Abstract
DNA aptamers generated by cell-SELEX against bacterial cells have gained increased interest as novel and cost-effective affinity reagents for cell labelling, imaging and biosensing. Here we describe the selection and identification of DNA aptamers for bacterial cells using a combined approach based on cell-SELEX, state-of-the-art applications of quantitative real-time PCR (qPCR), next-generation sequencing (NGS) and bioinformatic data analysis. This approach is demonstrated on Enterococcus faecalis (E. faecalis), which served as target in eleven rounds of cell-SELEX with multiple subtractive counter-selections against non-target species. During the selection, we applied qPCR-based analyses to evaluate the ssDNA pool size and remelting curve analysis of qPCR amplicons to monitor changes in pool diversity and sequence enrichment. Based on NGS-derived data, we identified 16 aptamer candidates. Among these, aptamer EF508 exhibited high binding affinity to E. faecalis cells (KD-value: 37 nM) and successfully discriminated E. faecalis from 20 different Enterococcus and non-Enterococcus spp. Our results demonstrate that this combined approach enabled the rapid and efficient identification of an aptamer with both high affinity and high specificity. Furthermore, the applied monitoring and assessment techniques provide insight into the selection process and can be highly useful to study and improve experimental cell-SELEX designs to increase selection efficiency.
Similar content being viewed by others
Introduction
DNA aptamers are short synthetic oligonucleotides (20–100 nucleotides) that can bind to a molecular target by their unique three-dimensional structure with high affinity and specificity. In contrast to their antibody-counterparts, aptamers offer many inherent advantages, which make them appealing alternatives in diverse analytical applications. First, aptamers can be readily synthesized in large quantities at relatively low costs and with low batch-to-batch variations. Due to chemical synthesis, they can be easily modified with chemical groups and fluorescence dyes in a targeted manner for immobilization and signalling purposes. Second, they are relatively small in terms of their molecular weight (10–30 kDa), which allows them to reach epitopes that antibodies (~ 150 kDa) cannot target due to steric hindrance1. Third, aptamers offer higher stability and can be regenerated by de/renaturation without losing their functionality, which is extremely useful, especially for biosensor development. However, the most appealing feature of aptamers is that they can be selected in vitro through an iterative process termed SELEX (Systematic Evolution of Ligands by EXponential enrichment)2,3, which can be tailored to the needs of the intended application. This process has been used over the past decades to select aptamers for a wide range of targets, from small molecules such as organic dyes4, cocaine5 or mycotoxins6,7 to numerous proteins such as thrombin8, tumour marker peptides9 and growth factors10 as well as complex targets such as mammalian cells11. More recently, bacterial cells have also become frequent targets in aptamer selection processes.
Currently, the discovery of aptamers for bacterial cells is mainly based on whole cell-SELEX, which is a modified form of the conventional SELEX procedure that uses purified target molecules12. In cell-SELEX, DNA aptamers typically evolve from large randomized single-stranded DNA (ssDNA) libraries (consisting of up to 1015 unique sequences) by repetitive binding to whole bacterial target cells and partitioning by centrifugation. Over the course of the selection process, cell-bound ssDNA fractions are repeatedly amplified by PCR and regenerated for the next round of cell-SELEX. After multiple rounds of selection (typically 6–20 rounds), the final ssDNA pool is traditionally cloned into plasmids followed by Sanger sequencing to identify aptamer candidates. In contrast to conventional SELEX using isolated cell-surface proteins that are immobilized on solid supports, whole cell-SELEX offers the inherent advantage that it allows selecting aptamers that interact directly with cellular epitopes in their native conformation with no prior knowledge of the molecular target12. In this way, aptamers have been selected for human and food-borne pathogens, such as Streptococcus spp.13,14, Listeria monocytogenes15, Salmonella spp.16,17, Staphylococcus aureus18, Vibrio parahaemolyticus19, Escherichia coli20 and Pseudomonas aeruginosa21.
Despite success in aptamer discovery, bacterial whole cell-SELEX suffers from methodological challenges. Many steps of the procedure are performed in a semi-blind fashion with limited control over and insight into how the ssDNA pool is evolving. This black box can make SELEX a time-consuming, labour-intensive and costly trial and error process. Also, the complex nature of bacterial cells with thousands of cell-surface molecules and structures leads to a much larger number of different aptamer sequences that are enriched in parallel compared to single and defined targets. Thus, traditional cloning and Sanger sequencing of 30–100 clones (e.g. Refs.22,23,24) from the last round of whole cell-SELEX can hinder the identification of aptamer candidates with desired binding characteristics. To address these limitations, next-generation sequencing (NGS) is increasingly replacing the low-throughput sequencing step in SELEX procedures25,26,27. NGS enables the analysis of millions of sequences and early identification of high-quality aptamers, which in turn can reduce the overall number of selection rounds required. Furthermore, complementary methods, such as quantitative real-time PCR (qPCR) have been proposed for SELEX monitoring and assessment on a round-to-round basis. For instance, qPCR was used to quantify the amount of bound ssDNA on human embryonic kidney cells28. In contrast to spectrophotometric measurements (e.g. Nanodrop) or fluorescence-based binding assays (e.g. flow cytometry), qPCR is highly sensitive and requires no labelling of ssDNA for the detection. Simultaneously, the quantification of cell-bound ssDNA enables the determination of the optimal (minimal) number of PCR cycles for amplifying the cell-bound fractions for the next round of SELEX28. This reduces the risk of PCR by-products and artefacts that often contaminate the enriched pool and render the selection ineffective28,29. Moreover, qPCR-based remelting curve analysis has been proposed to monitor changes in sequence diversity30,31,32. Based on the denaturation of double-stranded qPCR products after reannealing under stringent conditions, VanBrabant et al. applied remelting curve analysis to gain information on enrichment in steroid, peptide and protein targeting SELEX procedures, suggesting its general applicability as a monitoring tool31.
In the present study, we describe a combined approach that incorporates whole cell-SELEX, qPCR-based monitoring tools, NGS and custom-designed bioinformatic data analysis to select and identify DNA aptamers for Enterococcus faecalis (E. faecalis) cells. E. faecalis served primarily as a model organism, which we chose for the following reasons. E. faecalis is a commensal gut bacterium, which is widely used together with other enterococci as faecal indicator for assessing water quality33. Likewise, it is an opportunistic pathogen with increasing antibiotic resistance34. Based upon this health-relevant bacterium, we demonstrate the individual processes and the utility of this new and combined aptamer discovery approach for generating bacterial cell-recognizing aptamers.
Results and discussion
In vitro selection of aptamers specific for E. faecalis
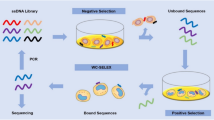
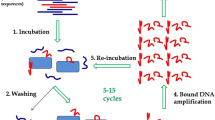
A general outline of the combined aptamer discovery approach developed to select DNA aptamers for whole bacterial cells is depicted in Fig. 1. Here we used a customized cell-SELEX protocol12 and performed eleven rounds of SELEX with E. faecalis type strain DSM-20478 as target cells. Following incubation with the random ssDNA library, cells were washed by centrifugation to remove unbound sequences. Sequences bound to the cells were amplified by PCR and rendered single stranded by Lambda-exonuclease digestion for the next round of SELEX. In each round, samples of the cell-bound ssDNA pool were subject to test-PCR reactions monitored in real-time. Based on the obtained amplification curves, we determined the minimum number of cycles needed for subsequent PCR amplification of the pool to prevent the formation of by-products and artefacts (see Supplementary Figure S1 and “Methods” for details).
