from malaria-endemic areas can carry long-lasting, transmissible, asymptomatic parasitaemia[36, 37]. Multiple introductions of distinct genetic sources could in part explain the high level of parasite diversity. However, the lack of genetic differentiation between parasites in different sites suggests that the prevailing epidemiological and demographical factors in Yemen are favorable to parasite dispersal. Imported malaria can, therefore, jeopardize the current success of control in the region despite the presence of a strong public health infrastructure. For example, countries such as Oman experience regular epidemics due to imported malaria[11]. To sustain malaria control and achieve elimination in the peninsula efforts should be directed to foci with on-going transmission, such as Yemen and Saudi Arabia.
The distribution of drug resistant genotypes seen in the present study fits with the history of anti-malarial usage, and possible migration of parasites from Africa and Asia into Yemen. The emergence of CQR in Yemen in 1989 Yemen[2, 7, 9, 38, 39], coincided with its appearance in close countries in East Africa[12, 16, 40] and Asia, Saudi Arabia[3], Iran and Pakistan[3, 41–46]. The presence of two distinct pfcrt genotype, CVIET (89%) and S VMNT (4%) suggests that CQR in Yemen has evolved from at least two different origins. Globally, there are five genotypes of pfcrt CVIET, S VMNT, S VIET, CVMNT and CVT NT[47]. The CVIET genotype is predominant in Africa[48], while the S VMNT is common in Asian countries close to the Arabian Peninsula, such as Iran[43], India[49] and Pakistan[41, 42]. This agrees with the hypothesis of frequent gene flow of African/Asian parasites into Yemen, and suggests that the CQR genotype (CVIET) is probably introduced via Africa, while genotype S VMNT has come from Asia. Analysis of microsatellites around pfcrt will allow further investigation to the origin of pfcrt resistance genotypes[29, 48].
Similar to pfcrt, high prevalence of pfmdr1 mutations was seen in Yemen. Mutations in codon 86Y has been linked to CQR, while those at codons 184, 1034, 1042 and 1246 are related to mefloquine (MQ), amodiaquine (AQ), halofantrine (HF) and quinine resistance[32, 50, 51]. In the present study, 86 (80%), 107 (99%) and 108 (100%) of the examined P. falciparum isolates carried alleles 86N and 184F and 1246D, respectively. Selection of pfmdr1 86N and 184F in recrudescent P. falciparum parasites following lumefantrine suggested a possible role of these alleles in the development of tolerance/resistance to lumefantrine[37, 52]. Increased prevalence of pfmdr1 86N, 184F and 1246D haplotype was seen in Zanzibar after several years of extensive use of artesunate-amodiaquine (ASAQ)[53]. A recent study has confirmed that P. falciparum parasites with the pfmdr1 N86/184F/D1246 haplotype can withstand 15-fold higher lumefantrine blood concentrations compared to those with the 86Y/Y184/1246Y haplotype[54]. The relatively high prevalence of the wild-type allele 86 N (80%) in the present study cannot be explained by withdrawal of CQ in Yemen, as the drug is still in use[55], and pfcrt 76T remains at a prevalence of 89%. It is therefore, more likely that the pattern of pfmdr1 polymorphisms, seen in Yemen, is due to resurgence in use of artesunate-SP combination. In addition to codon 86 and 184, mutations at the codons 1034 and 1042 were also high in Yemen; nonetheless no mutation was seen at codon 1246. Further longitudinal surveillance of pfmdr1 mutations, coupled with in vivo testing, should be considered to examine the impact of these polymorphisms on efficacy of ACT in Yemen.
