Abstract
Appropriate analytical methods are needed for the detection of food authentication. We investigated the applicability of a duplex real-time PCR assay targeting chromosome 1 and two singleplex real-time PCR assays targeting chromosome 9, both published recently, for the qualitative and quantitative determination of wild boar and domestic pig in processed food products. In addition, two singleplex real-time PCR assays targeting chromosome 7 were tested for their suitability to differentiate the two subspecies. Even by targeting the three genome loci, the probability of misclassification was not completely eliminated. Application of the real-time PCR assays to a total of 35 commercial meat products, including 22 goulash products, revealed that domestic pig DNA was frequently present, even in 14 out of 15 products declared to consist of 100% wild boar. Quantitative results obtained with the real-time PCR assays for wild boar (p < 0.001) and those for domestic pig (p < 0.001) were significantly different. However, the results obtained with the real-time PCR assays for wild boar (r = 0.673; p < 0.001) and those for domestic pig (r = 0.505; p = 0.002) were found to be significantly correlated. If the rules given in the paper are followed, the real-time PCR assays are applicable for routine analysis.
Similar content being viewed by others
Introduction
Food authentication is based on a variety of analytical methods, depending on the aspect of food adulteration one is interested in. Mislabeling of species in meat products is most commonly detected by DNA analysis1, 2. For the identification of pork, a number of methods based on polymerase chain reaction (PCR)—restriction fragment length polymorphism (RFLP) has been published3,4,5. Since PCR–RFLP is laborious and time consuming and not applicable to complex food products, it has mainly been replaced by singleplex6,7,8,9 and multiplex real-time PCR assays10,11,12,13,14. These assays usually allow the differentiation between pork and meat from other domesticated animals, e.g. beef, chicken or poultry, but are not applicable to distinguish pork from meat derived from wild boar. Differentiation between meat from domestic pig (Sus scrofa domesticus) and wild boar (Sus scrofa scrofa) is, however, necessary because in principle, both kinds of adulteration could occur: substitution of wild boar meat by pork and replacement of pork by wild boar meat. Since meat products derived from game animals are commonly more expensive than products from domesticated animals, some meat producers could be tempted to increase their profit by replacing (part of) wild boar meat by pork. However, at present some producers could also try to replace pork by wild boar meat due to an increase in pork prices, caused by slaughtering numerous pigs to reduce the chance of spreading of highly contagious African swine fever15.
Differentiation between pork and wild boar meat is a challenging task, because the genomes of wild boar and domestic pig are highly homologous16, 17. Thus, the number of subspecies-specific bases that can be targeted is very low. The common strategy is to make use of polymorphisms in genes that have been selected in the process of wild boar domestication. Genes associated with coat color, body composition, reproduction or behavior include melanocortin 1 receptor (MC1R), v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT), insulin like growth factor-2 (IGF2), ryanodine receptor 1 (RYR1) and nuclear receptor subfamily 6 group A member 1 (NR6A1)18.
Very recently, we developed a duplex and two singleplex real-time PCR assays for the differentiation between domestic pig and wild boar19. The assays are based on two nuclear DNA loci which had previously been reported to allow the differentiation between wild boar and domestic pig. The duplex real-time PCR assay, targeting the single nucleotide polymorphism (SNP) SNP g.299084751C>T in the NR6A1 gene on chromosome 1, consists of assayChr1W for wild boar and assayChr1D for domestic pig. The two singleplex real-time PCR assays for wild boar and domestic pig, assayChr9W and assayChr9D, target the SNP rs81416363 in an intergenic region on chromosome 9. Each of the assays was validated with regard to important analytical characteristics including selectivity, limit of detection (LOD), repeatability and robustness. By analyzing a total of 94 muscle tissue samples from 64 domestic pig and 30 wild boar individuals we found out that both the singleplex assays and the duplex assay led to some ambiguous results. However, we demonstrated that the assignment could be substantially improved by taking the results of both the duplex and the singleplex real-time PCR assays into account. With this approach, 91.5% of the 94 meat samples were classified correctly19.
Our previous study was restricted to unprocessed tissue samples from wild boar and domestic pig individuals. In the present study, we aimed at investigating the applicability of the real-time PCR assays for the qualitative and quantitative determination of wild boar and domestic pig meat in processed food products. Although the detection of undeclared meat species is sufficient to proof food adulteration, additional quantification allows the assessment if an undeclared species was added deliberately or is possibly contained due to contamination during food processing. A special case is Austria, where the proportion of wild boar meat and pork in commercial meat products is of interest because in “game meat” products, at least 38% of the meat content has to originate from game animals. In case the game species/subspecies is included in the product name, e.g. wild boar goulash, 38% of the meat content has to derive from the given species/subspecies20.
Results and discussion
In order to investigate the applicability of the duplex and the singleplex real-time PCR assays for processed food products, we analyzed 34 commercial meat products declared to contain wild boar and one declared to contain domestic pig. The set of meat products consisted of 22 goulash samples, two roasts, one burger, 5 sausages, one pastry, three hams and one chips sample.
