Abstract
This study developed a radiomics-based predictive model for radiation-induced pneumonitis (RP) after lung cancer stereotactic body radiation therapy (SBRT) on pretreatment planning computed tomography (CT) images. For the RP prediction models, 275 non-small-cell lung cancer patients consisted of 245 training (22 with grade ≥ 2 RP) and 30 test cases (8 with grade ≥ 2 RP) were selected. A total of 486 radiomic features were calculated to quantify the RP texture patterns reflecting radiation-induced tissue reaction within lung volumes irradiated with more than x Gy, which were defined as LVx. Ten subsets consisting of all 22 RP cases and 22 or 23 randomly selected non-RP cases were created from the imbalanced dataset of 245 training patients. For each subset, signatures were constructed, and predictive models were built using the least absolute shrinkage and selection operator logistic regression. An ensemble averaging model was built by averaging the RP probabilities of the 10 models. The best model areas under the receiver operating characteristic curves (AUCs) calculated on the training and test cohort for LV5 were 0.871 and 0.756, respectively. The radiomic features calculated on pretreatment planning CT images could be predictive imaging biomarkers for RP after lung cancer SBRT.
Similar content being viewed by others
Introduction
Stereotactic body radiotherapy (SBRT) is commonly administered for early stage non-small-cell lung cancer (NSCLC) to reduce treatment volumes and facilitate hypofractionation with delivery of large daily tumor doses1,2. The reported survival rate of SBRT is comparable to that of surgery2. However, radiation-induced pneumonitis (RP) is the most frequent acute pulmonary toxicity following SBRT for lung cancer. Although most patients develop asymptomatic grade 1 pneumonitis, clinically symptomatic pneumonitis is often observed3,4,5. Thus, previous studies have used clinical and dosimetric data to attempt to predict RP risk after radiation therapy (RT) for lung cancer6,7,8,9,10,11. Biological markers such as serum Krebs von den Lungen-6 (KL-6) and surfactant proteins-D (SP-D) levels are reportedly useful for the prediction of RP after SBRT treatment6,7. Other studies have concluded that RP incidence and grade are significantly related to various Vx (percentage lung volume receiving > x Gy) such as V20 and mean lung dose (MLD)8,9,10,11. Thus, the present study considered the dosimetric effects on RP development based on regions of interest (ROIs) segmented by dosimetric information.
Recent studies have reported improved RP prediction by applying machine learning. Cunliffe et al.12 investigated the relationship between radiation dose and changes in lung radiomic features on pre- and post-treatment computed tomography (CT) images in patients who received curative radiation doses for esophageal cancer, using multiple features in a classifier. Moran et al. reported that changes in radiomic features on pre- and post-SBRT CT images were significantly correlated with radiation oncologist-scored post-SBRT lung injury13. Cui et al.14 proposed a combination of handcrafted features with latent variables selected from 230 variables, including clinical factors and biomarkers in machine learning. However, to our knowledge, no studies have predicted the RP risk after lung cancer SBRT from the radiomic features obtained only from pretreatment planning CT images. Achieving the RP prediction with only pretreatment planning CT images prior to radiation delivery may be useful for selecting treatment options and creating treatment plans.
Previous studies have demonstrated the ability of radiomics to provide a quantitative evaluation of lung tissue reaction to radiation dose on pre- and post-RT CT images and assess the occurrence of RP12,13. We hypothesized that the radiomic features calculated from only pretreatment planning CT images could quantify RP texture patterns reflecting radiation-induced tissue reaction, thereby predicting RP. Therefore, the purpose of this study was to develop a predictive model for RP after lung cancer SBRT using radiomic features for lung ROI segmented by dosimetric information on pretreatment planning CT images.