With regard to genes controlling P. falciparum response to SP, a previous study has detected the mutant allele dhfr-59R among 4 out of 99 isolates in Lahj governorate, southeast of Yemen, and no information was given on other codons 51, 108 and 164[10]. However, the present study revealed high prevalence of mutations at codons 51 and 108 of the dhfr, while no mutation was seen at codon 59. This inconsistency can be attributed to the probability that some of the samples examined by Mubjer et al.[10] may have been collected from expatriates. If mutation 59R exists among local parasites in Yemen, it would have increase in frequency following recent shift to us of artesunate + sulphadoxine-pyrimethamine (SP) as first line therapy[55]. Epidemiological evidence suggests that dhfr mutations starts at codon 108, yielding low level resistance to pyrimethamine[38], then the resistance increases with acquisition of extra mutations at codons 51 or 59 and 164[56]. Analysis of evolution pattern of dhfr genotypes shows that the high resistance genotype (51I, 59R, 108 N) accumulates mutations in sequence. The most common orders of mutation are 108 N, 59R, 51I or 108 N, 51I, 59R[57]. These cumulative mutations in dhfr restore the parasite fitness and allow it to overcome the effect of the drug[58]. dhfr mutations appear earlier than dhps mutations in the development of resistance to SP combined effect[59], and P. falciparum has to gain mutations in dhfr and dhps genes to develop resistance to SP, thus absence of dhps mutation in Yemen suggest that the SP will be effective for some years to come. Nonetheless, the occurrence of high level resistance dhfr and dhps genotypes linked to SP failure in areas close to Yemen, such as Iran and Pakistan (100%), the Horn of Africa; Sudan (72.7%) and Ethiopia (84.5%)[60–64], necessitate the need for regular surveys to monitor possible emergence of these mutations into Yemen for timely change in drug policy.
Abbreviations
- ACT:
-
Artemisinin combination therapy
- SP:
-
Sulphadoxine-pyrimethamine
- Pfcrt:
-
P. falciparum chloroquine transport
- pfmdr1:
-
P. falciparum multidrug resistant
- dhfr:
-
Dihydrofolate reductase gene
- dhps:
-
Dihydropteroate synthase gene
- msp-2:
-
Merozoite surface protein-2
- pfg377:
-
P. falciparum gametocyte antigen
- PCoA:
-
Principle coordinate analysis
- He:
-
Expected heterozygosity
- Fst:
-
Fixation index.
References
Abdel-Hameed AA: Antimalarial drug resistance in the Eastern Mediterranean region. Eastern Mediterranean Health. 2003, 9: 492-508.
Al-Mekhlafi AM, Mahdy MA, Al-Mekhlafi HM, Azazy AA, Fong MY: High frequency of Plasmodium falciparum chloroquine resistance marker (Pfcrt T76 mutation) in Yemen: an urgent need to re-examine malaria drug policy. Parasit Vectors. 2011, 4: 94-10.1186/1756-3305-4-94.
Bin Dajem SM, Al-Qahtani A: Analysis of gene mutations involved in chloroquine resistance in Plasmodium falciparum parasites isolated from patients in the southwest of Saudi Arabia. Ann Saudi Med. 2010, 30: 187-192. 10.4103/0256-4947.62826.
Ismaeel AY, Senok AC, Jassim Al-Khaja KA, Botta GA: Status of Malaria in the Kingdom of Bahrain: a 10-year Review. J Travel Med. 2004, 11: 97-101.
Soucy A: Mixed migration from the Horn of Africa to Yemen. 2011, Kenya: Protection risks and challenges
WHO: World malaria report 2012. 2011, Geneva: World Health Organization
Alkadi HO, Al-Maktari MT, Nooman MA: Chloroquine-resistant Plasmodium falciparum local strain in Taiz Governorate, Republic of Yemen. Chemotherapy. 2006, 52: 166-170. 10.1159/000093592.
Mamser A: WHO/RA/MAL/005. Report on a visit to the Yemen Arab Republic from 14–2 to 13-3-1989. 1989, Geneva, EMRO: World Health Organization, 1-5.
Al-Shamahy H, Al-Harazy AH, Harmal NS, Al-Kabsi AM: The prevalence and degree of resistance of Plasmodium falciparum to first-line antimalarial drugs: an in vitro study from a malaria endemic region in Yemen. Ann Saudi Med. 2007, 27: 432-436. 10.4103/0256-4947.51457.
