Abstract
Perinatal piglet mortality is an important factor in pig production from economic and animal welfare perspectives; however, the statistical analysis of mortality is difficult because of its categorical nature. Recent studies have suggested that a binomial model for the survival of each specific piglet with a logit approach is appropriate and that recursive relationships between traits are useful for taking into account non-genetic relationships with other traits. In this study, the recursive binomial model is expanded in two directions: (1) the recursive phenotypic dependence among traits is allowed to vary among groups of individuals or crosses, and (2) the binomial distribution is replaced by the multiplicative binomial distribution to account for over or underdispersion. In this study, five recursive multiplicative binomial models were used to obtain estimates of the Dickerson crossbreeding parameters in a diallel cross among three varieties of Iberian pigs [Entrepelado (EE), Torbiscal (TT), and Retinto (RR)]. Records (10,255) from 2110 sows were distributed as follows: EE (433 records, 100 sows), ER (2336, 527), ET (942, 177), RE (806, 196), RR (870, 175), RT (2450, 488), TE (193, 36), TR (1993, 359), and TT (232, 68). Average litter size [Total Number Born (TNB)] and number of stillborns (SB) were 8.46 ± 2.27 and 0.25 ± 0.72, respectively. The overdispersion was evident with all models. The model with the best fit included a linear recursive relationship between TNB and the logit of \(\phi\) of the multiplicative binomial distribution, and it implies that piglet mortality increases with litter size. Estimates of direct effects showed small differences among populations. The analysis of maternal effects indicated that the dams whose mothers were EE had a larger SB, while dams with RR mothers reduced the probability of born dead. The posterior estimates of heterosis suggested a reduction in SB when the sow is crosbred. The multiplicative binomial distribution provides a useful alternative to the binomial distribution when there is overdispersion in the data. Recursive models can be used for modeling non-genetic relationships between traits, even if the phenotypic dependency between traits varies among environments or groups of individuals. Piglet perinatal mortality increased with TNB and is reduced by maternal heterosis.
Similar content being viewed by others
Introduction
Perinatal piglet mortality is a major issue in pig production from economic and animal welfare perspectives1. The number of stillborn (SB) is a categorical trait that includes a large proportion of zeros. Therefore, the statistical analysis of piglet mortality, as a maternal trait, is complicated because its distribution violates the Gaussian assumption of the standard mixed-model analysis. Varona and Sorensen2 calculated the degree of adjustment for several probability distributions (Poisson, Binomial, and Negative Binomial) in their standard version and with zero-inflated approaches. They concluded that a binomial distribution with logit link function to model the survival of each specific piglet within each litter was the most appropriate. However, one of the main limitations of models based on non-Gaussian distributions, such as the binomial, is the difficulty of accounting for non-genetic relationships between traits. Several studies have reported a strong relationship between litter size [Total Number Born (TNB)] and piglet mortality3,4,5,6, which, in some cases, is non-linear7,8,9. To address the relationship between litter size and piglet mortality, Varona and Sorensen8 suggested the use of recursive models10,11, because TNB is mainly determined by ovulation rate and embryo losses in early pregnacy12, while most stillborns take place around farrowing. Therefore, it is reasonable to postulate a recursive relationship between TNB and SB, under the assumption that mortality around farrowing is phenotypically determined by litter size.
The model proposed by Varona and Sorensen8 assumes a general recursive relationship for all data; however, a variety of genetic and non-genetic factors might influence the phenotypic dependence between traits, and it would be interesting to define a model that allows the phenotypic dependence among traits to vary among specific factors. Furthermore, the response variable (i.e., SB) may not strictly follow a binomial distribution, as it may show some degree of over or underdispersion. The binomial distribution can be generalized to the multiplicative binomial (MBN) distribution13,14, which can adequately consider over and under-dispersion with an additional parameter (\(\theta\)).
Therefore, the objective of this study was to develop a recursive model that allows specific recursive functions for a group of individuals or data, and that replaces the binomial distribution with the multiplicative binomial distribution. Specifically, the model was applied to TNB and SB within a full diallel cross15 among three varieties of Iberian pig [Entrepelado (EE), Retinto (RR), and Torbiscal (TT)]. The model of analysis had allowed up to nine specific recursive relationships, one for each diallel cross. In addition, the estimates of the recursive parameters were reparametrized to quantify the direct, maternal, and heterosis effects of the Dickerson model16 for TNB and SB.