Schematic illustration of the bacterial whole cell-SELEX process in combination with qPCR-based and NGS tools. A total of eleven SELEX rounds of positive selection with E. faecalis cells as target were performed. In rounds R03, R04, R05, R06 and R09 subtractive counter-selections were carried out with mixtures of enterococcal and non-enterococcal species, respectively, which were incubated with the enriched ssDNA pool prior to positive selection.
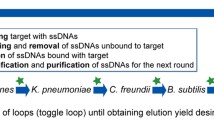
As outlined in Table 1, our selection strategy involved gradually increasing the selection pressure from round 2 (R02) onward by decreasing the concentration of input DNA (from 1.8 to 0.01 nmol), increasing the number of wash cycles (from one to five) and adding competitors in excess (BSA and salmon sperm DNA) to the binding reaction, while the number of target cells was kept constant (107 cells). To eradicate nonspecific binders from the pool that bind to common and highly conserved cell-surface molecules, we introduced subtractive counter-selections early in SELEX round R03 to R06 against mixtures of ten Enterococcus spp. (E. faecium, E. durans, E. hirae, E. gallinarum, E. casseliflavus, E. sulfureus, E. cecorum, E. avium, E. asini and E. mundtii) and later in SELEX R09 against a mixture of seven non-Enterococcus spp. (Streptococcus bovis, Streptococcus salivarius, Staphylococcus aureus, Escherichia coli, Citrobacter freundii, Klebsiella aerogenes and Pseudomonas aeruginosa). Sequences that bound to these counter-target cells were removed before the incubation with E. faecalis cells (no amplification step in-between). We refrained from using heat-elution and/or purification steps to detach the bound ssDNA from E. faecalis target cells prior to amplification in order to minimize the loss of potential aptamers associated with such additional steps35. We did not observe detrimental effects of cells or cell components on the efficiency of the employed PCR assays.
Monitoring the SELEX process by qPCR analyses
Most previous studies describing the selection of aptamers against bacterial cells applied spectrophotometric measurements (e.g., Nanodrop) to determine and monitor the proportion of ssDNA bound to the target cells and terminated the selection once a plateau was reached17,36,37. To monitor and assess the selection process in more detail, we used qPCR to quantify the cell-bound ssDNA pools from individual SELEX rounds and to determine the total amount of recovered ssDNA after incubation and washing. Cell-bound aptamer pools were then diluted and normalized (105 ssDNA molecules per reaction) for remelting curve analysis to estimate changes in pool diversity based on double-stranded qPCR products. In parallel, dilutions of the original random unselected ssDNA library (100% diversity at different ssDNA concentrations) were analysed to determine the average melting temperature and to compare the melting profiles. All SELEX rounds were subjected to these qPCR-based analyses, except for R01. R01 was excluded as the entire volume of cell-bound ssDNA pool was PCR amplified for the next SELEX round to recover all sequences, which are all unique at this point of the selection12,35.
Based on the qPCR results (Fig. 2), we found that the mean total amount of ssDNA recovered from R02 was 1.1 × 108 ssDNA molecules, which equals approximately 11 ssDNA molecules bound on average per E. faecalis cell. After eleven rounds of SELEX and with increased selection pressure and several counter-selections, the amount of cell-bound ssDNA and thus the number of ssDNA molecules bound to E. faecalis cells remained constant.
Total amounts of recovered ssDNA molecules from consecutive rounds of SELEX (R02–R11) measured by qPCR. To test for inhibitory effects, each sample was analysed undiluted and diluted 1:10. As no inhibitory effect was observed, the mean ssDNA quantity was calculated from both undiluted and diluted measurements. Error bars represent min. and max.
However, remelting curve analysis indicated that the diversity of these cell-bound sequences changed (Fig. 3). Based on the dilutions of the random ssDNA library we simulated enrichment to qualitatively assess the diversity of the ssDNA pools. As evident from Fig. 3A, highly diverse ssDNA pools with ≥ 105 different sequences showed continuous melting behaviour without distinct melting temperatures (ssLib dilutions + 7, + 6 and + 5). On the other hand, less diverse pools with < 105 different sequences (ssLib dilutions + 4, + 3) showed more discrete melting temperature ranges. This stems from a larger proportion of complementary sequences that form stable homo-duplexes after total denaturation and re-annealing at high temperatures (70 °C) when sequence diversity is low31. By comparing the remelting data of cell-bound ssDNA pools and the random (unselected) ssDNA library, we observed that distinct melting peaks appeared in SELEX R09 suggesting a drop in pool diversity and the emergence of potentially enriched sequences (Fig. 3B,C). From SELEX R09 to R11, melting peaks did not increase further, but changed their remelting shape, suggesting changes in sequence composition. In addition to melting peak height and shape, we also observed a shift towards higher melting temperatures. The average melting temperature of 77.6 °C (random ssDNA library, Fig. 3A) increased to 79.3 °C (R11, Fig. 3B). All these effects can be attributed to a change in pool composition and the accumulation of sequences31. Additional amplification and standard melting curves performed prior to remelting curve analysis (see “Methods” section for details) are given in Supplementary Figure S2. We ended the selection process after eleven rounds as further selection rounds are not only costly and time-consuming but can also make the selection prone to amplification errors or even the loss of aptamers38. To confirm the observation of sequence enrichment and to identify the sequences that bound to E. faecalis cells, we performed next-generation sequencing (NGS).
Monitoring the progression of the whole cell-SELEX process by qPCR-based remelting curve analysis. (A) Remelting curves from dilutions of the random ssDNA library ranging from 107 (Std + 7) to 103 (Std + 3) different molecules per reaction. (B) Remelting curves of cell-bound ssDNA pools from consecutive SELEX rounds (R02–R11) normalized to 105 molecules per reaction. (C) Individual remelting curves of the cell-bound ssDNA pools compared to random ssDNA library dilution 105 (Std + 5). Ascending melting peaks indicated the accumulation of sequences in the cell-bound ssDNA pools.
NGS data analysis and assessment of the SELEX process
We performed NGS sequencing on an Illumina MiSeq sequencer to analyse the ssDNA pools from SELEX round R02–R11 and to identify their sequence composition. A total of 2,948,294 reads were successfully demultiplexed by the MiSeq and used as input for our analysis workflow. Adding sequencing data of the starting ssDNA library (R00) from another run, our data analysis pipeline started with 3,055,932 reads. Filtering for intact 5′ and 3′ constant regions and correct insert length (i.e., removing too short and too long reads) left us with 2,686,568 reads (87%). Discarding low quality reads (Phred Q-score ≤ 30) reduced this number to 2,094,663 reads (69%). Paired-end reads were merged as an additional quality control but had only minor impact on the number of individual reads, resulting in a total of 2,089,419 reads (68%), which were used for further analysis. After data pre-processing, sequences were tracked over successive SELEX rounds to assess the progression of the SELEX process and to verify the sequence enrichment indicated by qPCR-based remelting curve analysis. To this end, sequences were grouped into different categories according to their read numbers (i.e. frequency: 1–9, 10–99, 100–999, 1,000–9,999) and normalized to the total number of reads in each SELEX round (% of total reads). Moreover, we performed nucleotide composition analysis of the 40-nt random region to check the quality of starting ssDNA library (equal distribution of 25% A, T, C, G) and to investigate if the composition changed over the selection process.