Materials and methods
Patient data
This retrospective study was performed with the ethical approval of the institutional review board of our hospital. 245 training cases and 30 test cases (a total of 275 patients) were selected as a training and test cohorts, respectively, from two different terms and planning CT scanners. The training cohort included 245 patients who underwent SBRT for NSCLC between August 2003 and July 2013 (median dose: 48 Gy; median age: 77 years; range: 52–91 years; TNM classification: T1–2, N0, M0). Twenty-two of these patients had grade 2 or higher RP. The test cohort consisted of 30 patients including eight RP cases and 22 non-RP cases who underwent SBRT for NSCLC between April 2014 and March 2018 (median dose: 48 Gy; median age: 74 years; range: 54–90 years; TNM classification: T1–2, N0, M0). The patients’ characteristics are summarized in Table 1. The patients of the training cohort were scanned using a planning CT (Mx 8000, Philips Healthcare, Amsterdam, The Netherlands) with a tube voltage of 120 kV, an in-plane pixel size of 0.98 mm, and a slice thickness of 2.0 mm for treatment planning. The patients of the test cohort were acquired on a different CT scanner for the training cohort (Aquilion Prime, Canon Medical Systems, Otawara, Japan) for validation under the same conditions as the training cohort. Three-dimensional conformal radiation therapy (3DCRT) plans with non-coplanar fields were created using a commercially available radiation treatment planning (RTP) system (Eclipse; Varian Medical Systems Inc., Palo Alto, USA). RP grades were scored using the Common Terminology Criteria for Adverse Events version 4.0 (CTCAE v.4.0) based on clinical assessment and imaging15.
Overall scheme
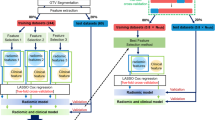
Figure 1 illustrates the overall workflow of the proposed scheme for RP prediction. First, the ROIs for calculating radiomic features were extracted from the treatment planning data. Four ROIs were created by extracting lung volumes excluding gross tumor volume (GTV) irradiated with more than 0, 5, 10, and 20 Gy for each patient, which were defined as LV0, LV5, LV10, and LV20, respectively. Second, a total of 486 radiomic features, which consisted of 54 original radiomic features (14 histogram-based and 40 texture features) and 432 wavelet-based radiomic features with 8 wavelet decompositions (54 features × 8 wavelet decomposition filters), were calculated from each ROI. Third, the significant features were selected as a signature (set of selected significant features) for each ROI, and then RP predictive models with signatures for each ROI were built to classify patients with and without grade ≥ 2 RP using a least absolute shrinkage and selection operator (LASSO) logistic regression. Finally, the constructed models for each ROI were evaluated with training and test cases by areas under the receiver operating characteristic (ROC) curve (AUC), sensitivity, specificity, and accuracy.
ROI extraction
The ROIs for calculating radiomic features were extracted using structure data for total lung volumes excluding the GTV and dosimetric data obtained from the treatment planning data. Four ROIs were created by extracting lung volumes irradiated with more than 0, 5, 10, and 20 Gy for each patient, which were defined as LV0, LV5, LV10, and LV20. The image processing was performed using in-house software with MATLAB 2019a (MathWorks).
Computation of radiomic features
A total of 486 radiomic features including 54 original features and 432 wavelet-based features were calculated from each ROI in pretreatment planning CT images for quantifying RP texture patterns using the MATLAB-based Radiomics tools package (implemented in MATLAB 2019a, MathWorks)16,17. The 54 original radiomic features consisted of 14 histogram-based and 40 texture features. The texture features were calculated from four texture-characterization matrices (i.e., a gray level co-occurrence matrix [GLCM]18, gray level run-length matrix [GLRLM]19, gray level size-zone matrix [GLSZM]20, and neighborhood gray-tone difference matrix [NGTDM]21). The 54 original radiomic features are listed in the Supplemental Information (Table S1). Then, 432 wavelet-based radiomic features were derived from the same 54 features as the original features on each of the eight wavelet decomposition images22. The wavelet transform can decompose multiscale local lung texture patterns related to RP and non-RP in an image into several low- and high-frequency components23. The decomposition was performed by applying either a low-pass filter (scaling function, L) or a high-pass filter (wavelet function, H) in the x, y, or z direction. The eight wavelet decomposition filters consisted of a combination of three using either a low-pass filter (L) or a high-pass filter (H) in each direction. Figure 2 shows the CT images with RP decomposed based on the wavelet analysis. The original image shows the texture patterns different from 7 (HLL to HHH) wavelet decomposition images, although the LLL image is similar to the original image. In this study, we assumed that radiomic features on original images could represent lung texture properties different from wavelet-based radiomic features provided by the wavelet decomposition images.