Methods
Animals and experimental design
The dataset comprised 10,255 records for TNB and SB from 2110 sows and is a survey derived from the purebred and crossbred populations generated in a full diallel cross carried out by the INGA FOOD S. A. and which included three varieties of Iberian pig [Retinto (RR), Torbiscal (TT), and Entrepelado (EE)] and their reciprocal crosses [Retinto × Torbiscal (RT), Torbiscal × Retinto (TR), Retinto × Entrepelado (RE), Entrepelado × Retinto (ER), Torbiscal × Entrepelado (TE), and Entrepelado × Torbiscal (ET)]. The three varieties are recognized in Spain’s official Iberian herd-book [Asociación Española de Criadores de Ganado Porcino Selecto Ibérico Puro y Tronco Ibérico (AECERIBER)]. A detailed description of their characteristics is provided by Ibáñez-Escriche et al.17. SB was recorded by counting piglets born dead in assisted deliveries and with clear indications of stillborn (remains of the placenta, condition of their hooves, general appearance of the piglet) in unattended births. For a detailed description of the experimental protocol of the diallel cross, see Noguera et al.18. The sows were in a commercial farm in which purebred and crossbred Iberian sows were mated with boars from a Duroc population following the standard production system of Iberian pigs under intensive management. The distribution of the data among crosses, and the average and standard deviation of TNB and SB for all crosses are presented in Table 1. Besides, the pedigree was extended back up to three generations and included 4609 individuals. The number of founders per population was 47 EE (13 sires and 34 dams), 80 RR (18 sires and 62 dams), and 107 TT (38 sires and 69 dams).
Statistical models
The multiplicative binomial (MBN) distribution is a generalization of the binomial distribution with two parameters (\(\phi \; {\text{and}} \;\theta )\) with the following probability density13,14:
where x is the number of positive or negative Bernoulli events (i.e. number of stillborn), nb is the number of Bernoulli events, and \(\phi \;{\text{and}} \;\theta\) are the parameters of the MBN distribution. The first parameter (\(\phi )\) ranged between 0 and 1 as in the binomial distribution, whereas the second (\(\theta\)) is greater than 0, and it is associated with the degree of over or underdispersion. The MBN distribution is equivalent to the binomial distribution with \(\theta\) = 1, and it presents overdispersion when θ < 1 and under-dispersion if θ > 1. The probability of success of each Bernoulli event with the MBN distribution depends on \(\phi \;{\text{and}} \;\theta\) and can be calculated as19:
with.
where a is 0 for Knb and 1 for Knb−1, and j is the loop variable in the summation. The effect of different values of the θ parameter in the MBN with probabilities of success of each Bernoulli event of 0.25 and 0.5 and with nb = 10 is presented in Fig. 1.
In this study, SB (\({\varvec{y}} = \left\{ {y_{i} } \right\}\)) and TNB (\({\varvec{t}} = \left\{ {t_{i} } \right\}\))) were measured at each of the 10,255 litters and they were analyzed under the assumption that the conditional distribution of y is the following product of multiplicative binomial distributions:
where \({\varvec{\phi}} = \left\{ {\phi_{i} } \right\}\) is the vector of the 10,255 parameters of the MBN distribution associated with each litter (i), N is the number of data, ti, and yiare the total number of piglets and stillborn in the ith litter, respectively. The \(\theta\) parameter was assumed to be the same for all litters and represented the general pattern of over or underdispersion.
Under a hierarchical Bayesian framework, a second hierarchy of the model postulated the following linear model for t:
where \({\varvec{b}}_{{\varvec{t}}}\)is the vector of the following systematic effects: type of cross (nine levels: EE, ER, ET, RE, RR, RT, TE, TR, and TT), order of parity (six levels: first, second, third, fourth, fifth, and sixth and beyond), and year-season (YS) (30 levels). In addition, ut and pt are the vectors of additive genetic (4609 levels) and dam permanent environmental effects (2110 levels), respectively, and et is the vector of gaussian residuals.
In addition, the logit of \(\phi\) was modeled linearly in five ways. Model I included analogous systematic (bl) and random effects (ul and pl) that were used in the model for litter size, as follows:
model II incorporated a recursive parameter (\(\lambda_{1} )\) with the vector of deviations (d) of TNB from the average litter size (\(\overline{t}\)) across all crosses and order of parities:
model III included an additional recursive parameter (\(\lambda_{2} )\):
model IV and model V included cross-specific linear (\(\lambda_{1k} )\) and quadratic (\(\lambda_{2k} )\) recursive parameters, where k has nine levels, which correspond to crosses EE, ER, ET, RE, RR, RT, TE, TR, and TT. Thus, the models were as follows:
the vector wk included the deviations of the litter size from the average litter size (\(\overline{t})\) if the individual belongs to the kth cross, and 0 otherwise.