NGS analysis revealed that sequence enrichment was first observable from SELEX round R06 to R07 when sequences with > 10 reads appeared (Fig. 4A). From SELEX R07 onward, the proportion of unique sequences and sequences with < 10 reads, respectively, steadily decreased while sequences with 10–99 and 100–999 reads increased. In ssDNA pools of SELEX R10 and R11, we also found sequences with > 1000 reads (Fig. 4A). Moreover, we determined that nucleotide composition changed over the course of the selection. As shown in Fig. 4B, the average proportion of nucleotide bases in the random region of the starting random ssDNA library (R00) was biased towards adenine-rich sequences, while cytosine and guanine were slightly underrepresented (A: 29.1%, C: 22.1%, G: 23.9%, T: 24.8%). After eleven rounds of SELEX, however, cytosine- and thymine-rich sequences had increased by 8.0% and 3.8%, respectively (R11; A: 21.6%, C: 30.1%, G: 19.7%, T: 28.6%). Likewise, the nucleotide distribution at each position of the 40-nt randomized regions was altered in the ssDNA population from SELEX R11 (Fig. 4C).
NGS analysis of the ssDNA population. (A) Proportion of reads (%) in the ssDNA pools from SELEX R02–R11 and the random ssDNA library (R00) that were counted 1–9, 10–99, 100–999, 1000–9999 times. (B) Changes in the nucleotide composition in the random regions over the selection rounds. (C) Distribution of nucleotides at each position of the 40-nt long random region of the random ssDNA library (R00) and the ssDNA pool of SELEX R11.
Candidate sequences for further testing were chosen from a compiled sequence table in which all sequences were ranked based on their frequency in R11 and tracked over the preceding rounds of SELEX. In addition, lists of the top 1000 most frequent sequences of each SELEX round were created (Supplementary Material S1). The following criteria were used for selecting sequences: sequence frequency, appearance in early rounds, sequence enrichment in rounds with counter-selection and increased selection pressure, as well as on predicted secondary structure and minimal free energy. Based on these criteria, we selected and chemically synthesized 16 aptamer candidate sequences for testing (Table 2). Their predicted secondary structures and minimal free energies are given in Supplementary Figure S3.
Aptamer identification
A series of binding assays based on qPCR analysis was performed (1) to screen the putative full-length candidate sequences in terms of their binding properties and (2) to select the most promising ones for further analysis. To this end, each candidate was incubated at a final concentration of 100 nM with 107 E. faecalis target cells in binding buffer. Several controls were carried out in parallel. First, blank reactions were performed without bacterial cells to determine the amount of non-specific background binding and to exclude differences in the observed binding that is possibly based on the sequence’s tendency to adhere to plastic tube surfaces. Measured aptamer quantities from these control samples were subtracted from positive samples (i.e., E. faecalis cells + ssDNA). Second, the random ssDNA library was assessed in parallel to compare the interaction behaviour. Furthermore, reactions with E. faecalis cells only (no ssDNA added) were performed to check for contamination. Two biological replicates were performed for each candidate tested. Results indicated that all tested candidates exhibited similar levels of binding to E. faecalis cells, which were on average 7-times higher than that of the random ssDNA library (Fig. 5). Among all 16 tested sequences, aptamer candidate EF508 displayed the highest mean cell-bound ssDNA quantity.
Results from initial screening of aptamer candidate sequences EF501 to EF516 based on qPCR analysis. Each aptamer was incubated with 107 E. faecalis cells in 100 µL binding buffer at a final aptamer concentration of 100 nM for 30 min at 21 °C and mild shaking. After washing three times by centrifugation, the amount of cell-bound aptamer was quantified via qPCR. Reactions with the random library (ssLib), without cells (blank samples) and without ssDNA (cells only, negative control) were run in parallel. Two biological replicates were performed for each sample. Reactions were judged free of contamination based on negative controls. Data are represented as the geometric mean of two biological replicates, after subtraction of blank samples. Error bars represent min. and max.
Based on the enrichment trajectories of the candidates, we tested the specificity of aptamer candidate EF508 and two other candidates EF504 and EF513 with higher and lower sequencing reads (Supplementary Table S1), respectively, on a small subset of non-target cells, comprising two Enterococcus spp. (E. faecium, E. mundtii) and two non-Enterococcus spp. (Gram-negative Escherichia coli NCTC9001 and Gram-positive Staphylococcus aureus). All four species were used for counter-selection in SELEX. The binding experiment was performed as described before using 100 nM aptamer and 107 bacteria cells. We found that all three aptamer candidates displayed higher binding to E. faecalis cells than to any of the tested non-target species (Fig. 6). However, we observed distinct differences in the level of specificity of the three aptamers. Based on these screening results and NGS data that showed that aptamer EF508 appeared early from R05 onwards (Supplementary Table S1), albeit at low read number, EF508 was further characterized in more detail.
Binding assay results from aptamer candidates EF504, EF508 and EF513 evaluating the specificity of the sequences towards non-target species Enterococcus faecium, Enterococcus mundtii, Escherichia coli NCTC 9001 and Staphylococcus aureus. 107 cells were used per reaction. All aptamers were tested in a final concentration of 100 nM. Blank reactions (without cells) and negative controls (without ssDNA) were run in parallel as a control. Two biological replicates were performed for each sample and the amount of cell-bound aptamer determined via qPCR. Based on negative controls, reactions were judged free of contamination. Data are represented as the geometric mean of two biological replicates, after subtraction of blank samples (reactions without cells). Error bars represent min. and max.
Aptamer binding affinity
The binding affinity of aptamer EF508 was assessed by incubating 107 E. faecalis cells with different aptamer concentrations ranging from 0 to 300 nM. Data obtained from qPCR analysis showed a typical saturation curve, which were fitted by non-linear regression analysis (Fig. 7A). This indicates that the aptamer interacts with E. faecalis cells in a concentration-dependent manner. The determined dissociation constant (KD) was 37 ± 4 nM, which is comparable to previously reported KD values of aptamers against bacteria cells that typically range between 10 and 200 nM36,37,39. The predicted most probable secondary structure of aptamer EF508 by RNAfold, with stable and defined stem-loop structures, is shown in Fig. 7B.
(A) Binding saturation curve of aptamer EF508 to E. faecalis cells. Cells were incubated with increasing concentrations of aptamer (0, 1, 3, 10, 30, 100 and 300 nM) and measured by qPCR. Data points represent the mean of three biological replicates. A non-linear regression curve was fitted to the data using SigmaPlot version 14.0. The dissociation constant (KD) is shown as the mean ± SD. (B) Predicted secondary structure and minimal free energy of aptamer EF508 using RNAfold software.