Qualitative analysis
In 29 out of the 35 commercial meat products, wild boar was detected with both assays for wild boar, assayChr9W and assayChr1W (Table 1). In two products the wild boar content was < LOD (0.2%, w/w19) of assayChr9W, in three products < LOD (2%, w/w19) of assayChr1W. For the pork chips sample, both assays yielded a negative result. Although 15 out of the 22 goulash samples were declared to contain 100% wild boar, we detected domestic pig DNA in 13 samples with both assayChr9D (LOD: 2%, w/w19) and assayChr1D (LOD: 5%, w/w19). In total, domestic pig DNA was identified in 29 of the 35 commercial meat products with both assays for domestic pig. Wild boar ham 3 was the only sample in which domestic pig was neither detected with assayChr9D nor with assayChr1D.
Results for wild boar ham 2 were ambiguous. According to the results obtained with the assays targeting chromosome 9, the sample should originate from pig, whereas with the assays targeting chromosome 1 it was classified as wild boar. Most probably, the individual the meat is derived from does not show the genotype typical for wild boars, as demonstrated for several individuals by high resolution melting (HRM) analysis in our previous study19.
Since a high proportion of meat products was found to contain domestic pig although most of the products were declared to consist of 100% wild boar, we verified our results by analyzing the samples with two further singleplex real-time PCR assays, assayChr7P and assayChr7D. AssayChr7P and assayChr7D are based on an insertion/deletion in a fragment of chromosome 7. AssayChr7P allows the detection of pig (wild boar and domestic pig), whereas assayChr7D is specific for domestic pig. Consequently, by taking the results of both assays into consideration, one is able to differentiate between wild boar and domestic pig. However, before the analysis of commercial meat products, we applied assayChr7P and assayChr7D to the analysis of DNA isolates from 64 domestic pig individuals, including 14 breeds and six cross-breeds, and a total of 30 wild boar samples from five different countries (Austria, Estonia, Germany, Romania and USA) (Suppl. Table S1). These DNA isolates had already been analyzed with assayChr9W, assayChr9D, assayChr1W and assayChr1D in our previous study19. As expected, with assayChr7P, a positive result was obtained for each of the 64 domestic pig and 30 wild boar individuals. With assayChr7D, almost all domestic pig breeds and cross-breeds led to an increase in the fluorescence signal. However, for three out of six Mangalica and all eight Turopolje individuals a negative result was obtained. The same Mangalica and Turopolje individuals had led to ambiguous results with the real-time PCR assays targeting chromosome 9 and chromosome 1, respectively.
For each of the 35 commercial meat products, a positive result was obtained with assayChr7P, indicating that DNA from either wild boar, domestic pig or both subspecies was present (Table 1). By applying assayChr7D, domestic pig DNA was detected in 24 out of the 35 products, including 10 goulash samples declared to contain 100% wild boar. This finding confirmed our results on the high frequency of domestic pig DNA in the set of commercial meat products obtained with assayChr9D and assayChr1D. Furthermore, assayChr7D confirmed that wild boar ham 3 was the only sample which did not contain any domestic pig DNA.
By comparing the overall result obtained for each of the commercial meat products (Table 1) with the overall result obtained for each domestic pig and wild boar individual (Suppl. Table S1) we found out, that even by targeting the three different gene loci, the probability of misclassification was not completely eliminated. However, a meat sample containing one of the domestic pig breeds/crossbreeds investigated would hardly be classified as wild boar. The overall result obtained for wild boar goulash 10 and 16 (assayChr9W, assayChr9D, assayChr1W and assayChr7P positive, assayChr1D and assayChr7D negative) was only identical with that obtained for one out of six Mangalica individuals (Mangalica 3). In addition, one wild boar individual from Austria (wild boar Austria 2) and two from Germany (Wiesbaden and Germany) led to the same overall result. The overall result obtained for wild boar goulash 11, 12, 13, 15 and 21 and wild boar roast 2 (assayChr9W, assayChr9D, assayChr1W, assayChr1D and assayChr7P positive, assayChr7D negative) was only identical with that obtained for one wild boar individual (Germany, Perleberg). Wild boar roast 1 led to the same overall result as five out of eight Turopolje individuals. Game sausage 1, wild boar sausage and wild boar pastry showed the same overall result as Bentheim Black Pied pig 2 and three out of five Krškopolje breeds. The overall result obtained for wild boar ham 1 and 2 was only identical with that obtained for the wild boar individual USA 1. At present, more than 90% of all pigs slaughtered in Germany are crossbreeds from predominantly three pig breeds: German Edelschwein, German Landrace and Pietrain21. Thus, non-specific positive classifications of old pig breeds which are usually rare and marketed as delicacies are not limiting the applicability of the assays.