Mubjer R, Adeel A, Chance M, Hassan A:Molecular markers of anti-malarial drug resistance in Lahj Governorate, Yemen: baseline data and implications. Malar J. 2011, 10: 245-10.1186/1475-2875-10-245.
WHO: World malaria report 2012. 2012, Geneva: World Health Organization
Menegon M, Severini C, Sannella A, Paglia MG, Sangaré D, Abdel-Wahab A, Abdel-Muhsin A-MA, Babiker H, Walliker D, Alano P: Genotyping of Plasmodium falciparum gametocytes by reverse transcriptase polymerase chain reaction. Mol Biochem Parasitol. 2000, 111: 153-161. 10.1016/S0166-6851(00)00314-5.
Kang J, Moon S, Kim J, Cho S, Lin K, Sohn W, Kim T, Na B: Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum field isolates from Myanmar. Malar J. 2010, 9: 131-10.1186/1475-2875-9-131.
Aubouy A, Migot-Nabias F, Deloron P: Polymorphism in two merozoite surface proteins of Plasmodium falciparum isolates from Gabon. Malar J. 2003, 2: 12-10.1186/1475-2875-2-12.
Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T: Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004, 305: 1124-1124. 10.1126/science.1098876.
Abdel-Muhsin AA, Mackinnon MJ, Awadalla P, Ali E, Suleiman S, Ahmed S, Walliker D, Babiker HA: Local differentiation in Plasmodium falciparum drug resistance genes in Sudan. Parasitology. 2003, 126: 391-400. 10.1017/S0031182003003020.
Djimdé A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourté Y, Dicko A, Su XZ, Nomura T, Fidock A, Wellems TE, Plowe CV: A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001, 344: 257-263. 10.1056/NEJM200101253440403.
Plowe CV, Cortese JF, Djimde A, Nwanyanwu OC, Watkins WM, Winstanley PA, Franco JGE, Mollinedo RE, Avila JC, Cespedes JL, Carter D, Doumbo OK: Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis. 1997, 176: 1590-1596. 10.1086/514159.
Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE: Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. AmJTrop Med Hyg. 1995, 52: 565-568.
Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LMB, bir Singh Sidhu A, Naudé B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE: Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000, 6: 861-871. 10.1016/S1097-2765(05)00077-8.
Cowman AF, Foote SJ: Chemotherapy and drug resistance in malaria. Int J Parasitol. 1990, 20: 503-513. 10.1016/0020-7519(90)90198-V.
Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ: Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988, 85: 9109-9113. 10.1073/pnas.85.23.9109.
Wang P, Read M, Sims PFG, Hyde JE: Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol Microbiol. 1997, 23: 979-986. 10.1046/j.1365-2958.1997.2821646.x.
Nei M: Molecular evolutionary genetics. Am J Physical Anthropology. 1988, 75: 428-429. 10.1002/ajpa.1330750317.
Hill WG, Babiker HA: Estimation of numbers of malaria clones in blood samples. Proc R Soc London Series B-Biol Sci. 1995, 262: 249-257. 10.1098/rspb.1995.0203.
Rousset F, Raymond M: Testing heterozygote excess and deficiency. Genetics. 1995, 140: 1413-1419.
Cockerham CC, Weir BS: Covariances of relatives stemming from a population undergoing mixed self and random mating. Biometrics. 1984, 40: 157-164. 10.2307/2530754.
Peakall R, Smouse PE: GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012, 28: 2537-2539. 10.1093/bioinformatics/bts460.
Anderson TJC, Haubold B, Williams JT, Estrada-Franco§ JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP: Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000, 17: 1467-1482. 10.1093/oxfordjournals.molbev.a026247.