Aptamer specificity and cross-reactivity towards bacteria species
In order to more broadly assess the specificity of the aptamer EF508, a total of 20 bacterial species were tested. The sample set consisted of ten Enterococcus spp., including E. faecium, E. durans, E. hirae, E. gallinarum, E. casseliflavus, E. sulfureus, E. cecorum, E. avium, E. asini and E. mundtii. Likewise, we tested five Gram-positive species, which are phylogenetically closely related to enterococci, namely Streptococcus bovis, Streptococcus salivarius, Lactococcus lactis and Staphylococcus aureus, as well as five Gram-negative species which are often found in fecally polluted waters, including three Escherichia coli strains (NCTC 9001, ATCC 25922, ATCC 8739), Citrobacter freundii, Klebsiella aerogenes and Pseudomonas aeruginosa. All these species had been used for counter-selection in SELEX, except for E. coli strains ATCC 25922 and ATCC 8739 and L. lactis. Each species was tested individually using 107 cells per reaction and 100 nM aptamer. Three biological replicates were performed and analysed with qPCR.
Based on qPCR results, aptamer EF508 showed a high degree of specificity (Fig. 8). It can not only discriminate E. faecalis from other Enterococcus spp., but also from both Gram-positive and Gram-negative species. Based on the calculated number of bound aptamers per cell, less than one aptamer molecule on average was bound to non-target cells. Likewise, no apparent binding to E. coli strains ATCC 25922 and ATCC 8739 as well as L. lactis DSM 20481 was observed, even though these strains were not used as counter-targets in SELEX. Comparing the total measured ssDNA quantities, we observed a more than 20- to 800-fold higher binding to E. faecalis target cells than to non-target cells, which to our knowledge is higher than previously reported for DNA aptamers that were selected in bacterial cell SELEX experiments. Typically, five- to ten-fold differences were observed in specificity tests that comprised smaller sample sets (three to twelve tested species)22, 36. These differences could stem from the high sensitivity of qPCR-based measurements, compared to e.g. fluorescence spectroscopy- or flow cytometry-based analyses. However, Marton et al.22 conducted qPCR-based specificity tests and also reported only a ten-fold higher binding of their aptamer to E. coli target cells than to twelve other bacterial species tested22.
Specificity of aptamer EF508 evaluated with (A) ten Enterococcus spp. and (B) ten non-Enterococcus spp. E. faecalis target cells were tested in parallel. Binding assays were performed with 100 nM of aptamer and 107 bacteria cells. Blank reactions (without cells, only aptamer) and negative controls (without ssDNA) were run in parallel as a control. Three biological replicates were performed for each sample and the amount of cell-bound aptamer determined via qPCR. Based on negative controls, reactions were judged free of contamination. Data are represented as the geometric mean of three biological replicates. Error bars represent min. and max. Based on the amount of recovered ssDNA, the average number of aptamers bound per cell was estimated.
Specific binding of the aptamer was also visually confirmed and qualitatively assessed by epifluorescence microscopy on a subset of bacterial stains, including E. faecalis target cells and six non-target cells (E. faecium, E. hirae, E. gallinarum, S. bovis, E. coli NCTC 9001 and C. freundii). Each species was tested individually using 107 cells that were incubated with 100 nM of 5′-FAM labelled aptamer EF508. In parallel, experiments were performed without aptamer (binding buffer used instead of aptamer). These blank reactions were either subjected to SYBR Gold staining serving as control and reference for direct cell detection approaches or were left untreated to control for any fluorescence signals caused by autofluorescence of the bacterial cells. As shown in Fig. 9, E. faecalis target cells incubated with the aptamer induced fluorescence reflecting the morphology of the bacteria, albeit at lower intensity than the non-selective SYBR Gold fluorescence dye. In contrast, non-target cells incubated with 5′-FAM-labelled aptamer showed no fluorescence (Fig. 9).
Epifluorescence microscopy image of E. faecalis target cells and six bacterial non-target species stained with SYBR Gold (left column), 100 nM of 5′-FAM labelled aptamer EF508 (middle column) and without any staining (right column). Experiments were carried out with 107 cells collected on a 0.2 µm membrane filter for microscopic observation. Magnification: 1000 ×, Exposure time: 100 ms. White scale bars denote 10 µm.
Overall, the binding preference of aptamer EF508 suggests that the selected candidate binds to a molecular target that is predominantly expressed on the cell surface of type strain E. faecalis. However, further studies would be needed to identify this unknown cell-surface epitope, e.g. using pull-down experiments from the cell followed by LC–MS/MS analysis40. Likewise, comprehensive studies including environmental isolates, establishing limits of detection and testing different sample matrices would be necessary to ensure the functionality and robustness of the aptamer in future downstream diagnostic applications. Depending on the field of application (e.g. water quality testing), signal enhancement strategies might be needed for establishing future aptamer-based epifluorescence microscopy methods (e.g. using quantum dot labelling41). Based on our results, however, we anticipate that aptamer EF508 is a promising candidate for any such endeavours.
Conclusion
In this study, we used a customized aptamer discovery approach to select and identify DNA aptamers for E. faecalis cells. We applied state-of-the-art applications of qPCR as tools for optimizing the PCR amplification step of the SELEX procedure and for assessing the overall selection process in-line while it is progressing. We also showed that measuring the ssDNA pools size during bacterial cell-SELEX may not always be informative. Solely based on amounts of cell-bound ssDNA measured by qPCR, we were unable to observe a selection progress. Yet, sequence diversity changed as indicated by remelting curve analysis of qPCR products and confirmed by NGS analysis. Thus, we conclude that remelting curve analysis is a highly useful complementary tool to gain information on sequence enrichment and to rationally decide whether a bacterial cell-SELEX experiment should be continued, sequenced or rather be aborted. Due to next-generation sequencing and data analysis, we were able to trace the population dynamics over the selection procedure based on ssDNA pools from several rounds that were barcoded and sequenced in parallel. The obtained data illustrated that NGS data are vital in identifying bacterial cell-aptamer candidates. Based on this new and combined approach, we identified aptamer EF508, which made up 0.394% of the final pool population and which displayed high binding affinity in the nanomolar range and high specificity to E. faecalis target cells. Even though future work will be required to fully characterize and test this novel binding ligand, we conclude that the aptamer has significant potential to be used for diagnostic applications.
Methods
Bacterial strains
Type strain Enterococcus faecalis (E. faecalis) DSM-20478 was used as target for the in vitro aptamer selection process. For counter-selection and characterization studies the following organisms were used: E. faecium (DSM-20477), E. durans (DSM-20633), E. hirae (DSM-20160), E. gallinarum (DSM-20628), E. casseliflavus (DSM-20680), E. mundtii (DSM-4838), E. sulfureus (DSM-6905), E. cecorum (DSM-20682), E. avium (DSM-20679), E. asini (DSM-11492), Streptococcus bovis (DSM-20480), Streptococcus salivarius (DSM-20560), Lactococcus lactis (DSM-20481), Staphylococcus aureus (DSM-20232), three strains of Escherichia coli (NCTC 9001, ATCC 25922, ATCC 6633), Citrobacter freundii (DSM-30039), Klebsiella aerogenes (NCTC 9528) and Pseudomonas aeruginosa (NCTC 10662). The bacterial strains were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ), Braunschweig, Germany and were cultivated under aerobic conditions until exponential growth phase. Enterococcus spp. and Streptococcus spp. were grown in tryptic soy broth, whereas all other species were cultivated in Brain Heart Infusion. Detailed information on the preparation of stock solutions and the cultivation conditions are given in the Supplementary Information S1.