Quantitative analysis
Next, we investigated the applicability of the duplex real-time PCR assay (assayChr1W and assayChr1D) and the two singleplex real-time PCR assays (assayChr9W and assayChr9D) for the determination of the content of wild boar and domestic pig in processed food products. (The assays targeting chromosome 7 are not suitable for obtaining quantitative information.) We pursued a relative quantification approach, based on relating the concentration of wild boar DNA (determined with assayChr9W or assayChr1W) or domestic pig DNA (determined with assayChr9D or assayChr1D) to the concentration of total meat DNA (determined with a reference real-time PCR assay published previously22). However, before we could apply the real-time PCR assays to the analysis of commercial meat products, we had to validate the assays regarding limit of quantification (LOQ) and recovery.
Limit of quantification (LOQ)
We determined the LOQ by performing two series of experiments. In one series we used herring sperm DNA as background DNA. Herring sperm DNA is frequently used as non-target DNA in meat species authentication19, 22, 23. In the second series, we used domestic pig DNA as background DNA for assayChr9W and assayChr1W and wild boar DNA as background DNA for assayChr9D and assayChr1D. The LOQ was defined as the lowest DNA concentration which could be determined with a relative standard deviation (RSD) ≤ 25%24. In one of our previous studies, LOQ values determined by using DNA mixtures were found to be similar to those determined by using DNA isolated from meat mixtures25. The advantage of using DNA mixtures is that mixtures with low proportions can be prepared without the necessity to weigh out high meat amounts. In addition, one can avoid inaccuracy due to the water content in low meat amounts.
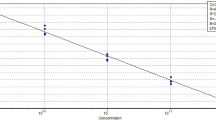
When we determined the LOQ in herring sperm DNA as background DNA, the LOQ of assayChr9W (Fig. 1A) and assayChr1W (Fig. 1B) was found to be 0.4% (w/w) wild boar DNA, that of assayChr9D (Fig. 1C) and assayChr1D (Fig. 1D) 2% (w/w) and 1% (w/w) domestic pig DNA, respectively. In the presence of a surplus of domestic pig DNA, the LOQ of assayChr9W (Fig. 1E) and assayChr1W (Fig. 1F) was 0.5% (w/w) and 3.0% (w/w), respectively. However, since at 3.0% (w/w), recovery of assayChr1W was only 41%, we set the LOQ of assayChr1W to 5.0%. In the presence of an excess of wild boar DNA, the LOQ of assayChr9D (Fig. 1G) and assayChr1D (Fig. 1H) was found to be 2% (w/w) and 20.0% (w/w), respectively.
Determination of the LOQ by analyzing serial dilutions of wild boar or domestic pig DNA. (A)–(D) herring sperm DNA as background DNA, (E)–(F) domestic pig DNA as background DNA, (G)–(H) wild boar DNA as background DNA. (A),(E) assayChr9W, (B),(F) assayChr1W, (C),(G) assayChr9D, and (D),(H) assayChr1D. Column plots show mean recovery and relative standard deviation (RSD). Horizontal blue lines indicate a RSD of 25%.
These data indicate that in the presence of DNA of the non-target subspecies, the LOQ of the assays targeting chromosome 1 was ten times higher than the LOQ of the assays targeting chromosome 9. The higher LOQ of assayChr1W and assayChr1D was most probably caused by signal suppression19. In our previous study, we demonstrated that at the same target DNA concentration, pure DNA extracts (either from wild boar or domestic pig) resulted in higher fluorescence signals (ΔRn values) than DNA mixtures containing both species (25:75 v/v)19.
Recovery in meat extract mixtures
In order to determine the recovery of the duplex and singleplex real-time PCR assays, we analyzed DNA isolates from nine meat extract mixtures containing 0–70% (w/w) wild boar, 0–70% (w/w) domestic pig and 0–96% (w/w) cattle (Table 2). PCR was carried out in six replicates.
Recoveries of assayChr9W were in the range from 62 to 97%. Low recoveries of 62% and 72% were obtained for meat extract mixtures containing 5% or 2% wild boar DNA, respectively. In case of assayChr9D, a domestic pig content of 2% (w/w) was < LOQ. For the meat extract mixture containing 5% (w/w) domestic pig, a recovery of 60% was obtained, mixtures containing from 10 to 70% domestic pig resulted in recoveries between 84 and 124%. In meat extract mixtures containing 2% and 5% (w/w) wild boar, the wild boar DNA concentration was < LOQ of assayChr1W. For meat extract mixtures containing 10–70% (w/w) wild boar, recoveries ranged from 87 to 116%. With assayChr1D, domestic pig DNA could not be quantified in a meat extract mixture containing 10% (w/w) domestic pig (< LOQ). For meat extract mixtures containing 25–70% (w/w) domestic pig, we obtained recoveries between 84 and 114%. A significant difference between recoveries was neither found for assayChr9W and assayChr1W (p = 0.356) nor for assayChr9D and assayChr1D (p = 0.653).