Construction of signatures and building RP predictive model using LASSO logistic regression
Although we assumed that original image features could represent lung texture properties different from wavelet-based image features, some features could be linearly dependent, especially features from original and LLL images. However, according to the LASSO theory24, that linear dependence does not imply dispensability, and individual dispensability does not imply pairwise dispensability, we decided to employ the original CT images. Nevertheless, to avoid the risk of overfitting on the RP prediction model, the significant features among 486 radiomic features were reduced to a number of features using a LASSO logistic regression with MATLAB 2019a (MathWorks)25. This process was repeated 1000 times for each ROI. The radiomic features with the highest frequency were extracted from the 486 radiomic features to build the RP predictive model for each ROI26. The RP grades were annotated by 1 for RP = 2 or above and 0 for otherwise, as the teacher data to be inputted into the logistic regression models. The logistic regression models were constructed with the radiomic signatures for each ROI to classify patients with and without grade ≥ 2 RP.
RP predictive model with dose-volume histogram parameters
A logistic regression model with four dose-volume histogram (DVH) parameters of the lung volumes receiving more than 5, 10, and 20 Gy (V5, V10, V20) and MLD was also constructed for comparisons between DVH and the radiomics models. The most frequently selected significant DVH parameter combination using a LASSO logistic regression, which was similar to the signature construction mentioned above, was used for the RP predictive model.
Construction of an ensemble averaging model with imbalanced datasets adjustment strategy
In this study, only 22 (9%) of the 245 training patients had grade ≥ 2 RP. Imbalanced datasets cause performance loss in the classification model27. To address the issue of imbalanced data, this study used an imbalance adjustment strategy adapted from that described by Schiller et al.28. As shown in Fig. 3a, the data were partitioned into a collection of balanced subsets. Thus, 10 subsets consisting of all 22 RP cases and 22 or 23 randomly extracted non-RP cases were created from the imbalanced training dataset of 245 patients. The recommended number of features should be generally smaller than around one-tenth of the number of training cases to avoid the overfitting problem29,30. Additionally, in leave-one-out cross-validation performed beforehand for each subset with an increasing number of top features for RP prediction, the predictive models with the top four features showed the highest performance (Fig. S1). Therefore, for each subset with each ROI, the top four significant features were selected for the construction of signatures, and 10 predictive models were built with the signatures using LASSO logistic regression. Significant DVH parameter combination was also selected for each subset using LASSO logistic regression for the RP predictive model. Finally, as shown in Fig. 3a, an ensemble averaging model was newly built by averaging the RP probabilities of the 10 predictive models constructed from 10 different subsets made from 245 training cases for each ROI.
RP predictive model training and testing
The ensemble averaging model for the RP prediction was considered to be trained with 245 training cases. Four significant features were selected in each predictive model for each ROI using LASSO logistic regression. As shown in Fig. 3b, the built ensemble averaging model was tested with 30 test cases in the same manner as the model training. The RP predictive model was evaluated according to the AUC, sensitivity, specificity, and accuracy. The sensitivity, specificity, and accuracy are given by
and
where TP, FP, TN, and FN are the numbers of true positives, false positives, true negatives, and false negatives, respectively. The AUC was obtained from the area under the ROC curve, which was a plot of sensitivity against (1-specificity) by changing the discrimination threshold of a classifier system.
Ethical statement
This retrospective study was performed with the ethical approval of the institutional review board of our hospital. Written informed consent was obtained from all subjects within the dataset collected in our hospital. All of the methods were carried out in accordance with the Declaration of Helsinki.
Results
Table 2 shows AUCs, sensitivity, specificity, and accuracy of the ensemble averaging model for 245 training and 30 test cases. The AUCs of the RP predictive model for the training cohort with DVH parameters and radiomic signatures for LV0, LV5, LV10, and LV20 were 0.703, 0.868, 0.871, 0.905, and 0.890, respectively. The AUCs for the test cohort were 0.290, 0.557, 0.756, 0.602, and 0.608, respectively. All radiomic models showed higher performance than DVH model.
Table 3 shows the top four radiomic features selected most frequently for 10 subsets for each ROI. The radiomic feature of “correlation” computed with GLCM on the original images was selected as the signature for each ROI.