Unless otherwise noted, aliquots of the prepared stock solutions were used for SELEX and binding assay experiments. Before use, the cells were washed two times with 1 mL of 0.2 µm-sterile filtered 1 × PBS, pH 7.4 (137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4; Thermo Fisher Scientific, AM9625) by centrifugation at 13,000×g for 10 min. The cell pellet was finally resuspended in aptamer binding buffer (1 × PBS, pH 7.4 with 0.05% [v/v] Tween20; 0.2 µm-sterile filtered).
ssDNA library and primers
The synthetic ssDNA library consisted of a central 40-nt randomized region flanked on either side with 23-nt constant primer binding regions for PCR (5′-TAG GGA AGA GAA GGA CAT ATG AT—N40—TTG ACT AGT ACA TGA CCA CTT GA-3′). It was chemically synthesized and HPLC-purified by Integrated DNA Technologies (Coralville, USA). Unmodified forward primer (5′-TAG GGA AGA GAA GGA CAT ATG AT), unmodified reverse primer (5′-TCA AGT GGT CAT GTA CTA GTC AA-3′) and 5′-phosphorylated reverse primer (5′-Phos-TCA AGT GGT CAT GTA CTA GTC AA-3′) were purchased from Eurofins MWG Operon (Ebersberg, Germany). The synthetic ssDNA library and the primers were dissolved in nuclease-free water (Roth, Germany) to a final concentration of 100 µM.
Bacterial whole cell-SELEX process
In total, eleven SELEX rounds were performed (Table 1). For the first round of SELEX, 1.8 nmol of the ssDNA library (~ 1015 molecules) were prepared in 50 µL binding buffer, heated at 95 °C for 5 min and kept at 21 °C for 30 min to allow the formation of folded DNA structures. The folded ssDNA library was mixed and incubated with 107 washed E. faecalis cells in a final reaction volume of 250 µL binding buffer at 21 °C for 30 min with moderate shaking at 550 rpm on an Eppendorf thermomixer. After the binding reaction, the cells were collected by centrifugation for 10 min at 13,000×g. The supernatant containing unbound sequences was discarded and the cell pellet washed once by resuspending the cells in 1 mL binding buffer. After centrifugation for 10 min at 13,000×g, the bacteria-aptamer complexes were recovered by discarding the binding buffer and resuspending the cell pellet in 50 µL nuclease-free water. This fraction is referred to as “cell-bound ssDNA pool”. To eliminate sequences that bind to the tube walls, the mixture was transferred into a fresh 1.5 mL Eppendorf DNA LoBind tube (VWR, Germany). Heat-elution and/or purification steps of cell-bound ssDNA were omitted to minimize the loss of potential aptamers. Negative controls consisting of cells without added DNA were run in parallel throughout the entire procedure of incubation, washing and resuspension to check for cross-contamination.
In the first SELEX round, the entire cell-bound ssDNA pool (50 µL) was subject to preparative PCR, whereas in subsequent rounds approximately 15 µL of the cell-bound ssDNA pool was amplified by preparative PCR. PCR amplification of the cell-bound ssDNA pool was performed in a reaction volume of 25 µL containing 1 × Q5 High-Fidelity Master Mix (New England Biolabs, Germany), 1 µM forward primer, 1 µM phosphorylated reverse primer, and 1.0 µL of DNA template (cell-bound ssDNA; in the first round 5.0 µL DNA template were used for each reaction). All PCR reactions were run on a T100 Thermal Cycler (BioRad, Germany) according to the following thermal cycling conditions: an initial step of 3 min at 95 °C, followed by 15 s at 95 °C, 15 s at 55 °C, 15 s at 72 °C and a final extension step at 2 min at 72 °C. To avoid by-product formation, the number of PCR cycles was optimized for every round of SELEX as follows: prior to preparative PCR, a real-time monitored test-PCR reaction including 1 × EvaGreen fluorescence dye (Biotium/VWR, Germany) was performed on a Mastercycler ep realplex Real-time PCR system (Eppendorf, Hamburg, Germany). The cycle number before the amplification curve reached the fluorescence maximum was then used for subsequent preparative PCR amplification of cell-bound ssDNA fractions (typically between 20 and 25 cycles, Supplementary Figure S1).
To recover the ssDNA pool for subsequent selection rounds, PCR products were rendered single-stranded by Lambda-Exonuclease digestion42. To that end, all preparative amplification reactions were pooled, and PCR products were separated from primers and other reaction components using the Monarch PCR and DNA Cleanup Kit (New England Biolabs, Ipswich, USA). Two µg of purified dsDNA were then incubated with 5 U of Lambda Exonuclease (New England Biolabs, Ipswich, USA) in a total reaction volume of 50 µL at 37 °C for 60 min (four reactions with 2 µg of purified dsDNA each). The reactions were terminated by heat inactivation of the enzyme at 80 °C for 10 min. After ssDNA clean-up and concentration with the Monarch PCR and DNA Cleanup Kit, the recovered ssDNA was quantified via the NanoDrop One (Thermo Fisher Scientific, Germany). Aliquots of the preparative PCR product, the final enzyme digestion mixture and the purified ssDNA were analysed on a 4% agarose gel stained with SYBR Gold to check the size of the dsDNA and ssDNA products of each round, respectively.
Subsequent selection rounds were performed with decreasing amounts of input ssDNA (from 1.8 to 0.01 nmol), increasing number of washing cycles (from one to five washing cycles) and the addition of competitors, such as 0.5 µg µL−1 BSA (bovine serum albumin) and 0.25 µg µL−1 salmon sperm DNA in the binding buffer during incubation. To eliminate non-specific binding sequences from the ssDNA pool, counter-selections with mixtures of non-target bacteria were carried out before incubation with E. faecalis target cells as follows: ssDNA pools were first incubated with counter-target cells in binding buffer for 30 min at 21 °C and then the unbound ssDNA in the supernatant was withdrawn by centrifugation and incubated with 107 E. faecalis cells. In rounds R03 to R06, ssDNA pools were counter-selected with a defined mixture of ten enterococcal non-target cells, namely E. faecium, E. durans, E. hirae, E. gallinarum, E. casseliflavus, E. sulfureus, E. cecorum, E. avium, E. asini and E. mundtii (107 cells each, total of ~ 108 cells). In round 9, ssDNA pools were counter-selected with a defined mixture of seven non-enterococcal non-target cells, namely S. bovis, S. salivarius, S. aureus, E. coli (NCTC 9001), C. freundii, K. aerogenes and P. aeruginosa (107 cells each, ~ 8*107 cells). The SELEX conditions used to isolate aptamers binding to E. faecalis cells are summarized in Table 1.