Recovery in model game sausages
In addition to meat extract mixtures, we analyzed seven model sausages, including five wild boar and two game sausages. Six out of the seven sausages contained only meat from wild boar and domestic pig, whereas the seventh game sausage additionally contained meat from deer species (Table 2). The DNA isolates were analyzed in six PCR replicates.
In general, wild boar and domestic pig meat contents > 10% led to recoveries between 70 and 130%. The presence of low amounts of the target subspecies in the surplus of the non-target subspecies led to higher deviations from the theoretical value.
With assayChr9W, we obtained recoveries between 69 and 130% for sausages with wild boar contents ranging from 25 to 50% (w/w). For the game sausage containing 7% (w/w) wild boar, 30% (w/w) domestic pig and 63% (w/w) deer meat, a recovery of 73% was obtained. For sausages consisting of 2% and 10% (w/w) wild boar and an excess of domestic pig (98% and 90% (w/w), respectively), the recoveries for wild boar were drastically too high (recovery 671% and 257%, respectively). High deviations from theoretical contents may be caused by differences in the composition, in particular the fat content, of meat tissues. We commonly try to overcome this problem by using calibration mixtures, carefully adjusted to composition and concentration of the samples25,26,27. The high recoveries obtained for wild boar in the presence of an excess of domestic pig is most probably caused by cross-reactivity of assayChr9W. For five sausages, assayChr9D led to recoveries between 77 and 122% for domestic pig contents between 30 and 98% (w/w). For the sausage containing 61.5% (w/w) domestic pig and 38.5% (w/w) wild boar, the domestic pig content was determined to be 14% (w/w) (recovery 23%). For the sausage containing 30% (w/w) domestic pig and 7% (w/w) wild boar, the recovery was 63%. Surprisingly, in both cases, the domestic pig content was underestimated. Due to the cross-reactivity with wild boar19, we actually expected the content to be overestimated. With assayChr1W, recoveries between 51 and 101% were obtained for sausages with wild boar contents ranging from 25 and 50% (w/w). In the wild boar sausages containing 2% or 10% (w/w) wild boar in domestic pig, the concentration of wild boar DNA was < LOD and < LOQ, respectively. In addition, in the game sausage containing 7% (w/w) wild boar, 30% (w/w) domestic pig and 63% (w/w) deer, the wild boar concentration was < LOQ. With assayChr1D, sausages with domestic pig contents ranging from 50 to 98% (w/w) resulted in recoveries between 78 and 121%. The domestic pig content in the sausage containing 7% (w/w) wild boar, 30% (w/w) domestic pig and 63% (w/w) deer was < LOQ. A significant difference between recoveries was neither found for assayChr9W and assayChr1W (p = 0.150) nor for assayChr9D and assayChr1D (p = 0.513).
Analysis of commercially available meat products
Quantitative results obtained by analyzing the 35 commercial meat samples are summarized in Table 3. Figure 2A shows the mean values ± 30% error ranges for wild boar, determined with assayChr9W and assayChr1W, Fig. 2B for domestic pig, determined with assayChr9D and assayChr1D. Statistical analyses revealed significant differences between the results obtained for wild boar with assayChr9W and assayChr1W (p < 0.001) and also between the results for domestic pig obtained with assayChr9D and assayChr1D (p < 0.001). However, results obtained for wild boar with assayChr9W were significantly correlated with results obtained with assayChr1W (r = 0.673; p < 0.001). In addition, results obtained for domestic pig with assayChr9D significantly correlated with results obtained with assayChr1D (r = 0.505; p = 0.002).
Quantitative results obtained for 35 commercial meat products. Mean content determined ± 30% error range (n ≥ 4). (A) wild boar (blue: assayChr9W; red: assayChr1W; green: overall mean) and (B) domestic pig (blue: assayChr9D; red: assayChr1D; green: overall mean). For results < LOD and < LOQ, LOD/2 and LOQ/2, respectively, are shown and were used for calculations. The horizontal blue line indicates a wild boar content of 38% (w/w).
Application in routine analysis
For this task, the following strategy should be pursued. To obtain qualitative information, each sample should be analysed with the six assays (assayChr9W, assayChr9D, assayChr1W, assayChr1D, assayChr7P and assayChr7D). If at least two of the three assays for the same subspecies lead to a positive result, the sample should be considered to contain the respective subspecies. If only one assay for this subspecies yields a positive result, the sample should be considered to not contain the subspecies. To determine the content of the detected subspecies, the application of the assay(s) is based on the qualitative results. If, e.g. domestic pig was detected with assayChr9D and assayChr7D, quantification should be done with assayChr9D (because the assays targeting chromosome 7 are not applicable for quantification). If, e.g. domestic pig was detected with both quantitative assays (assayChr9D and assayChr1D), the mean value of the concentrations obtained with the assays should be used for authentication. If the concentration was determined to be < LOQ, it was substituted with half the LOQ.