Mobegi VA, Loua KM, Ahouidi AD, Satoguina J, Nwakanma DC, Amambua-Ngwa A, Conway DJ: Population genetic structure of Plasmodium falciparum across a region of diverse endemicity in West Africa. Malar J. 2012, 11: 223-10.1186/1475-2875-11-223.
Awadalla P, Walliker D, Babiker H, Mackinnon M: The question of Plasmodium falciparum population structure. Trends Parasitol. 2001, 17: 351-353. 10.1016/S1471-4922(01)02034-7.
Hayton K, Su XZ: Drug resistance and genetic mapping in Plasmodium falciparum<. Curr Genet. 2008, 54: 223-239. 10.1007/s00294-008-0214-x.
Koepfli C, Timinao L, Antao T, Barry AE, Siba P, Mueller I, Felger I: A large Plasmodium vivax reservoir and little population structure in the South Pacific. PLoS One. 2013, 8: e66041-10.1371/journal.pone.0066041.
Sultan DM, Khalil MM, Abdouh AS, Doleh WF, Al Muthanna AAM: Imported malaria in United Arab Emirates: Evaluation of a new DNA extraction technique using nested PCR. Korean J Parasitol. 2009, 47: 227-233. 10.3347/kjp.2009.47.3.227.
Al Harthi SA: Detection of drug resistance markers for chloroquine and pyrimethamine-sulfadoxine in Jazan area, Saudi Arabia using PCR and restriction digestion. J Egypt Soc Parasitol. 2007, 37: 17-30.
Nassir E, Abdel-Muhsin AMA, Suliaman S, Kenyon F, Kheir A, Geha H, Ferguson HM, Walliker D, Babiker HA: Impact of genetic complexity on longevity and gametocytogenesis of Plasmodium falciparum during the dry and transmission-free season of eastern Sudan. Int J Parasitol. 2005, 35: 49-55. 10.1016/j.ijpara.2004.10.014.
Nwakanma D, Kheir A, Sowa M, Dunyo S, Jawara M, Pinder M, Milligan P, Walliker D, Babiker HA: High gametocyte complexity and mosquito infectivity of Plasmodium falciparum in the Gambia. Int J Parasitol. 2008, 38: 219-227. 10.1016/j.ijpara.2007.07.003.
Al-Mekhlafi AMQ, Mahdy MAK, Azazy AA, Fong MY: Clinical situation of endemic malaria in Yemen. Trop Biomed. 2010, 27: 551-558.
Dlamini SV, Beshir K, Sutherland CJ: Markers of anti-malarial drug resistance in Plasmodium falciparum isolates from Swaziland: identification of Pfmdr1-86F in natural parasite isolates. Malar J. 2010, 9: 68-10.1186/1475-2875-9-68.
Al-Saai S, Kheir A, Abdel-Muhsin AMA, Al-Ghazali A, Nwakanma D, Swedberg G, Babiker HA: Distinct haplotypes of dhfr and dhps among Plasmodium falciparum isolates in an area of high level of sulfadoxine–pyrimethamine (SP) resistance in eastern Sudan. Infect Genet Evol. 2009, 9: 778-783. 10.1016/j.meegid.2009.04.010.
Rawasia WF, Sridaran S, Patel JC, Abdallah J, Ghanchi NK, Barnwell JW, Escalante AA, Udhayakumar V, Beg MA: Genetic backgrounds of the Plasmodium falciparum chloroquine resistant transporter (Pfcrt) alleles in Pakistan. Infect Genet Evol. 2012, 12: 278-281. 10.1016/j.meegid.2011.11.008.
Ghanchi NK, Ursing J, Beg MA, Veiga MI, Jafri S, Mårtensson A: Prevalence of resistance associated polymorphisms in Plasmodium falciparum field isolates from southern Pakistan. Malar J. 2011, 10: 18-10.1186/1475-2875-10-18.