Discussion
Using the radiomic features for lung ROIs dosimetrically segmented from the pretreatment planning CT images of 275 NSCLC patients, we found that the radiomic predictive models to classify patients with and without grade ≥ 2 RP performed well. In the training cohort, the AUC for the ensemble averaging model with LV10 signatures using the top four radiomic features reached the maximum value of 0.905. In the test cohort, the radiomic predictive model for LV5 reached the highest AUC of 0.756. This model for LV5 also showed a high AUC of 0.871 in the training cohort. Based on these results, the radiomic predictive model for LV5 was considered the best model.
The prediction results of the test cohort were lower than those of the training cohort. In particular, in terms of low sensitivity, the number of RP cases in the training and test cohorts might be insufficient. The difference between CT equipment and breathing methods during image acquisition may also have affected the radiomic features. The CT scans were performed on free breathing in the training cohort, while breath-hold techniques were used in the test cohort.
The “correlation” computed with GLCM on the original images was selected as one of the frequently selected features for each ROI in Table 3. Correlation is a measure of how correlated a pixel is to its neighbor over the whole image. Figure 4 shows a bar graph of “correlation” values on the original images for LV5 of RP and non-RP cases in the training cohort and an example of pretreatment planning CT images of RP and non-RP cases. The values of “correlation” of RP cases were significantly higher than those of non-RP cases. These results indicate that the “correlation” on the original images could quantify the RP characteristics different from the one on the wavelet decomposition images and might be one of the imaging biomarkers for RP after lung cancer SBRT.
Previous studies on RP prediction are summarized in Table 4. The previous studies often used DVH parameters such as V20 and MLD as risk factors for RP prediction8,9,10,11,14,31. Various clinical factors and biomarkers such as cytokines, single nucleotide polymorphisms (SNPs), and microRNA have also been used for RP prediction14,31. In the field of radiomics, Cunliffe et al. proposed that dose-dependent texture changes between pre- and post-RT CT images could classify patients with and without grade ≥ 2 RP. When multiple features were combined in a classifier, AUC increased significantly (from 0.59 to 0.84)12. Moran et al.13 found that changes in radiomic features calculated from follow-up CT images after SBRT for 14 patients were significantly correlated with post-SBRT lung injury scores provided by a radiation oncologist, and that the AUCs using GLCM texture features ranged from 0.689 to 0.750.
Previous studies used differences in radiomic features between pre- and post-treatment CT images for RP prediction12,13. However, this study predicted RP risk using only pretreatment planning CT images. Therefore, before new patients receive radiation therapy, it may be possible to determine the RP risk by applying treatment planning data to our RP predictive models. In addition, this method is reasonable in terms of clinical application as it requires only treatment planning data without additional clinical examinations.
This study has two limitations. First, only 22 (9%) of the 245 training patients included in this study had grade ≥ 2 RP. The imbalanced data was also a factor reducing the predictive model performance. To address these issues, the balanced subsets were sampled, and the ensemble averaging model was constructed using the 10 predictive models obtained from each subset. Second, we did not evaluate the repeatability and reproducibility of the radiomic features since we used only pretreatment planning CT. Traverso et al. reported that only radiomic features with high repeatability and reproducibility should be used in predictive models to reduce the risk of false-positive associations32. Therefore, to reduce the influence of radiomic feature variation on RP prediction as much as possible, we calculated the radiomic features under the same conditions for image acquisition settings, image reconstruction algorithm, digital image preprocessing, and software used to extract radiomic features in the training cohort. Moreover, the constructed models were tested in a separate test cohort, which was scanned on another equipment to validate repeatability and reproducibility. Nevertheless, these problems should be considered a limitation because texture features were less reproducible than histogram features32 and 11 of the 16 radiomic features selected as signatures for 10 subsets with four ROIs, shown in Table 3, were texture features.
In conclusion, the results of this study demonstrated the potential of RP predictive models after lung cancer SBRT using radiomic features for lung ROIs segmented by dosimetric information on pretreatment planning CT images. All radiomic models showed higher performance than the DVH model. The radiomic predictive model for LV5 was considered as the best model with a high AUC of 0.871 and 0.756 in both the training and test cohorts. Radiomic features calculated from pretreatment planning CT images can be used as imaging biomarkers for RP prediction in SBRT treatment planning for lung cancer.