By following these rules, the final qualitative (Table 1) and quantitative (Table 3) results were obtained. For three out of 35 samples (wild boar goulash 18, wild boar sausage and wild boar pastry) the wild boar content could not be determined. Only in one out of 15 goulash samples declared to contain only wild boar, domestic pig could not be detected. Thirteen goulash samples contained both wild boar and domestic pig, and in one goulash sample, only domestic pig was detected. According to the Codex Alimentarius Austriacus, in the goulash samples declared to contain wild boar, 38% (w/w) of the meat content should originate from wild boar. By taking into account a 30% error range, 13 out of the 22 goulash samples met the requirement of the Codex. In the wild boar pastry, wild boar could not be detected. We assume that the DNA was too degraded to be amplified. All burger, roast and ham samples met the requirement, whereas the wild boar sausage did not meet the requirement of the Codex.
Conclusions
Both qualitative and quantitative determination of wild boar and domestic pig in processed food products turned out to be challenging. Even by targeting three different genome loci, the probability of misclassification was not completely eliminated. This is, however, not mainly caused by cross-reactivity of the real-time PCR assays. The major problem is that special pig breeds and cross-breeds show one or even two alleles commonly found in wild boar, whereas certain wild boar individuals are heterozygous or even homozygous for the allele usually occurring in domestic pig19. This obviously may also lead to discrepancies in quantitative results obtained by real-time PCR assays targeting different genome loci. Another well-known difficulty in the quantification of meat species in food by real-time PCR is the calculation of the meat content (w/w) from the target DNA concentration determined. Our strategy to overcome this problem is to use calibration mixtures similar to the analyzed sample in both the composition and concentration of the meat species of interest. In previous studies, this strategy was very successful25,26,27.
Materials and methods
Samples
Meat from wild boar individuals was collected in Austria and Germany, meat from domestic pig and cattle was purchased from local suppliers. All samples originated from lean muscle meat.
Meat extract mixtures were prepared by mixing the extracts from meat flesh of the respective animal species prior to DNA isolation. Model sausages were produced according to the guidelines of the Codex Alimentarius Austriacus20. “Wild boar sausages” containing different amounts of wild boar [2%, 10%, 25%, 38.5% or 50% (w/w)] and domestic pig [[98%, 90%, 75%, 61.5% or 50% (w/w), respectively] were produced at the Austrian Agency for Health and Food Safety (AGES, Vienna, Austria). Two other game sausages were produced at the Higher Technical College for Food Technology Hollabrunn (Hollabrunn, Austria). One game sausage contained 65% (w/w) domestic pig (30% bacon and 35% meat) and 35% (w/w) wild boar. The other game sausage consisted of 21% red deer, 21% fallow deer, 21% roe deer, 7% wild boar and 30% (w/w) bacon (domestic pig). Both sausages also contained typical sausage ingredients like nitrite curing salt (28 g/kg sausage), dextrose (3 g/kg sausage) and sucrose (2 g/kg sausage). In addition, the first one contained 54 g and the latter one 27 g of an allergen mix (celery, white/brown/black mustard, sesame, soy, wheat, egg powder and milk powder) per 15 kg.
34 food products declared to contain wild boar meat were purchased from supermarkets in Austria and Germany, comprising goulash (n = 22), roast (n = 2), burger (n = 1), sausage (n = 5), pastry (n = 1) and ham (n = 3). Furthermore, chips (n = 1) declared to contain domestic pig were analyzed.
DNA isolation
Sausages were homogenized in a knife mill (Retsch GM 200, Retsch, Haan, Germany). Muscle meat samples were cut into small pieces and used as-is. Commercial meat products were homogenized or cut into small pieces. 1–2 g of the sample material (meat, model sausage or commercial game product) were weighed out and 10 mL CTAB extraction solution (2% (w/v) CTAB, 1.4 M NaCl, 0.1 M Tris, 0.02 M EDTA, adjusted to pH 8.0 with 4 M HCl, autoclaved) and 80 µL proteinase K solution (1 mg/mL) were added. Lysis was carried out in an incubator (Unihood 750, Uniequip, Martinsried, Germany) at 50 °C under shaking (Intelli-Mixer RM-2L, LTF Labortechnik, Wasserburg, Germany) until the starting material was dissolved (at least 4 h). DNA was isolated at least twice from each lysis batch according to a previously published CTAB protocol28. To determine the DNA concentration and purity of the obtained DNA isolates, the absorbance was measured at 260 nm and 280 nm using a QIAxpert spectrophotometer (Qiagen, Hilden, Germany). The DNA isolates were stored at − 20 °C.
Real-time PCR
The two singleplex assays, assayChr9W and assayChr9D, and the duplex real-time PCR assay, consisting of assayChr1W and assayChr1D, were developed and validated previously19. Primer and probe sequences, target genes, accession numbers (NCBI GenBank accession number), final concentrations of primers and probes used in the PCR assays and amplicon lengths are given in Suppl. Table S2. Primers and probes were synthesized by Sigma Aldrich and Eurogentec (Seraing, Belgium), respectively.
