Abstract
Cytochrome P450 monooxygenases (CYPs/P450s), heme thiolate proteins, are well known for their role in organisms’ primary and secondary metabolism. Research on eukaryotes such as animals, plants, oomycetes and fungi has shown that P450s profiles in these organisms are affected by their lifestyle. However, the impact of lifestyle on P450 profiling in bacteria is scarcely reported. This study is such an example where the impact of lifestyle seems to profoundly affect the P450 profiles in the bacterial species belonging to the phylum Firmicutes. Genome-wide analysis of P450s in 972 Firmicutes species belonging to 158 genera revealed that only 229 species belonging to 37 genera have P450s; 38% of Bacilli species, followed by 14% of Clostridia and 2.7% of other Firmicutes species, have P450s. The pathogenic or commensal lifestyle influences P450 content to such an extent that species belonging to the genera Streptococcus, Listeria, Staphylococcus, Lactobacillus, Lactococcus and Leuconostoc do not have P450s, with the exception of a handful of Staphylococcus species that have a single P450. Only 18% of P450s are found to be involved in secondary metabolism and 89 P450s that function in the synthesis of specific secondary metabolites are predicted. This study is the first report on comprehensive analysis of P450s in Firmicutes.
Similar content being viewed by others
Introduction
Among the bacteria that inhabit the human gut, species belonging to the bacterial phylum Firmicutes and Bacteroides are dominant1,2,3. Firmicutes species are gram-positive microorganisms with rod or sphere shapes and reproduce via binary fission. This phylum contains bacteria possessing diverse characteristics that are adapted to diverse ecological niches. Some members are saprophytes that live in soil and aquatic environments, performing mainly decomposition and recycling of organic matter, some members are commensals of humans and some members are pathogens of animals, including humans, and plants4. Some members have been subjected to thorough investigation to gain understanding of endospore formation and survival5 and the biotechnological potential of these organisms has been explored for the production of dairy products6, enzymes7 and antibiotics8.
The Firmicutes phylum is further divided into seven subphyla, namely Bacilli, Clostridia, Erysipelotrichia, Limnochordia, Negativicutes, Thermolithobacteria, and Tissierellia4. Species belonging to the subphyla Bacilli are well known for the production of secondary metabolites valuable to humans, organic compounds that have medicinal properties (Table 1). Clostridia consist of species that produce short chain fatty acids in the human gut, such as butyrate, which is essential fuel for colonocytes (enterocytes referred to as colonocytes in the colon)9 and also species causing botulism, tetanus, gas gangrene, food poisoning, pseudomembranous colitis, and antibiotic-associated diarrhea in humans10,11. Quite a number of studies have explored the relation between the changes in the percentage of Firmicutes species in the human gut and human conditions such as obesity, inflammatory bowel disease, systemic lupus erythematosus and psoriasis but results are not conclusive2,12,13.
Because of the potential biotechnological applications of secondary metabolites produced mostly by Bacillus species (Table 1), comprehensive analysis of cytochrome P450 monooxygenases (CYPs/P450s) in the species belonging to the genus Bacillus has recently been carried out and their association with secondary metabolism has been unraveled30. P450s are heme thiolate proteins that play an important role in organisms’ primary and secondary metabolism. Because of their diverse enzymatic reactions, P450s are found to play key roles such as conferring diversity on secondary metabolites31,32. The study revealed the presence of 507 P450s belonging to 13 P450 families and 28 P450 subfamilies in 128 Bacillus species and 112 P450s were found to be part of secondary metabolite biosynthetic gene clusters (BGCs)30. BGCs are groups of genes clustered together that are responsible for producing secondary metabolites in organisms33. However, this study was limited to the genus Bacillus. None of the species belonging to other genera or a comprehensive analysis of P450s in Firmicutes species has been reported.
Studies on the role of P450s in organism’s adaptations or changes in P450 profiles according to an organism’s life have been conducted on eukaryotic organisms such as fungi34,35,36,37,38,39,40,41, oomycetes42, plants43, and animals44. However, this phenomenon is scarcely reported in prokaryotes; currently data are available only for bacterial species belonging to the genera Streptomyces45,46, Mycobacterium47,48 and the phylum Cyanobacteria49, suggesting that more research is needed to unravel P450s’ role in other bacterial species. The presence of species with diverse characteristics that are adapted to diverse ecological niches, such as in Firmicutes, will enhance understanding of the role of P450s in bacterial species, especially with respect to their role in those organism’s adaptations. Since Firmicutes species have diverse lifestyles, it would be interesting to see if there are any changes in P450 profiles with respect to their lifestyle, as studies have shown that organisms lose a considerable number of P450s owing to their lifestyle. Examples are adapting to utilize simple carbon sources, as observed in Saccharomyces species37, or readily available abundant carbon sources in the host, as observed in mycobacterial species47, or making more copies of specific P450s both at P450 family and subfamily level in their genomes (P450 blooms) owing to adaptation to different ecological niches34,36,38,39,40,41,42,44.