References
Nagata, Y. et al. Clinical outcomes of a phase I/II study of 48 Gy of stereotactic body radiotherapy in 4 fractions for primary lung cancer using a stereotactic body frame. Int. J. Radiat. Oncol. Biol. Phys. 63, 1427–1431 (2005).
Onishi, H. et al. Stereotactic body radiotherapy (SBRT) for operable Stage I non-small-cell lung cancer: can SBRT be comparable to surgery?. Int. J. Radiat. Oncol. Biol. Phys. 81, 1352–1358 (2011).
Marks, L. B. et al. Radiation dose-volume effects in the lung. Int. J. Radiat. Oncol. Biol. Phys. 76, 70–76 (2010).
Timmerman, R. et al. Extracranial stereotactic radioablation: results of a phase I study in medically inoperable stage I non-small cell lung cancer. Chest 124, 1946–1955 (2003).
Yamashita, H. Radiation pneumonitis after stereotactic radiation therapy for lung cancer. World J. Radiol. 6, 708 (2014).
Hara, R., Itami, J., Komiyama, T., Katoh, D. & Kondo, T. Serum levels of KL-6 for predicting the occurrence of radiation pneumonitis after stereotactic radiotherapy for lung tumors. Chest 125, 340–344 (2004).
Yamashita, H. et al. Prescreening based on the presence of CT-scan abnormalities and biomarkers (KL-6 and SP-D) may reduce severe radiation pneumonitis after stereotactic radiotherapy. Radiat. Oncol. 5, 1–9 (2010).
Graham, M. V. et al. Clinical dose-volume histogram analysis for pneumonitis after 3D treatment for non-small cell lung cancer (NSCLC). Int. J. Radiat. Oncol. Biol. Phys. 45, 323–329 (1999).
Tsujino, K. et al. Predictive value of dose-volume histogram parameters for predicting radiation pneumonitis after concurrent chemoradiation for lung cancer. Int. J. Radiat. Oncol. Biol. Phys. https://doi.org/10.1016/S0360-3016(02)03807-5 (2003).
Fay, M. et al. Dose-volume histogram analysis as predictor of radiation pneumonitis in primary lung cancer patients treated with radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. https://doi.org/10.1016/j.ijrobp.2004.08.025 (2005).
Palma, D. A. et al. Predicting radiation pneumonitis after chemoradiation therapy for lung cancer: an international individual patient data meta-analysis. Int. J. Radiat. Oncol. Biol. Phys. 85, 444–450 (2013).
Cunliffe, A., Iii, S. G. A., Castillo, R., Pham, N. & Guerrero, T. Lung texture in serial thoracic computed tomography scans: correlation of radiomics-based features with radiation therapy dose and radiation pneumonitis development. Radiat. Oncol. Biol. 91, 1048–1056 (2015).
Moran, A., Daly, M. E., Yip, S. S. F. & Yamamoto, T. Radiomics-based assessment of radiation-induced lung injury after stereotactic body radiotherapy. Clin. Lung Cancer 18, e425–e431 (2017).
Cui, S., Luo, Y., Tseng, H. H., Ten Haken, R. K. & El Naqa, I. Combining handcrafted features with latent variables in machine learning for prediction of radiation-induced lung damage. Med. Phys. 46, 2497–2511 (2019).
Cancer Therapy Evaluation Program NCI. Common terminology criteria for adverse events (CTCAE) Version 4.0. 2009.
Vallières, M., Freeman, C. R., Skamene, S. R. & El Naqa, I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys. Med. Biol. 60, 5471–5496 (2015).
Vallières M. Radiomics: MATLAB programming tools for radiomics analysis. GitHub; 2015. https://github.com/mvallieres/radiomics; 2015 (accessed 30 April 2020).
Blondel, P. & Gómez Sichi, O. Textural analyses of multibeam sonar imagery from Stanton Banks, Northern Ireland continental shelf. Appl. Acoust. 70, 1288–1297 (2009).