The prior distributions for additive genetic and permanent environmental effects were assumed as follows:
where A is the numerator relationship matrix, and
where \(\sigma_{al}^{2}\) and \(\sigma_{at}^{2}\) are the additive genetic variances for the logit of \({\varvec{\phi}}\) and TNB, and \(\sigma_{alt}\) is the covariance between them. Furthermore, \(\sigma_{pl}^{2}\) and \(\sigma_{pt}^{2}\) are the permanent environmental variances for the logit of \({\varvec{\phi}}\) and TNB, and \(\sigma_{plt}\) is the covariance between them. The prior distribution for systematic effects (bt and bl), G, P, and the residual variance of litter size (\(\sigma_{et}^{2} )\) were assumed uniform within appropriate bounds.
All models were implemented with a Gibbs sampler20 with a Metropolis-Hasting step21 to sample for the logit and using own software developed in FORTRAN90. The analysis was performed with a single long chain of 550,000 iterations after the first 50,000 were discarded. Convergence was confirmed by visual inspection of the chains and by the CODA package in R software22. The Gibbs sampler provides the marginal posterior distributions of all unknowns in the models of analysis that include systematic, additive genetic and permanent environmental effects, as well as variance components and parameters of the MBN distribution.
Calculation of the Dickerson model parameters
The posterior distributions of the parameters of the Dickerson model were obtained by transforming the output of the cross effects in each iteration of the Gibbs Sampler using a property of the Bayesian analysis that allows to calculate the posterior distribution of any linear transformation of the parameters of the model. Therefore, the direct-line (Dx(E), Dx(R), Dx(T)), maternal (Mx(E), Mx(R), Mx(T)) for Entrepelado (EE), Retinto (RR) and Torbiscal (TT), and the heterosis for Entrepelado and Retinto (Hx(ER)), Entrepelado and Torbiscal (Dx(ET)) and Retinto and Torbiscal (Dx(RT)) were calculated for TNB (x = t) and the logit of the \(\phi\) parameter of the multiplicative binomial (x = l). The samples for the Dickerson model parameters at each iteration of the Gibbs sampler were obtained by solving the following system of equations:
where bx(EE), bx(ER), bx(ET), bx(RE), bx(RR), bx(RT), bx(TE), bx(TR) and bx(TT) were the samples for the nine levels of the systematic effects for litter size (x = t) or the logit of the \(\phi\) parameter of the multiplicative binomial (x = l).
Model comparison
The models were compared by the logarithm of the conditional predictive ordinate (LogCPO)23. If we consider the vector \({\varvec{y}} = \left( {y_{i} ,{\varvec{y}}_{ - i} } \right)\), where yi is the ith datum, and \({\varvec{y}}_{ - i}\) is the vector of the data with the ith datum deleted, the conditional predictive distribution has the following probability density:
where \(\theta\) is the vector of the unknown parameters in the model. Therefore, \(p\left( {y_{i} {|}{\varvec{y}}_{ - i} } \right)\) is the probability of each datum given the rest of the data, which is the conditional predictive ordinate (CPO) for the ith datum. The pseudo log-marginal probability of the data is \(\mathop \sum \limits_{i} lnp\left( {y_{i} {|}{\varvec{y}}_{ - i} } \right)\).
A Monte Carlo approximation of the CPO suggested by Gelfand22 is \(\mathop \sum \limits_{i} ln\hat{p}\left( {y_{i} {|}{\varvec{y}}_{ - i} } \right),\) where \(\hat{p}\left( {y_{i} {|}{\varvec{y}}_{ - i} } \right) = N\left[ {\mathop \sum \limits_{j = 1}^{Nm} \frac{1}{{p\left( {y_{i} {|}\theta^{j} } \right)}}} \right]^{ - 1}\) , and Nm is the number of Markov Chain Monte Carlo (MCMC) draws, and \(\theta^{j}\) is the jth draw from the posterior distribution of the corresponding parameter. The higher the value of the LogCPO, the better the fit of the model to the data.
Ethics approval and consent to participate
The research ethics committee of the Institute of Agrifood Research and Technology (IRTA) approved all of the management and experimental procedures involving live animals, which were performed in accordance with the Spanish Policy of Animal Protection RD1201/05, which complies with the European Union Directive 86/609 for the protection of animals used in experimentation.