In order to address the above fascinating research gaps on the impact of lifestyle on Firmicutes species P450s, if any, in this study comprehensive analysis of P450s and their association with secondary metabolism in 972 Firmicutes species belonging to 158 genera has been carried out.
Results and discussion
Only a few Firmicutes species have P450s
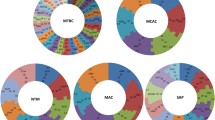
Comprehensive genome-wide data mining and annotation of P450s in 972 Firmicutes species (Table S1) revealed that only 229 species have P450s in their genomes (Fig. 1A). This indicates that most of the Firmicutes species do not have P450s in their genomes (Fig. 1A). Subphylum level analysis revealed that 38% of Bacilli species have P450s (Fig. 1B). In contrast to Bacilli, only 14% and 2.7% of Clostridia and other Firmicutes species have P450s (Fig. 1B). Owing to the availability of few species genomes that belong to the other five Firmicutes subphyla (Erysipelotrichia, Limnochordia, Negativicutes, Thermolithobacteria, and Tissierellia), these species were kept under “Others” at KEGG50. Thus, we indicated the P450 profiles in these species under the name “other species”. Only one species (Limnochorda pilosa) among 37 species in this category had P450s. Analysis of the Firmicutes genera disclosed that of the 158 genera, species belonging to 37 genera have P450s in their genomes (Fig. 1C). Most of the species belonging to the genus Bacillius have P450s, followed by Paenibacillus and Clostridium (Fig. 1C). Based on the number of species used in the study, we can safely conclude that species belonging to the genera Streptococcus, Lactobacillus, Listeria, Geobacillus, Lactococcus and Leuconostoc do not have P450s (Fig. 1C). Furthermore, among 86 Staphylococcus species only nine species have P450s (Fig. 1C). Information on genera, species and the number of P450s in a species is presented in Table S1. On average three P450s were found in 229 species, whereas 92 species had a single P450 in their genome (Table S1). Among the Firmicutes, species belonging to the genus Paenibacillus had the highest number of P450s in their genomes. P. mucilaginosus 3016 had the highest number of P450s (11 P450s) in its genome, followed by P. mucilaginosus K02 and P. mucilaginosus KNP414 (10 P450s each) (Table S1). Comparative analysis of different bacterial species revealed that Firmicutes species had the lowest average number of P450s in their genomes compared to the bacterial species belonging to the genera Streptomyces and Mycobacterium and the phylum Cyanobacteria (Table 2). The absence of P450s in species belonging to the genera Streptococcus, Lactobacillus, Listeria, Lactococcus, Leuconostoc or the presence of a single P450 in a handful of Staphylococcus species strongly suggests that the impact of lifestyle profoundly influenced the P450 repertoire in the species, as also observed in mycobacterial species47 and in Saccharomyces species37. The pathogenic lifestyle of species belonging to the genera Streptococcus, Listeria and Staphylococcus led them to adapt readily available carbon sources in the host and thus possibly led to the loss of P450s, similarly to mycobacterial species 47. Species belonging to the genera Lactobacillus, Lactococcus and Leuconostoc are involved in fermentation processing (industrial scale or inside the human gut), indicating that adaption to thrive on simple carbon sources led to the loss of P450s, just as observed in Saccharomyces species37.
Analysis of P450s in Firmicutes species. Comparative analysis of P450s at species level (A), subphylum level (B) and genera level (C) is presented in the figure. Owing to the availability of few species genomes belonging to the other five subphyla of Firmicutes (Erysipelotrichia, Limnochordia, Negativicutes, Thermolithobacteria, and Tissierellia), these species were kept under “Others” at KEGG50. Thus, we indicated the P450 profiles in these species under the name Other species. Numbers next to the bars indicate the number of species. Detailed information on Firmicutes species P450 profiles is presented in Table S1.