Quantitative PCR
The total ssDNA quantities during SELEX and characterization studies, respectively, were measured via quantitative PCR (qPCR). All qPCR reactions were performed in a total reaction volume of 15 µL containing 1 × Kapa SYBR Fast (PeqLab, Germany), 500 nM unmodified primers, and 1.0 µL of cell-bound ssDNA pool. The amplification reactions were run on a Mastercycler ep realplex Real-time PCR system (Eppendorf, Germany) according to the following thermal cycling conditions: an initial step of 3 min at 95 °C, followed by 35 cycles of 15 s at 95 °C and 20 s at 62 °C. All amplification reactions were performed in duplicates, including no-template controls (NTC) in each run to check for contamination. Quantification was based on dilutions of the random synthetic ssDNA library. Standard dilutions were prepared in an unspecific background of 500 µg L−1 poly(dI-dC) (Roche Diagnostics, Germany) to avoid adsorption of ssDNA onto the reaction tubes at lower concentrations. A total of six tenfold serial dilutions of standard (103–108 ssDNA copies per reaction) were included in each qPCR run. Unless otherwise noted, samples were measured undiluted and diluted 1:10 in 10 mM of Tris–EDTA buffer (pH 8.5) and were judged free of inhibition if concentrations in the dilutions matched.
Remelting curve analysis
To monitor the progress of the in vitro selection process, remelting curve analysis of PCR amplicons was performed as described earlier31,32 with several modifications. Reactions were performed in a total volume of 25 µL containing 1 × Q5 High-Fidelity Master Mix, 1 µM unmodified primers, 1 × EvaGreen dye and 1.0 µL of cell-bound ssDNA pool (normalized based on qPCR results to 105 ssDNA molecules per reaction). Cycling conditions were 3 min at 95 °C, followed by 35 cycles at 15 s at 95 °C, 15 s at 55 °C and 15 s at 72 °C. After amplification, two melting curve analyses were performed (referred to as “standard melting curves” and “remelting curves”). The first standard melting curve protocol consisted of 15 s at 95 °C and reannealing for 15 s at 60 °C before heating to 95 °C in increments of 0.03 °C s−1. The second remelting curve protocol followed the same conditions, except reannealing was performed for 15 s at 70 °C before heating to 90 °C. Remelting curve analysis was performed with all cell-bound ssDNA pools from SELEX (R02-R11) and dilutions of the synthetic ssDNA library (107–103 ssDNA molecules per reaction). The ssDNA library dilution 105 was used as a diversity standard representing maximum diversity to estimate sequence enrichment via remelting curves.
Next-generation sequencing
The cell-bound ssDNA pools from SELEX R02 – R11 were prepared for next-generation sequencing (NGS) analysis following a modified 16S rRNA amplicon sequencing library preparation procedure43. In brief, a two-step PCR procedure was used to attach sequencing primer regions, barcodes and flow-cell adapters (Supplementary Figure S6). Primer sequences are given in Supplementary Table S3 and an overview of the resulting amplicons is shown in Supplementary Figure S7. The first NGS-PCR used primers (NGS_PCR#1_forward and NGS_PCR#1_reverse) that consisted of SELEX-specific sequences (complementary to the constant primer binding regions of the ssDNA pools) and overhang sequences that included Illumina’s sequencing primer sites. PCR reactions were carried out in a total volume of 25 µL containing 1 × Q5 High-Fidelity Master Mix, 0.5 µM of NGS_PCR#1 primers and equal amounts of ssDNA (106 molecules per reaction). Cycling conditions were 3 min at 95 °C, followed by 22 cycles of 15 s at 95 °C, 15 s at 55 °C, 15 s at 72 °C and a final extension step for 2 min at 72 °C. The optimal number of cycles for the first NGS-PCR (22 cycles) was also determined by test-PCR reactions but using NGS_PCR#1 primers. After the first NGS-PCR, the PCR products were checked on a 3% agarose gel and were purified with the Agencourt AMPure XP Purification system (Beckman Coulter). The second NGS-PCR used primers (NGS_PCR#2_forward and NGS_PCR#2_reverse) that consisted of complementary overlapping sequences, barcodes (i7 and i5 indexes) and adapter sequences to attach the oligonucleotides onto the flow cell. PCR reactions were carried out again in a total volume of 25 µL containing 1 × Q5 High-Fidelity Master Mix, 1 µM of NGS_PCR#2 primers and 2.5 µL of the purified first NGS-PCR product. Cycling conditions were 30 s at 95 °C, followed by 6 cycles of 10 s at 95 °C, 30 s at 55 °C, 30 s at 72 °C and a final extension step for 2 min at 72 °C. The second NGS-PCR was performed with a very limited number of cycles. Six cycles were sufficient to attach barcodes and flow cell adapters and to visualize the products on an agarose gel. The PCR products were purified as described before with the Agencourt AMPure XP Purification system. The DNA concentration of the final products was then determined with the QuantiFluor dsDNA system (Promega, Germany) and the GloMax-Multi + fluorometer (Promega, Germany) according to the manufacturer’s instructions. All ten samples (R02–R11) were mixed in equimolar ratio to a final concentration of 4 nM. The final NGS library was clustered at 8 pM with 10% of 8 pM PhiX added to the run. Sequencing was done on the Illumina MiSeq platform using the MiSeq Reagent Micro Kit v2 (300 cycles) in paired-end mode. Sequencing data were demultiplexed using bcl-2fastq2 v2.20.
Sequencing data analysis
Analysis of the raw NGS-data was accomplished with a workflow implemented in Nextflow 19.10.044 (illustrated in Supplementary Figure S8). Data were pre-processed using cutadapt 2.445 to identify and cut the constant 5′ and 3′ SELEX primer binding regions. Every sequence had to consist of the 5′ end constant region ‘TAGGGAAGAGAAGGACATATGAT’, a random region of 40 ± 3 nt length, and the 3′ end constant region ‘TTGACTAGTACATGACCACTTGA’. An error rate of 20% (≤ 4 nt mismatches) within the constant regions was allowed. Sequences not fulfilling these conditions were discarded. Next, fastp 0.20.046 was used to filter sequences by quality score, discarding all sequences of an overall Phred quality score ≤ 30. Fastp was also used for merging forward and reverse NGS-reads using the default parameters (5 mismatches allowed, min. 30 nt overlap). Log files from cutadapt and fastp were graphically processed using MultiQC v1.947.
The pre-processed sequences were dereplicated using a custom counting script SELEXderep. The output of SELEXderep was used to determine every sequence’s reads per million total sequences (RPM) per round and to determine the ratio of unique to duplicate sequences of every round. Aptamers for further analysis were chosen based on RPM counts, enichment and structural properties of the aptamers, which were determined using RNAfold webserver v2.4.13 (https://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi)48. The custom script SELEXntcomposition was programmed to calculate the nucleotide composition of every round and for every position. Analysis was done using R 3.6.2 and Python 3.7. The following R libraries were used: dplyr49, ggplot250, Biostrings51, tidyr52. For Python, library Pandas53 was used.