Results and discussion
The estimates of the variance components and the differences between the LogCPOs with respect to the best model are presented in Table 2. Comparison of the models indicated that the model with the best fit was model II, which proposes a linear phenotypic dependence between TNB and the logit of Φ. Under this model, the slope of the recursive parameter has a posterior mean of 0.257 logit units per piglet with a posterior standard deviation of 0.007. This positive slope implies that the probability of stillborn increases with litter size, confirming the results of previous studies7,8,24. It can be caused by a prolonged farrowing, which increases the risk of hypoxia25 or influenced by litter weight or average piglet weight24,26,27. The differences in LogCPO among the models that included a recursive relationship between TNB and the logit of the \(\it \Phi\) parameter of the MBN distribution (models II, III, IV, and V) were small (17.93 LogCPO units). However, there is no evidence of non-linearity (models III and V) or variability of this phenotypic dependence among crosses (models IV and V). The posterior mean estimates of the recursive relationship in more complex models, such as model V (see Supplementary Fig. 1a), may suggest a cross specific non-linear relationship between litter size and the logit of Φ. Nevertheless, the differences with the posterior estimate of the general recursive relationship defined in model II (see Supplementary Fig. 1b) were small. Therefore, the logCPO method for model comparison weights the goodness of fit and complexity of the proposed models and it applied Occam’s razor criteria for model comparison and selected a less parameterized model as the best. Furthermore, the ability of the recursive models to capture the non-genetic relationship between traits is illustrated by the large difference in LogCPO (302.92 units) between model II and model I, which considers only additive genetic and permanent environmental covariance between traits.
The posterior mean estimates of the θ parameter of the MBN distribution ranged between 0.816 (model II) to 0.892 (model I), and, in all models, the posterior distributions were different from 1. Therefore, we found strong evidence of overdispersion that reinforces the adequacy of the MBN distribution rather than the binomial distribution suggested by Varona and Sorensen8. The binomial distribution postulates the independence of the series of Bernoulli events that compose the response variable (SB), and this assumption is far from reality in the perinatal mortality of pigs. In this case, the Bernoulli events (survivors or not) are not independent due to a long list of shared biological causes such as, among others, infectious diseases, gestation and farrowing length or stress24,26. This phenomenon causes overdispersion with respect to the standard binomial distribution and matches with the postulates argued in the development of the MBN distribution13,14, where the values of θ < 1 indicate a positive dependence of the events and lead to overdispersion, while values of θ > 1 reflect a negative dependence among them and generate under-dispersion with respect the standard binomial distribution.
The results of the variance component estimates were similar for all models (Table 2). For TNB, and with the model with the best fit (model II), the estimates of the posterior mean ± posterior standard deviation of the additive genetic, permanent environmental and residual variance were 0.397 ± 0.090, 0.368 ± 0.080 and 4.497 ± 0.071, respectively. The relative importance of the additive genetic variation was similar to the estimates available from white6,28,29 and Iberian18,30 pig populations. The estimates of the posterior mean (± posterior standard deviation) for the additive genetic variation of the logit of the \(\phi\) parameter was 0.050 ± 0.044, which was substantially less than the posterior mean of the permanent environmental variance (0.418 ± 0.056). The low maternal additive genetic variation in pig mortality was similar to the results of previous studies in white pigs5,31,32 and the Iberian population33. However, estimates of the additive variances should be taken with caution as the sows come from three different populations. In addition, the posterior estimates of the permanent environmental variances were higher and consistent with the results of Vanderhaeghe et al.34, which reported an increase of the risk for stillborn if the sow has had more than one stillborn piglet in a previous farrowing.
The genetic and the permanent environmental correlations between the logit of \(\phi\) and TNB were different among the models (Table 2). In model I, the correlations were positive (0.507 and 0.524, respectively) and similar to other estimates35,36. The posterior estimates in all of the models that included some type of recursive relationship between TNB and piglet mortality (models II to V) were closer to zero. They ranged from 0.203 to 0.238 for the genetic correlation and from 0.022 to 0.031 for the permanent environmental correlation. However, the Highest Posterior Density at 95% (HPD95) of those estimates always included zero. Those differences among the models were expected because, in model I, the correlation between TNB and logit of \(\phi\) can be interpreted directly but, in models II to V, it must be understood as the correlation between TNB and the logit of \(\phi\) after the correction for TNB. Furthermore, model I assumed independence between the errors of the two response variables (TNB and SB) and, therefore, the additive and permanent environmental correlations may be higher to compensate for the possible positive environmental correlation between them. In contrast, the remaining models modeled a linear or non-linear dependence between traits and included some degree of residual relationship.