Real-time PCR was performed in an optical 96-well reaction plate (0.2 mL, Applied Biosystems, Foster City, CA, USA) sealed with optical adhesive film (Applied Biosystems) on the ABI 7500 Real-time PCR System (Applied Biosystems). The reaction mix (total volume 25 µL) consisted of 12.5 µL of QuantiTect Multiplex PCR NoROX Master Mix (Qiagen), 2.5 µL of ultrapure water, 5 µL of 5 × primer/probe mix and 5 µL DNA isolate (DNA concentration between 5 and 20 ng/µL). For using the QuantiTect Multiplex PCR NoROX Master Mix on the ABI 7500 Real-time PCR System, 1.8 mL of the mix was pre-mixed with 2 µL ROX Reference Dye (25 µM) (Invitrogen by Life Technologies, Carlsbad, CA, USA). The final ROX concentration per PCR reaction was 14 nM. The temperature program was initiated with a denaturation step at 95 °C for 15 min, followed by 40 cycles at 94 °C for 1 min and 60 °C for 1 min.
DNA isolates from the 35 commercial food products were diluted with water to a concentration of 10 ng/µL. In addition to the samples, positive controls were added to each PCR plate. The positive control for assayChr9W and assayChr9D contained 0.2% (w/w) wild boar DNA, 0.5% (w/w) domestic pig DNA and 99.3% (w/w) herring sperm DNA. In case of assayChr1W and assayChr1D, a mixture consisting of 2% (w/w) wild boar DNA, 5% (w/w) domestic pig DNA and 93% (w/w) herring sperm DNA was used.
Limit of quantification (LOQ)
The LOQ was determined as recommended by the European Network of GMO Laboratories24. The LOQ was defined as the lowest concentration which could be determined with a RSD ≤ 25%. AssayChr9W and assayChr1W were calibrated with a mixture (20 ng/µL) of 25% (w/w) wild boar DNA and 75% (w/w) herring sperm DNA (non-target DNA), assayChr9D and assayChr1D with a mixture (20 ng/µL) of 25% (w/w) domestic pig DNA and 75% (w/w) herring sperm DNA (non-target DNA). The mixtures were analyzed both undiluted and serially diluted (1:4; 1:16; 1:64; 1:256; 1:1,024; in water) in triplicates. The following DNA mixtures (5 ng/µL) were used: assayChr9W and assayChr1W: 1%, 0.5%, 0.4%, 0.3%, 0.2% and 0.1% (w/w) wild boar DNA in herring sperm DNA; assayChr9D: 5%, 4%, 3%, 2%, 1% and 0.5% (w/w) domestic pig DNA in herring sperm DNA and assayChr1D: 1%, 0.5%, 0.4%, 0.3%, 0.2% and 0.1% (w/w) domestic pig DNA in herring sperm DNA.
In addition, the LOQ of the assays for wild boar and domestic pig was determined by using domestic pig and wild boar DNA as background, respectively.
AssayChr9W and assayChr1W were calibrated with a mixture (20 ng/µL) containing 25% (w/w) wild boar DNA and 75% (w/w) domestic pig DNA, assayChr9D with a mixture (20 ng/µL) containing 25% (w/w) domestic pig DNA and 75% (w/w) wild boar DNA and assayChr1D with a mixture (20 ng/µL) containing 50% (w/w) domestic pig DNA and 50% (w/w) wild boar DNA. The mixtures were analyzed undiluted and serially diluted (1:4; 1:16; 1:64; 1:256; 1:1,024; in water) in triplicates. The following DNA mixtures (5 ng/µL) were used: assayChr9W: 1%, 0.5%, 0.4%, 0.3%, 0.2% and 0.1% (w/w) wild boar DNA in domestic pig DNA; assayChr9D: 5%, 4%, 3%, 2%, 1% and 0.5% (w/w) domestic pig DNA in wild boar DNA; assayChr1W: 20%, 10%, 5%, 4%, 3% and 2% (w/w) wild boar DNA in domestic pig DNA; assayChr1D: 40%, 20%, 10%, 5%, 4% and 3% (w/w) domestic pig DNA in wild boar DNA.
Quantitative analysis of samples
The content of wild boar and domestic pig in meat extract mixtures, model game sausages and commercial meat products was determined relatively. The DNA isolates were analyzed with the respective sub-species specific real-time PCR assay (assayChr9W, assayChr9D, assayChr1W or assayChr1D) and a reference real-time PCR assay published previously22. The reference real-time PCR assay allowed the determination of the total DNA concentration of mammalian and poultry species in the isolate.