Galloway, M. M. Texture analysis using gray level run lengths. Comput. Graph. Image Process. https://doi.org/10.1016/s0146-664x(75)80008-6 (1975).
Thibault, G. et al. Shape and texture indexes application to cell nuclei classification. Int. J. Pattern Recognit. Artif. Intell. 27, 1–23 (2013).
Amadasun, M. & King, R. Textural features corresponding to textural properties. IEEE Trans. Syst. Man. Cybern. 19, 1264–1274 (1989).
Soufi, M., Arimura, H. & Nagami, N. Identification of optimal mother wavelets in survival prediction of lung cancer patients using wavelet decomposition-based radiomic features. Med. Phys. 45, 5116–5128 (2018).
Mallat, S. G. A theory for multiresolution signal decomposition: the wavelet representation. IEEE Trans. Pattern Anal. Mach. Intell. 11, 674–693 (1989).
Tibshirani, R. J. The lasso problem and uniqueness. 7, 1456–1490 (2013).
Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B 58, 267–288 (1996).
Van Der Ploeg, T. & Steyerberg, E. W. Feature selection and validated predictive performance in the domain of Legionella pneumophila: a comparative study. BMC Res. Notes 9, 1–7 (2016).
Akbani, R., Kwek, S. & Japkowicz, N. Applying support vector machines to imbalanced datasets. In Machine Learning: ECML (eds Boulicaut, J.-F. et al.) 39–50 (Springer, Berlin, 2004).
Schiller, T. W., Chen, Y., El Naqa, I. & Deasy, J. O. Modeling radiation-induced lung injury risk with an ensemble of support vector machines. Neurocomputing 73, 1861–1867 (2010).
Peduz, P., Concato, J., Kemper, E., Holford, T. R. & Feinstein, A. R. Study of the number of events per variable in logistic regression analysis. J. Clin. Epidemiol. 49, 1373–1379 (1996).
Jain, A. K., Duin, R. P. W., Mao, J. & Member, S. Statistical pattern recognition: a review. IEEE Trans. Pattern Anal. Mach. Intell. 22, 4–37 (2000).
Luna, J. M. et al. Predicting radiation pneumonitis in locally advanced stage II–III non-small cell lung cancer using machine learning. Radiother. Oncol. 133, 106–112 (2019).
Traverso, A., Wee, L., Dekker, A. & Gillies, R. Repeatability and reproducibility of radiomic features: a systematic review. Radiat. Oncol. Biol. 102, 1143–1158 (2018).
Acknowledgements
This work was supported by JSPS KAKENHI Grant Number 19K16803 and 20K08113. The authors are grateful to all members of Arimura Laboratory (http://web.shs.kyushu-u.ac.jp/~arimura), whose comments contributed significantly to this study. We would also like to thank Editage (www.editage.com) for English language editing.
Author information
Authors and Affiliations
Contributions
T.H. conducted and designed this study. T.H. wrote the manuscript. H.A., J.F., and Y.S. supervised this study. T.H. and K.N. collected and analyzed the image data. T.Y. collected medical information data and scored radiation pneumonitis. H.A. and K.N. provided programming scripts for analysis and knowledge of machine learning and data science. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hirose, Ta., Arimura, H., Ninomiya, K. et al. Radiomic prediction of radiation pneumonitis on pretreatment planning computed tomography images prior to lung cancer stereotactic body radiation therapy. Sci Rep 10, 20424 (2020). https://doi.org/10.1038/s41598-020-77552-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-77552-7
- Springer Nature Limited
This article is cited by
-
Radiation pneumonitis prediction after stereotactic body radiation therapy based on 3D dose distribution: dosiomics and/or deep learning-based radiomics features
Radiation Oncology (2022)
-
Using deep learning to predict radiation pneumonitis in patients treated with stereotactic body radiotherapy (SBRT) for pulmonary nodules: preliminary results
Journal of the Korean Physical Society (2022)
-
Risk factors for radiation-induced lung injury in patients with advanced non-small cell lung cancer: implication for treatment strategies
World Journal of Surgical Oncology (2021)
-
Biological dosiomic features for the prediction of radiation pneumonitis in esophageal cancer patients
Radiation Oncology (2021)