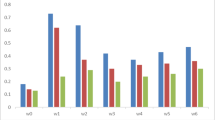
The posterior estimates of systematic cross effects under the model that had the best fit (model II) are presented in Fig. 2 and 3. Figure 2a shows the posterior estimates for TNB. The RE and TE crosses were the best and had estimates of the posterior mean ± posterior standard deviation of 0.526 ± 0.128 and 0.401 ± 0.214 piglets above the average, respectively (Fig. 2a). TT was the worst cross and had an estimate of the posterior mean (± posterior standard deviation) of − 0.982 ± 0.209 below the average line effect. Those estimates of cross effects can be re-parameterized in terms of the Dickerson model in the direct, maternal, and heterosis effects (Fig. 2b). The posterior mean estimates of the direct effects ranged from − 0.391 (Torbiscal) to 0.492 (Retinto); the maternal effects ranged from − 0.193 (Torbiscal) to 0.282 (Entrepelado). The posterior mean estimates (± posterior standard deviation) of the effects of heterosis were 0.532 ± 0.109, 0.681 ± 0.169, and 0.584 ± 0.118 between Entrepelado and Retinto, Entrepelado and Torbiscal, and Retinto and Torbiscal, respectively. As expected, the results were similar to those of Noguera et al.18, who applied a mixed linear model to the same populations.
Posterior mean (and Highest Posterior Density at 95%) estimates of the deviation of the cross effects (EE—Entrepelado, ER—Entrepelado-Retinto-, ET—Entrepelado-Torbiscal-, RE—Retinto-Entrepelado-, RR—Retinto, RT—Retinto-Torbiscal-, TE—Torbiscal-Entrepelado-, TR—Torbiscal-Retinto and TT—Torbiscal) with respect to their average (a) and of the direct line (LE, LR, and LT), maternal (ME, MR and MT), and heterosis (HER, HET, HRT) effects of the Dickerson model (b) for total number born (TNB) under model II.
Posterior mean (and highest posterior density at 95%) estimates of the deviation of the cross effects (EE—Entrepelado, ER—Entrepelado-Retinto-, ET—Entrepelado-Torbiscal-, RE—Retinto-Entrepelado-, RR—Retinto, RT—Retinto-Torbiscal-, TE—Torbiscal-Entrepelado-, TR—Torbiscal-Retinto and TT—Torbiscal) with respect to their average (a) and of the direct line (LE, LR, and LT), maternal (ME, MR and MT), and heterosis (HER, HET, HRT) effects of the Dickerson model (b) for the logit of the Φ parameter of the multiplicative binomial distribution under model II.
Figure 3a shows the posterior mean and standard deviation estimates of the line effects for the logit of Φ. It should be noted that altought model II proposes the same slope for all crosses, the procedure was able to detect differences among crosses at the intercept, which was set at the average TNB. The cross with the lowest posterior mean ± posterior standard deviation was the TR with − 0.196 ± 0.080, followed by ET and ER with − 0.143 ± 0.095 and − 0.135 ± 0.709, respectively. The EE and TE crosses had the highest posterior mean ± posterior standard deviation with 0.237 ± 0.122 and 0.136 ± 0.166. Those estimates were re-parameterized in terms of the Dickerson model in the direct, maternal, and heterosis effects (Fig. 3b). Estimates of direct effects ranged between − 0.051 ± 0.159 (Torbiscal) to 0.069 ± 0.112. The results of the maternal effects suggest that the dams that had an Entrepelado mother had more stillborn offspring (0.150 ± 0.077). In contrast, the dams that had a Retinto mother showed an effect in the opposite direction (− 0.120 ± 0.047).
The posterior estimates of the heterosis effects for piglet mortality were always negative and indicates a positive effect on heterosis (a reduction of the probability of stillborn), as expected from previous studies37. They ranged between − 0.196 ± 0.091 (Entrepelado and Retinto) to − 0.130 ± 0.147 (Entrepelado and Torbiscal), and the posterior probabilities of a negative heterosis effect were 98.34, 81.05 and 90.14% for Entrepelado and Retinto, Entrepelado and Torbiscal and Retinto and Torbiscal, respectively.
The heterosis in piglet survival observed in our study reinforces the suggestion of Noguera et al.18, who recommended using a pyramidal scheme in the production of crossbred Iberian sows. Previous studies30,38 have reported the presence of heterosis for litter size in the Iberian pig populations and, our study has shown that crossbred Iberian sows also may provide a lower chance of stillborns. Prolificacy is lower in the Iberian pig populations than it is in the white pig populations39. However, efforts to improve the reproductive efficiency of the former are possible given the estimates of genetic variation in the population18,29. The success of selection for litter size, however, is limited by maternal ability to rear the extra piglets40. Therefore, the exploitation of heterosis for piglet survival can play an important role in ensuring an acceptable survival rate in sows that produce large litters in Iberian pig production.