Zakeri S, Afsharpad M, Kazemzadeh T, Mehdizadeh K, Shabani A, Djadid ND: Association of Pfcrt but not Pfmdr1 alleles with chloroquine resistance in Iranian isolates of Plasmodium falciparum. Am J TropMed Hyg. 2008, 78: 633-640.
Esmaeili Rastaghi AR, Nateghpour M, Assmar M, Razavi MR, Kanbara H, Uemura H, Naddaf SR, Keshavarz H, Raeisi A, Mohebali M: Detection of K76T mutation inPfcrt gene as an applicable genetic marker for prediction of chloroquine resistant falciparum malaria in isolates from an endemic district of Iran. Iran J Parasitol. 2008, 3: 48-56.
Zakeri S, Farahani MS, Afsharpad M, Salehi M, Raeisi A, Djadid ND: High prevalence of the 437G mutation associated with sulfadoxine resistance among Plasmodium falciparum clinical isolates from Iran, three years after the introduction of sulfadoxine–pyrimethamine. Int J Infect Dis. 2010, 14: e123-e128.
Jafari S, Le Bras J, Asmar M, Durand R: Molecular survey of Plasmodium falciparum resistance in south-eastern Iran. Ann Trop Med Parasitol. 2003, 97: 119-124. 10.1179/000349803235001552.
Awasthi G, Satya GBK, Das A: Pfcrt haplotypes and the evolutionary history of chloroquine-resistant Plasmodium falciparum. Mem Inst Oswaldo Cruz. 2012, 107: 129-134. 10.1590/S0074-02762012000100018.
Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ: Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002, 418: 320-323. 10.1038/nature00813.
Pati SS, Mishra S, Mohanty S, Mohapatra DN, Sahu PK, Priyadarshi N, Kumar S, Sharma SK, Tyagi PK, Chitnis CE, Das BS: Pfcrt haplotypes and in-vivo chloroquine response in Sundergarh district, Orissa, India. Trans R Soc Trop Med Hyg. 2007, 101: 650-654. 10.1016/j.trstmh.2007.01.008.
Reed MB, Sallba KJ, Caruana SR, Kirk K, Cowman AF: Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000, 403: 906-909. 10.1038/35002615.
Danquah I, Coulibaly B, Meissner P, Petruschke I, Müller O, Mockenhaupt FP: Selection of Pfmdr1 and Pfcrt alleles in amodiaquine treatment failure in north-western Burkina Faso. Acta Trop. 2010, 114: 63-66. 10.1016/j.actatropica.2009.12.008.
Sisowath C, Ferreira PE, Bustamante LY, Dahlström S, Mårtensson A, Björkman A, Krishna S, Gil JP: The role of Pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop Med Int Health. 2007, 12: 736-742. 10.1111/j.1365-3156.2007.01843.x.
Fröberg G, Jörnhagen L, Morris U, Shakely D, Msellem M, Gil J, Björkman A, Mårtensson A: Decreased prevalence of Plasmodium falciparum resistance markers to amodiaquine despite its wide scale use as ACT partner drug in Zanzibar. Malar J. 2012, 11: 321-10.1186/1475-2875-11-321.
Malmberg M, Ferreira PE, Tarning J, Ursing J, Ngasala B, Björkman A, Mårtensson A, Gil JP: Plasmodium falciparum drug resistance phenotype as assessed by patient antimalarial drug levels and its association with Pfmdr1 polymorphisms. J Infect Dis. 2013, 207: 842-847. 10.1093/infdis/jis747.
Bin Ghouth AS: Availability and prescription practice of anti-malaria drugs in the private health sector in Yemen. J Infect Dev Ctries. 2013, 7: 404-412.
Hyde JE: The dihydrofolate reductase-thymidylate synthetase gene in the drug resistance of malaria parasites. Pharmacol Ther. 1990, 48: 45-59. 10.1016/0163-7258(90)90017-V.
Lozovsky ER, Chookajorn T, Brown KM, Imwong M, Shaw PJ, Kamchonwongpaisan S, Neafsey DE, Weinreich DM, Hartl DL: Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc Natl Acad Sci USA. 2009, 106: 12025-12030. 10.1073/pnas.0905922106.