Binding assays
All binding assays were performed in a total reaction volume of 100 µL of binding buffer. Prior to incubation, the aptamer was folded (by heating at 95 °C for 5 min and subsequent incubation at 21 °C for 30 min) and the cells were washed twice in binding buffer. For testing the binding affinity (KD determination), 107 E. faecalis cells were incubated with various concentrations of aptamer (final aptamer concentrations: 0, 1, 3, 10, 30, 100, and 300 nM) for 30 min at 21 °C while shaking at 550 rpm. After incubation, unbound aptamers were removed by centrifugation for 10 min at 13,000×g and washing the cell pellet three times with 1 mL fresh binding buffer (centrifugation steps in between). Cell pellets were subsequently resuspended in 50 µL nuclease-free water. For specificity testing, the aptamer was incubated with ten different Enterococcus spp. (107 cells each) and ten different non-Enterococcus spp. (107 cells each) at a final aptamer concentration of 100 nM. The amount of cell-bound aptamer was finally measured via qPCR. Three biological replicates were performed for each tested aptamer concentration and bacterial species, respectively. The KD (dissociation constant) was determined by a non-linear regression analysis using SigmaPlot v14.0 (Systat Software, Inc.)54.
Epifluorescence microscopy
For fluorescence microscopy studies, 107 bacterial cells were incubated with 100 nM of 5′-FAM-labelled aptamer (binding buffer for blank reactions) in 100 µL binding buffer for 30 min at 21 °C and 550 rpm shaking. After the binding reaction, the aptamer-cell complexes were washed with 1 mL binding buffer and resuspended in a final volume of 500 µL binding buffer and the suspension filtered onto a 0.2 µm black polycarbonate filter membrane (25 mm diameter; Merck Isopore, GTBP02500). For SYBR Gold staining the filters were placed in a petri dish on a drop (30 µL) of SYBR Gold (10,000 × concentrate in DMSO, diluted 400-fold in sterile deionized water) and kept in the dark for 15 min. After incubation, the filters were washed three times with a few drops of sterile 1 × PBS. All filters were dried in the dark at room temperature for approximately 30 min and then mounted onto a microscopic slide with one drop of anti-fading mounting solution (Citifluor). Another drop of mounting solution was directly placed on the filter before adding the cover slip. To remove excess liquid, the prepared slide was carefully pressed between two layers of absorbent paper. Slides were examined with immersion oil using a Nikon Eclipse Ni microscope at 1000 × magnification (Ex ~ 470, Em ~ 515) equipped with a Nikon DS-Qi2 camera.
Data availability
The Nextflow script and additional script files are located in the github repository https://github.com/hovercat/aptaflow/releases/tag/1.0. Raw FASTQ files were uploaded to the Sequence Read Archive (SRA) at NCBI and can be downloaded using the accession number PRJNA615076. Results of the analysis workflow are given in the Supplementary Material but can also be reproduced using the workflow on the raw NGS data.
References
Yüce, M., Kurt, H., Hussain, B. & Budak, H. in Biomedical Applications of Functionalized Nanomaterials (eds Bruno Sarmento & José das Neves) 211–243 (Elsevier, 2018).
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505 (1990).
Ellington, A. D. & Szostak, J. W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 355, 850–852 (1992).
Stojanovic, M. N., de Prada, P. & Landry, D. W. Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 123, 4928–4931. https://doi.org/10.1021/ja0038171 (2001).
McKeague, M. et al. Screening and initial binding assessment of fumonisin B1 aptamers. Int. J. Mol. Sci. 11, 4864–4881 (2010).
McKeague, M. et al. Selection and characterization of a novel DNA aptamer for label-free fluorescence biosensing of ochratoxin A. Toxins 6, 2435–2452 (2014).
Bock, L. C., Griffin, L. C., Latham, J. A., Vermaas, E. H. & Toole, J. J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355, 564–566 (1992).
Ferreira, C. S. M., Matthews, C. S. & Missailidis, S. DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA aptamers. Tumor Biology 27, 289–301 (2006).
Green, L. S. et al. Inhibitory DNA ligands to platelet-derived growth factor B-chain. Biochemistry 35, 14413–14424. https://doi.org/10.1021/bi961544+ (1996).
Blank, M., Weinschenk, T., Priemer, M. & Schluesener, H. Systematic evolution of a DNA aptamer binding to rat brain tumor microvessels: selective targeting of endothelial regulatory protein pigpen. J. Biol. Chem. 276, 16464–16468 (2001).
Sefah, K., Shangguan, D., Xiong, X., O’Donoghue, M. B. & Tan, W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 5, 1169–1185 (2010).
Hamula, C. L. A., Le, X. C. & Li, X.-F. DNA Aptamers Binding to Multiple Prevalent M-Types of Streptococcus pyogenes. Anal. Chem. 83, 3640–3647. https://doi.org/10.1021/ac200575e (2011).
14Alfavian, H., Mousavi Gargari, S. L., Rasoulinejad, S. & Medhat, A. Development of a DNA aptamer that binds specifically to group A Streptococcus serotype M3. Can. J. Microbiol. 63, 160–168. doi:https://doi.org/10.1139/cjm-2016-0495 (2016).
Duan, N. et al. Selection, identification and application of a DNA aptamer against Listeria monocytogenes. Food Control 33, 239–243. https://doi.org/10.1016/j.foodcont.2013.03.011 (2013).
Park, H.-C., Baig, I. A., Lee, S.-C., Moon, J.-Y. & Yoon, M.-Y. Development of ssDNA aptamers for the sensitive detection of Salmonella typhimurium and Salmonella enteritidis. Appl. Biochem. Biotechnol. 174, 793–802 (2014).
Lavu, P. S. R., Mondal, B., Ramlal, S., Murali, H. S. & Batra, H. V. Selection and characterization of aptamers using a modified whole cell bacterium SELEX for the detection of Salmonella enterica serovar typhimurium. ACS Combin. Sci. 18, 292–301. https://doi.org/10.1021/acscombsci.5b00123 (2016).
Cao, X. et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 37, 4621–4628. https://doi.org/10.1093/nar/gkp489 (2009).
Duan, N., Wu, S., Chen, X., Huang, Y. & Wang, Z. Selection and identification of a DNA aptamer targeted to Vibrio parahemolyticus. J. Agric. Food Chem. 60, 4034–4038 (2012).
Savory, N. et al. Selection of DNA aptamers against uropathogenic Escherichia coli NSM59 by quantitative PCR controlled Cell-SELEX. J. Microbiol. Methods 104, 94–100. https://doi.org/10.1016/j.mimet.2014.06.016 (2014).
Wang, K. Y., Zeng, Y. L., Yang, X. Y., Li, W. B. & Lan, X. P. Utility of aptamer-fluorescence in situ hybridization for rapid detection of Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 30, 273–278 (2011).
22Marton, S., Cleto, F., Krieger, M. A. & Cardoso, J. Isolation of an aptamer that binds specifically to E. coli. PloS one 11, e0153637 (2016).
23Song, M. Y., Nguyen, D., Hong, S. W. & Kim, B. C. Broadly reactive aptamers targeting bacteria belonging to different genera using a sequential toggle cell-SELEX. Sci. Rep. 7 (2017).
Yu, Q. et al. Selection and characterization of ssDNA aptamers specifically recognizing pathogenic Vibrio alginolyticus. J. Fish Dis. 42, 851–858. https://doi.org/10.1111/jfd.12985 (2019).
Schütze, T. et al. Probing the SELEX process with next-generation sequencing. PLoS ONE 6, e29604 (2011).