Finally, it is worth noting that recursive methods are useful means of overcoming one of the main limitations of the use of non-Gaussian distributions, i.e., the difficulty in modeling non-genetic relationships between traits8. However, the phenotypic relationship between traits might differ among environments or groups of individuals. In this study, we extended the approach of Varona and Sorensen8 with two important novelties: (a) by including several recursive functions that, in this study, were assigned to nine diallel crosses, although the recursive relationships might be defined as nested within each systematic effect in the model, and (b) by replacing the binomial distribution by a multiplicative binomial distribution that is able to consider for the overdispersion. Nevertheless, the results of the statistical analysis did not show that the variability in the recursive relationships was relevant, although we found strong evidence of overdispersion.
The proposed approach has been developed under a hierarchical Bayesian scheme that included: (1) the multiplicative binomial probability of SB given TNB and the parameters of the MBN distribution, (2) The conditional distributions of the logit of the \(\phi\) parameters of the MBN distribution and TNB given the additive genetic and systematic effects, the recursive parameters and the residual variance for TNB, (3) The prior distribution of the additive genetic and systematic effects given the additive and permanent environmental variance components, and (4) the prior distributions of the variance components and of the \(\theta\) parameter of the MBN distribution. Nevertheless, alternative approaches could be developed by the implementation of a likelihood-based inference using a maximization algorithm such as Expectation–Maximization41.
Finally, it is worth to mention that the model used in this study still had several assumptions that should be considered. First, it was assumed that the systematic effects associated with the diallel crosses (EE, ER, ET, RE, RR, RT, TE, TR, and TT) were independent a-priori, although they were related as the sows presents at each cross were genetically linked. Therefore, the additive genetic effects and the cross effect may be somewhat confused. However, we decided to include the additive genetic effects in the models in order to correct for the covariances between individuals within and between crosses. Theoretically, inference for cross effect or Dickerson model parameters must be made from unrelated observations between crosses, that it was not feasible with the available information from the survey. Nevertheless, the genetic relationships among the sows between crosses should have a minimal influence on the results as the additive variance for TNB and SB was very low. Secondly, only linear and quadratic relationships between TNB and the logit of Φ were allowed, and, in the literature, some interesting alternative proposals used change-point techniques42 to model non-linear recursive relationships7,43 that may be used in future studies.
Conclusions
The multiplicative binomial distribution provides a useful alternative to the binomial distribution when there is overdispersion in the data. Recursive models can be applied for modeling non-genetic relationships between traits across environments or groups of individuals. In the Iberian pig populations, the perinatal piglet mortality measured as the SB increased with TNB and the effects of maternal heterosis on piglet survival are also revealed.
Data availability
The software and dataset used during the current study are available from the corresponding author on reasonable request.
Abbreviations
- EE:
-
Entrepelado
- RR:
-
Retinto
- TT:
-
Torbiscal
- EE:
-
Entrepelado × Entrepelado
- ER:
-
Entrepelado × Retinto
- ET:
-
Entrepelado × Torbiscal
- RE:
-
Retinto × Entrepelado
- RR:
-
Retinto × Retinto
- RT:
-
Retinto × Torbiscal
- TE:
-
Torbiscal × Entrepelado
- TR:
-
Torbiscal × Retinto
- TT:
-
Torbiscal × Torbiscal
- TNB:
-
Total number born
- SB:
-
Stillborn
- CPO:
-
Conditional predictive ordinate
- HPD95:
-
Highest posterior density at 95%
- MBN:
-
Multiplicative binomial distribution
- HYS:
-
Herd-year-season
References
Edwards, S. A. & Baxter, E. M. Piglet mortality: causes and prevention. In The Gestating and Lactanting Sow (ed. Farmer, C.) 253–328 (Wageningen Academic Publishers, Wageningen, 2015).
Varona, L. & Sorensen, D. A genetic analysis of mortality in pigs. Genetics 184, 277–284 (2010).
Roehe, R. & Kalm, E. Estimation of genetic and environmental risk factors associated with pre-weaning mortality in piglets using generalized linear mixed models. Anim. Sci. 70, 227–240 (2000).
Lund, M. S., Puonti, M., Rydhmer, L. & Jensen, J. Relationship between litter size and perinatal and pre-weaning survival in pigs. Anim. Sci. 74, 217–222 (2002).