Costanzo MS, Hartl DL: The evolutionary landscape of antifolate resistance in Plasmodium falciparum. J Genetics. 2011, 90: 187-190. 10.1007/s12041-011-0072-z.
Pearce RJ, Drakeley C, Chandramohan D, Mosha F, Roper C: Molecular determination of point mutation haplotypes in the dihydrofolate reductase and dihydropteroate synthase of Plasmodium falciparum in three districts of Northern Tanzania. Antimicrob Agents Chemother. 2003, 47: 1347-1354. 10.1128/AAC.47.4.1347-1354.2003.
Afsharpad M, Zakeri S, Pirahmadi S, Djadid ND: Molecular monitoring of Plasmodium falciparum resistance to antimalarial drugs after adoption of sulfadoxine–pyrimethamine plus artesunate as the first line treatment in Iran. Acta Trop. 2012, 121: 13-18. 10.1016/j.actatropica.2011.09.010.
Khatoon L, Baliraine FN, Bonizzoni M, Malik SA, Yan G: Prevalence of antimalarial drug resistance mutations in Plasmodium vivax and P. falciparum from a malaria-endemic area of Pakistan. Am J TropMed Hyg. 2009, 81: 525-528.
Menegon M, Talha AA, Severini C, Elbushra SM, Mohamedani AA, Malik EM, Mohamed TA, Wernsdorfer WH, Majori G, Nour BYM: Frequency distribution of antimalarial drug resistance alleles among Plasmodium falciparum isolates from Gezira State, Central Sudan, and Gedarif State, Eastern Sudan. Am J TropMed Hyg. 2010, 83: 250-257.
Eshetu T, Berens-Riha N, Fekadu S, Tadesse Z, Gürkov R, Hölscher M, Löscher T, Miranda IB: Different mutation patterns of Plasmodium falciparum among patients in Jimma University Hospital, Ethiopia. Malar J. 2010, 9: 226-10.1186/1475-2875-9-226.
Jelinek T, Peyerl-Hoffmann G, Mühlberger N, Wichmann O, Wilhelm M, Schmider N, Grobusch MP, von Sonnenburg F, Gascon J, Laferl H, Hatz C, Alifrangis M, Burchard G, McWhinney P, Schulze M, Kollaritsch H, da Cunha S, Beran J, Kern P, Gjørup I, Cuadros J: Molecular surveillance of drug resistance through imported isolates of Plasmodium falciparum in Europe. Malar J. 2002, 1: 11-10.1186/1475-2875-1-11.
Acknowledgements
We acknowledge the support of technical staff of Biochemistry Department, College of Medicine and Health Sciences, Sultan Qaboos University, Oman, and staff of the Children Sweden Hospital Taiz, Yemen. This study was supported by Sultan Qaboos University, Oman, Project IG/MED/BIOC/09/03.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SA-H, carried out the laboratory work, acquired the data, performed analysis and contributed to preparation of the manuscript. ZA-H, HA-F, AB-P and HB contributed to study design, analysis and interpretation of data, supervised laboratory work and revised the manuscript for important intellectual content. AMA-M, MAKM, MI and HB contributed to concept development and study design, edited and revised the manuscript critically for content. All authors read and approved the final manuscript.
Electronic supplementary material
12936_2013_2871_MOESM1_ESM.docx
Additional file 1: Alleles of 3 microsatellites (MS) on chromosomes 8, msp-2 and pfg377 genes among 108 Plasmodium falciparum isolates in Yemen.(DOCX 101 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Al-Hamidhi, S., Mahdy, M.A., Al-Hashami, Z. et al. Genetic diversity of Plasmodium falciparum and distribution of drug resistance haplotypes in Yemen. Malar J 12, 244 (2013). https://doi.org/10.1186/1475-2875-12-244
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-12-244