For obtaining quantitative results, the real-time PCR assays had to be calibrated. For the analysis of meat extract mixtures, a meat extract mixture containing 25% (w/w) wild boar, 50% (w/w) domestic pig and 25% (w/w) cattle was used for calibration. PCR was carried out in six replicates. For the quantitative analysis of commercial meat products, the assays were calibrated with a meat extract mixture consisting of 30% (w/w) wild boar and 70% (w/w) domestic pig. PCR was carried out in four replicates.
The calibration mixtures were adjusted to a DNA concentration of 20 ng/µL and serially diluted in water (1:4; 1:16; 1:64; 1:256 and 1:1,024). DNA isolates from samples were diluted to a DNA concentration of 5 ng/µL. The content of wild boar and domestic pig was assessed by using the following equations:
where cDNA wild boar/domestic pig, cDNA total meat are the concentrations of wild boar DNA, domestic pig DNA and total meat DNA, respectively; Ctwild boar/domestic pig, Ctref. are the Ct values obtained with the wild boar-/domestic pig-specific assay and the reference real-time PCR assay, respectively; dwild boar/domestic pig, dref. are the intercepts of the standard curves of the wild boar-/domestic pig-specific assay and the reference real-time PCR assay, respectively; and slopewild boar/domestic pig, sloperef. are the slopes of the calibration curves of the wild boar-/domestic pig-specific assay and the reference real-time PCR assay, respectively.
In case of meat extract mixtures and model sausages, the recovery was calculated by referring the wild boar/domestic pig content determined by real-time PCR to the theoretical wild boar/domestic pig content using the following equation:
Qualitative analysis carried out with a real-time PCR targeting a fragment of chromosome 7
Experimental details are given in the Supplementary Material.
Statistical analysis
Statistical analyses were carried out with IBM SPSS Statistics Version 26.0. Concentrations < LOD and < LOQ were substituted with default values, namely half the LOD and half the LOQ, respectively, as proposed previously29. The T test was used to test for significant differences between recoveries of PCR assays. Pearson´s correlation coefficient was used to assess the relationship between results obtained with different PCR assays. In all cases, a p value < 0.05 (two-sided) was considered significant.
Use of experimental animals
We did not perform experiments on live vertebrates, we only analysed muscle meat which we obtained from dead/slaughtered animals.
References
Ballin, N. Z. Authentication of meat and meat products. Meat Sci. 86, 577–587 (2010).
Ballin, N. Z., Vogensen, F. K. & Karlsson, A. H. Species determination—Can we detect and quantify meat adulteration? Meat Sci. 83, 165–174 (2009).
Fajardo, V. et al. Differentiation of European wild boar (Sus scrofa scrofa) and domestic swine (Sus scrofa domestica) meats by PCR analysis targeting the mitochondrial D-loop and the nuclear melanocortin receptor 1 (MC1R) genes. Meat Sci. 78, 314–322 (2008).
Fontanesi, L. et al. Differentiation of meat from European wild boars and domestic pigs using polymorphisms in the MC1R and NR6A1 genes. Meat Sci. 98, 781–784 (2014).
Klomtong, P., Chaweewan, K., Phasuk, Y. & Duangjinda, M. MC1R, KIT, IGF2, and NR6A1 as markers for genetic differentiation in Thai native, wild boars, and Duroc and Chinese Meishan pigs. Genet. Mol. Res. 14, 12723–12732 (2015).
Laube, I. et al. Methods for the detection of beef and pork in foods using real-time polymerase chain reaction. Int. J. Food Sci. Tech. 38, 111–118 (2003).
Ali, M. E. et al. Analysis of pork adulteration in commercial meatballs targeting porcine-specific mitochondrial cytochrome b gene by TaqMan probe real-time polymerase chain reaction. Meat Sci. 91, 454–459 (2012).
Farrokhi, R. & Jafari Joozani, R. Identification of pork genome in commercial meat extracts for Halal authentication by SYBR green I real-time PCR. Int. J. Food Sci. Technol. 46, 951–955 (2011).
Rodríguez, M. A., García, T., González, I., Hernández, P. E. & Martín, R. TaqMan real-time PCR for the detection and quantitation of pork in meat mixtures. Meat Sci. 70, 113–120 (2005).
Köppel, R., Zimmerli, F. & Breitenmoser, A. Heptaplex real-time PCR for the identification and quantification of DNA from beef, pork, chicken, turkey, horse meat, sheep (mutton) and goat. Eur. Food Res. Technol. 230, 125–133 (2009).
Köppel, R., Ruf, J., Zimmerli, F. & Breitenmoser, A. Multiplex real-time PCR for the detection and quantification of DNA from beef, pork, chicken and turkey. Eur. Food Res. Technol. 227, 1199–1203 (2008).
Köppel, R., Ruf, J. & Rentsch, J. Multiplex real-time PCR for the detection and quantification of DNA from beef, pork, horse and sheep. Eur. Food Res. Technol. 232, 151–155 (2011).
Iwobi, A. et al. A multiplex real-time PCR method for the quantification of beef and pork fractions in minced meat. Food Chem. 169, 305–313 (2015).