Ibáñez-Escriche, N., Varona, L., Casellas, J., Quintanilla, R. & Noguera, J. L. Bayesian threshold analysis of direct and maternal genetic parameters for piglet mortality at farrowing in Large White, Landrace, and Pietrain populations. J. Anim. Sci. 87, 80–87 (2009).
Bidanel, J. P. Biology and genetics of reproduction. In The Genetics of the Pig (eds Rotshchild, M. A. F. & Ruvinsky, A.) 218–241 (CABI, Wallingford, 2011).
Ibáñez-Escriche, N., de Maturana, E. L., Noguera, J. L. & Varona, L. An application of change-point recursive models to the relationship between litter size and number of stillborns in pigs. J. Anim. Sci. 88, 3493–3503 (2010).
Varona, L. & Sorensen, D. Joint analysis of binomial and continuous traits with a recursive model: a case study using mortality and litter size of pigs. Genetics 196, 643–651 (2014).
Mulder, H. A., Hill, W. G. & Knol, E. F. Heritable environmental variance causes nonlinear relationships between traits: application to birth weight and stillbirth of pigs. Genetics 199, 1255–1269 (2015).
Gianola, D. & Sorensen, D. Quantitative genetic models for describing simultaneous and recursive relationships between phenotypes. Genetics 167, 1407–1424 (2004).
Varona, L., Sorensen, D. & Thompson, R. Analysis of litter size and average litter weight in pigs using a recursive model. Genetics 177, 1791–1799 (2007).
Blasco, A., Bidanel, J. P., Bolet, G., Haley, C. S. & Santacreu, M. A. The genetics of prenatal survival of pigs and rabbits: a review. Livest. Prod. Sci. 37, 1–21 (1993).
Altham, P. Two generalizations of the binomial distribution. Appl. Statist. 27, 162–167 (1978).
Lovison, G. An alternative representation of Altham’s multiplicative-binomial distribution. Stat. Probabil. Lett. 36, 415–420 (1998).
Hayman, B. I. The theory and analysis of diallel crosses. Genetics 39, 789–809 (1954).
Dickerson, G. E. Experimental approaches in utilizing breed resources. Anim. Breed. Abstr. 37, 191–202 (1969).
Ibáñez-Escriche, N., Magallón, E., Gonzalez, E., Tejeda, J. F. & Noguera, J. L. Genetic parameters and crossbreeding effects of fat deposition and fatty acid profiles in Iberian pig lines. J. Anim. Sci. 94, 28–37 (2016).
Noguera, J. L., Ibáñez-Escriche, N., Casellas, J., Rosas, J. P. & Varona, L. Genetic parameters and direct, maternal and heterosis effects on litter size in a diallel cross among three commercial varieties of Iberian pig. Animal 13, 2765–2772 (2019).
Lawal, B. H. Employing the double, multiplicative and the com-poisson binomial distributions for modeling over and under-dispersed binary data. J. Adv. Math. Comput. Sci. 23, 1–17 (2017).
Gelfand, A. E. & Smith, A. F. M. Sampling-based approaches to calculating marginal densities. J. Am. Stat. Assoc. 85, 398–409 (1990).
Hastings, W. K. Monte Carlo sampling methods using markov chains and their applications. Biometrika 57, 97–109 (1970).
Plummer, M., Best, N., Cowles, K. & Vines, K. CODA: convergence diagnosis and output analysis for MCMC. R News. 6, 7–11 (2006).
Gelfand, A. E. Model determination using sampling-based methods. In Markov chain Monte Carlo in Practice (eds Gilks, W. R. et al.) 145–161 (Chapman & Hall, London, 1996).
Rangstrup-Christensen, L., Krogh, M. A., Pedersen, L. J. & Sørensen, J. T. Sow-level risk factors for stillbirth of piglets in organic sow herds. Animal 11, 1078–1083 (2017).
Herpin, P. et al. Effects of the level of asphyxia during delivery on viability at birth and early postnatal vitality of newborn pigs. J. Anim. Sci. 74, 2067–2075 (1996).
Le Cozler, Y., Guyomarc’h, C., Pichodo, X., Quinio, P. Y. & Pellois, H. Factors associated with stillborn and mummified piglets in high-prolific sows. Anim. Res. 51, 261–268 (2002).
Vanderhaeghe, C., Dewulf, J., de Kruif, A. & Maes, D. Non-infectious factors associated with stillbirth in pigs: a review. Anim. Reprod. Sci. 139, 76–88 (2013).