Civit, L. et al. Systematic evaluation of cell-SELEX enriched aptamers binding to breast cancer cells. Biochimie 145, 53–62. https://doi.org/10.1016/j.biochi.2017.10.007 (2018).
Hassan, E. M., Willmore, W. G., McKay, B. C. & DeRosa, M. C. In vitro selections of mammaglobin A and mammaglobin B aptamers for the recognition of circulating breast tumor cells. Sci. Rep. 7, 14487. https://doi.org/10.1038/s41598-017-13751-z (2017).
Avci-Adali, M. et al. Absolute quantification of cell-bound DNA aptamers during SELEX. Nucleic Acid Ther. 23, 125–130. https://doi.org/10.1089/nat.2012.0406 (2013).
Musheev, M. U. & Krylov, S. N. Selection of aptamers by systematic evolution of ligands by exponential enrichment: Addressing the polymerase chain reaction issue. Anal. Chim. Acta 564, 91–96. https://doi.org/10.1016/j.aca.2005.09.069 (2006).
Charlton, J. & Smith, D. Estimation of SELEX pool size by measurement of DNA renaturation rates. RNA (New York, N.Y.) 5, 1326–1332. doi:https://doi.org/10.1017/s1355838299991021 (1999).
Vanbrabant, J., Leirs, K., Vanschoenbeek, K., Lammertyn, J. & Michiels, L. reMelting curve analysis as a tool for enrichment monitoring in the SELEX process. Analyst 139, 589–595. https://doi.org/10.1039/C3AN01884A (2014).
Luo, Z., He, L., Wang, J., Fang, X. & Zhang, L. Developing a combined strategy for monitoring the progress of aptamer selection. Analyst 142, 3136–3139. https://doi.org/10.1039/C7AN01131H (2017).
Boehm, A. B. & Sassoubre, L. M. in Enterococci: from commensals to leading causes of drug resistant infection [Internet] (Massachusetts Eye and Ear Infirmary, 2014).
Fiore, E., Van Tyne, D. & Gilmore, M. S. Pathogenicity of Enterococci. Microbiol. Spectr. 7, 1. https://doi.org/10.1128/microbiolspec.GPP3-0053-2018 (2019).
Wang, T. et al. A detailed protein-SELEX protocol allowing visual assessments of individual steps for a high success rate. Hum. Gene Therapy Methods 30, 1–16. https://doi.org/10.1089/hgtb.2018.237 (2019).
Kim, Y. S., Song, M. Y., Jurng, J. & Kim, B. C. Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell-systematic evolution of ligands by exponential enrichment approach. Anal. Biochem. 436, 22–28 (2013).
Soundy, J. & Day, D. Selection of DNA aptamers specific for live Pseudomonas aeruginosa. PLoS ONE 12, e0185385 (2017).
Ozer, A., Pagano, J. M. & Lis, J. T. New technologies provide quantum changes in the scale, speed, and success of SELEX methods and aptamer characterization. Mol. Ther. Nucleic Acids 3, e183. https://doi.org/10.1038/mtna.2014.34 (2014).
Ramlal, S., Mondal, B. & Lavu, P. S. Capture and detection of Staphylococcus aureus with dual labeled aptamers to cell surface components. Int. J. Food Microbiol. 265, 74–83. https://doi.org/10.1016/j.ijfoodmicro.2017.11.002 (2018).
Bing, T., Shangguan, D. & Wang, Y. Facile discovery of cell-surface protein targets of cancer cell aptamers. Mol. Cell. Proteomics 14, 2692–2700 (2015).
Wen, L., Qiu, L., Wu, Y., Hu, X. & Zhang, X. Aptamer-modified semiconductor quantum dots for biosensing applications. Sensors 17, 1736. https://doi.org/10.3390/s17081736 (2017).
Citartan, M., Tang, T.-H., Tan, S.-C. & Gopinath, S. C. B. Conditions optimized for the preparation of single-stranded DNA (ssDNA) employing lambda exonuclease digestion in generating DNA aptamer. World J. Microbiol. Biotechnol. 27, 1167–1173. https://doi.org/10.1007/s11274-010-0563-8 (2011).
Amplicon, P., Clean‐Up, P. & Index, P. 16s metagenomic sequencing library preparation, https://emea.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (2013).
Di Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319. https://doi.org/10.1038/nbt.3820 (2017).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 17(3), 2011. https://doi.org/10.14806/ej.17.1.200 (2011).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. https://doi.org/10.1093/bioinformatics/bty560 (2018).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. https://doi.org/10.1093/bioinformatics/btw354 (2016).
WebServer, R. Available online: https://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi. RNAfold. cg (Accessed on May 2020).
dplyr: A Grammar of Data Manipulation v. R package version 0.8.4. (2020).
Wickham, H. ggplot2: elegant graphics for data analysis. (Springer, 2016).
Biostrings: Efficient manipulation of biological strings v. R package version 2.52.0 (2019).
tidyr: Tidy Messy Data v. R package version 1.0.2 (2020).
McKinney, W. in Proceedings of the 9th Python in Science Conference. 51–56 (Austin, TX).
SigmaPlot, V. 14.0. Systat Software, San Jose, CA, USA (2018).
Acknowledgements
Claudia Kolm is a recipient of the DOC Fellowship of the Austria Academy of Science at the Institute of Chemical, Environmental and Bioscience Engineering (TU Wien). This study was part of the Life Science Call 2017 Project LSC17-007 funded by the Niederösterreichische Forschungs- und Bildungsgesellschaft (NFB). This study was a joint collaboration of the Interuniversity Cooperation Centre Water and Health (ICC Water and Health, www.waterandhealth.at).
Author information
Authors and Affiliations
Contributions
C.K., A.K., A.F. and G.R. conceived and designed the experiments. I.C. and C.K. performed all experiments. U.A. developed the bioinformatic analysis pipeline and analysed the NGS data together with C.K. C.K., N.B., and R.S. cultivated and prepared the bacterial strains. S.J. assisted in flow cytometry and microscopy analysis. M.D., R.M. and R.K. provided scientific advice and resources. C.K. wrote the manuscript and prepared Figs. 1 and 7 and supplementary figures S3 and S7. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kolm, C., Cervenka, I., Aschl, U.J. et al. DNA aptamers against bacterial cells can be efficiently selected by a SELEX process using state-of-the art qPCR and ultra-deep sequencing. Sci Rep 10, 20917 (2020). https://doi.org/10.1038/s41598-020-77221-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-77221-9
- Springer Nature Limited
This article is cited by
-
Aptamer selection against cell extracts containing the zoonotic obligate intracellular bacterium, Anaplasma phagocytophilum
Scientific Reports (2024)
-
Aptamer decorated PDA@magnetic silica microparticles for bacteria purification
Microchimica Acta (2024)
-
In vitro selection of aptamers and their applications
Nature Reviews Methods Primers (2023)
-
Advances in aptamer-based biosensors for monitoring foodborne pathogens
Journal of Food Science and Technology (2023)
-
Ultrasensitive colorimetric strategy for Hg2+ detection based on T-Hg2+-T configuration and target recycling amplification
Analytical and Bioanalytical Chemistry (2021)