Iwobi, A. et al. A multiplex real-time PCR method for the quantitative determination of equine (horse) fractions in meat products. Food Control 74, 89–97 (2017).
Mason-D’croz, D. et al. Modelling the global economic consequences of a major African swine fever outbreak in China. Nat. Food 1, 221–228 (2020).
Goedbloed, D. J. et al. Genome-wide single nucleotide polymorphism analysis reveals recent genetic introgression from domestic pigs into Northwest European wild boar populations. Mol. Ecol. 22, 856–866 (2013).
Dzialuk, A., Zastempowska, E., Skórzewski, R., Twarużek, M. & Grajewski, J. High domestic pig contribution to the local gene pool of free-living European wild boar: A case study in Poland. Mammal Res. 63, 65–71 (2018).
Rubin, C. J. et al. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA 109, 19529–19536 (2012).
Kaltenbrunner, M. et al. Differentiation between wild boar and domestic pig in food by targeting two gene loci by real-time PCR. Sci. Rep. 9, 9221 (2019).
Codex Alimentarius Austriacus, Österreichisches Lebensmittelbuch, Codexkapitel/B14/Fleisch und Fleischerzeugnisse. (2005).
Dahlke, J. (February 6, 2018). Gefährdete Schweinerassen in Deutschland. https://www.agrarheute.com/tier/schwein/gefaehrdete-schweinerassen-deutschland-451303. Accessed April 25, 2019.
Druml, B., Kaltenbrunner, M., Hochegger, R. & Cichna-Markl, M. A novel reference real-time PCR assay for the relative quantification of (game) meat species in raw and heat-processed food. Food Control 70, 392–400 (2016).
Köppel, R., Daniels, M., Felderer, N. & Brünen-Nieweler, C. Multiplex real-time PCR for the detection and quantification of DNA from duck, goose, chicken, turkey and pork. Eur. Food Res. Technol. 236, 1093–1098 (2013).
ENGL. (2015). European Network of GMO Laboratories. Definition of Minimum Performance Requirements for Analytical Methods of GMO Testing. https://gmo-crl.jrc.ec.europa.eu/guidancedocs.htm. Accessed 31 Oct 2019.
Kaltenbrunner, M., Hochegger, R. & Cichna-Markl, M. Development and validation of a fallow deer (Dama dama)-specific TaqMan real-time PCR assay for the detection of food adulteration. Food Chem. 243, 82–90 (2018).
Kaltenbrunner, M., Hochegger, R. & Cichna-Markl, M. Red deer (Cervus elaphus)-specific real-time PCR assay for the detection of food adulteration. Food Control 89, 157–166 (2018).
Kaltenbrunner, M., Hochegger, R. & Cichna-Markl, M. Sika deer (Cervus nippon)-specific real-time PCR method to detect fraudulent labelling of meat and meat products. Sci. Rep. 8, 7236 (2018).
Druml, B., Mayer, W., Cichna-Markl, M. & Hochegger, R. Development and validation of a TaqMan real-time PCR assay for the identification and quantification of roe deer (Capreolus capreolus) in food to detect food adulteration. Food Chem. 178, 319–326 (2015).
Office of Pesticide Programs, U.S. Environmental Protection Agency Washington, DC. Assigning values to non-detected/non-quantified pesticide residues in human health food exposure assessments 2000. https://archive.epa.gov/pesticides/trac/trac/science/trac3b012.pdf. Accessed 2 May 2020.
Acknowledgements
We would like to thank the Research Institute of Wildlife Ecology (Vienna, Austria), the Wildpark Ernstbrunn (Ernstbrunn, Austria) and the University of Veterinary Medicine Vienna (Vienna, Austria) for providing meat samples from different animal species. In addition, we thank the treasure project partners from Slovenia (Marjeta Čandek-Potokar, Kmetijski Inštitut Slovenije, Agricultural Institute of Slovenia), Italy (Carolina Pugliese, Università Degli Studi Firenze) and Croatia (Danijel Karolyi, Department of Animal Science, Faculty of Agriculture, University of Zagreb) (https://treasure.kis.si/home/partner/) for providing sample material from various domestic pig breeds.
Funding
This article was supported by the Open Access Publishing Fund of the University of Vienna.
Author information
Authors and Affiliations
Contributions
R.H., M.C.-M. and M.K. planned the experiments. K.K., R.E., H.R., R.H. and M.K. collected sample material. W.M., M.K, K.K., R.E. and H.R. designed the primers. M.K., K.K., R.E. and H.R. performed the experiments and analysed the data; M.K. and M.C.-M. drafted the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kaltenbrunner, M., Mayer, W., Kerkhoff, K. et al. Applicability of a duplex and four singleplex real-time PCR assays for the qualitative and quantitative determination of wild boar and domestic pig meat in processed food products. Sci Rep 10, 17243 (2020). https://doi.org/10.1038/s41598-020-72655-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-72655-7
- Springer Nature Limited