Noguera, J. L., Varona, L., Babot, D. & Estany, J. Multivariate analysis of litter size for multiple parities with production traits in pigs: I. Bayesian variance component estimation. J. Anim. Sci. 80, 2540–2547 (2002).
Ogawa, S. et al. Estimation of genetic parameters for farrowing traits in purebred Landrace and Large White pigs. Anim. Sci. J. 90, 23–28 (2019).
Fernández, A., Rodrigáñez, J., Zuzúarregui, J., Rodríguez, M. C. & Silió, L. Genetic parameters for litter size and weight at different parities in Iberian pigs. Span. J. Agric. Res. 6, 98–106 (2008).
Su, G., Sorensen, D. & Lund, M. S. Variance and covariance components for liability of piglet survival during different periods. Animal 2, 184–189 (2008).
Wolf, J., Žáková, E. & Groeneveld, E. Within-litter variation of birth weight in hyperprolific Czech Large White sows and its relation to litter size traits, stillborn piglets and losses until weaning. Livest. Sci. 115, 195–205 (2008).
Muñoz, M. et al. Direct and maternal additive effects are not the main determinants of Iberian piglet perinatal mortality. J. Anim. Breed. Genet. 134, 512–519 (2017).
Vanderhaeghe, C. et al. Longitudinal field study to assess sow level risk factors associated with stillborn piglets. Anim. Reprod. Sci. 120, 78–83 (2010).
Hellbrügge, B. et al. Genetic aspects regarding piglet losses and the maternal behaviour of sows. Part 2. Genetic relationship between maternal behaviour in sows and piglet mortality. Animal 2, 1281–1288 (2008).
Zhang, T. et al. Heritabilities and genetic and phenotypic correlations of litter uniformity and litter size in Large White sows. J. Integr. Agric. 15, 848–854 (2016).
Johnson, R. K. & Omtvedt, I. T. Maternal heterosis in swine: reproductive performance and dam productivity. J. Anim. Sci. 40, 29–37 (1975).
García-Casco, J. M., Fernández, A., Rodríguez, M. C. & Silió, L. Heterosis for litter size and growth in crosses of four strains of Iberian pig. Livest. Sci. 147, 1–8 (2012).
Silio, L., Rodriguez, M. C., Rodrigañez, J. & Toro, M. A. La selección de cerdos ibéricos in Porcino Ibérico. Aspectos Claves (ed. Buxade, C. & Daza, A.) 125–149 (Mundi-Press, 2001).
Knol, E. F., Leenhouwers, J. I. & Van der Lende, T. Genetic aspects of piglet survival. Livest. Prod. Sci. 78, 47–55 (2002).
Dempster, A. P., Laird, N. M. & Rubin, D. B. Maximum Likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser.. B Stat. Methodol. 39, 1–38 (1977).
Carlin, B. P., Gelfand, A. E. & Smith, A. F. M. Hierarchical Bayesian analysis of change points problems. Appl. Statist. 41, 389–405 (1992).
López de Maturana, E., Wu, X. L., Gianola, D., Weigel, K. A. & Rosa, G. J. M. Exploring biological relationships between calving traits in primiparous cattle with a Bayesian recursive model. Genetics 181, 277–287 (2009).
Acknowledgements
The authors gratefully acknowledge INGA FOOD S. A. and its veterinarians Emilio Magallón, Lourdes Muñoz, Pilar Diaz, and Sara Negro for their cooperation with the experimental protocol and their technical support. The authors also wish to acknowledge the helpful comments of the reviewers with a special emphasis on the recommendation to considering over-dispersion.
Funding
The work was partially funded by the Center for Industrial Technological Development (CDTI) via grant IDI-20170304 and by grant CGL-2016–80155 from the Ministry of Economy, Industry and Competitiveness (MINECO), Spain.
Author information
Authors and Affiliations
Contributions
L.V. proposed the model of analysis, developed the FORTRAN90 code, analyzed the data and wrote the first draft. J.L.N. and N.I.E. designed and generated the experimental population. J.P.R., N.I.E., and J.L.N. supervised the generation of data. J.C. and M.M.H. collaborated in writing the final manuscript. All of the authors discussed the results, made suggestions and corrections. All of the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Varona, L., Noguera, J.L., Casellas, J. et al. A cross-specific multiplicative binomial recursive model for the analysis of perinatal mortality in a diallel cross among three varieties of Iberian pig. Sci Rep 10, 21190 (2020). https://doi.org/10.1038/s41598-020-78346-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-78346-7
- Springer Nature Limited