Firmicutes species have the lowest P450 diversity
Firmicutes species P450s were grouped into different P450 families and P450 subfamilies following the International P450 Nomenclature Committee rules that include phylogenetic analysis of P450s (Fig. 2)51,52,53. Based on the percentage identity of > 40% for a family and > 55% for a subfamily and following the evolutionary analysis where P450s belonging to the same family grouped together (Fig. 2), all 712 P450s found in 229 Firmicutes species were grouped into 14 P450 families and 53 P450 subfamilies (Table 3). Firmicutes species P450s identified in this study, along with their protein sequences and species, are presented in Supplementary Dataset 1. The number of P450 families found in Firmicutes species is very low compared to other bacterial species (Table 2), indicating the lowest P450 diversity. As predicted, P450 families such as CYP107, CYP102, CYP152, CYP109 and CYP106 were expanded in Firmicutes species, as the percentage contribution of these families to the total number of P450s was higher compared to the rest of the P450 families (Table 3). Among P450 families, CYP107 had the highest number of P450s (199 P450s), contributing 28% of 712 P450s, followed by CYP102 (179 P450s), CYP152 (110 P450s), CYP109 (95 P450s) and CYP106 (57 P450s) (Table 3). Blooming of certain P450 families in species is a common phenomenon and is observed in species belonging to different biological kingdoms as a potential indication of adaptation to an ecological niche34,36,38,39,40,41,42,44. Comparative analysis of dominant P450 families across different bacterial species revealed that the CYP107 family is dominantly present in Firmicutes species and Streptomyces species and CYP125 and CYP110 are dominantly present in mycobacterial and cyanobacterial species, respectively (Table 2).
Phylogenetic tree of Firmicutes species P450s. Different P450 families are indicated with different colours. A high-resolution phylogenetic tree is provided in Supplementary Dataset 2.
Analysis of P450 subfamilies revealed subfamily-level blooming in Firmicutes species as some subfamilies were expanded in a family (Table 3). CYP107 had the highest number of P450 subfamilies (12 subfamilies), followed by CYP152 (11 subfamilies), CYP109 (10 subfamilies), CYP197 (6 subfamilies), CYP106 (3 subfamilies) and CYP134 and CYP1731 and (Table 3). Seven of 14 P450 families had a single subfamily (Table 3). The dominant subfamilies in expanded P450 families were the following: CYP107 family had the subfamily “J” as the dominant subfamily; subfamily “B” was dominant in P450 families CYP106 and CYP109 and subfamily “A” was dominant in the CYP152 family. It is interesting to note that the CYP102 P450 family, despite contributing the second largest number of P450s, had a single subfamily “A” (Table 3). Blooming of certain P450 subfamilies in a family was also observed in species of different biological kingdoms and it is assumed that the P450 blooms possibly bestow certain advantages on organisms in adapting to particular ecological niches34,36,38,39,40,41,42,44. Analysis of conservation of P450 families in 229 Firmicutes species revealed that none of the 14 P450 families was conserved in these species (Fig. 3). However, based on the heat-map profile of P450 families, the P450 families CYP152, CYP107, CYP012 and CYP109 were found to be a co-presence in most Firmicutes species (Fig. 3). Cyanobacterial species also had no P450 family conserved, but P450 families CYP110 and CYP120 were found to be a co-presence in most of these species49. the large CYP107 family was found to be conserved in Streptomyces species46 and quite a number of P450 families were found to be conserved in mycobacterial species47.
Analysis of presence of P450 family (red) or its absence (green) in 229 Firmicutes species. Two hundred and twenty-nine Firmicutes species form the horizontal axis and P450 family numbers form the vertical axis. The data used in the generation of this figure are presented in Supplementary Dataset 3.
A small proportion of P450s are involved in secondary metabolism in Firmicutes species
Among 712 P450s identified in 229 Firmicutes species, 125 P450s (18%) of 62 Firmicutes species were found to be part of secondary metabolite BGCs (Fig. 4 and Table S2). P450s that were part of BGCs were from species belonging to the subphylum Bacilli and most of these species belonged to the genera Bacillus (49 species) (Table S2). Most of the Firmicutes species P450 families were found to be part of BGCs (Fig. 4A). Among 14 P450 families, ten families, namely CYP107, CYP113, CYP134, CYP152, CYP102, CYP109, CYP1706, CYP106, CYP1179 and CYP197, were found to be part of different secondary metabolite BGCs (Fig. 4A). Among these families, P450s belonging to the CYP107 family were dominantly present in BGCs with more than half of the P450s (55%) were part of BGCs (Fig. 4A). A point to be noted is that P450 families such as CYP107, CYP152, CYP102 and CYP109 are expanded in Firmicutes and part of the BGCs also clearly support a previous hypothesis that “species populate specific P450s if they are useful in their adaptation to certain ecological niches or useful in their physiology”34,36,38,39,40,41,42,44. Interestingly, some P450 families such as CYP113, CYP134, CYP197, CYP1706 and CYP1179 that are scarcely present in Firmicutes species but they are also part of BGCs, indicating their important role in producing these metabolites.
Comparative analysis of P450s associated with secondary metabolism in Firmicutes species. (A) Comparative analysis of P450 families that are part of secondary metabolite biosynthetic gene clusters (BGCs). The P450 family name, number of P450s and their percentage of the total number of P450s part of BGCs are presented in the figure. (B) Comparative analysis of types of BGCs. The number at the top of each bar represents the number of P450s in the type of BGC. (C) Comparative analysis of most similar known clusters that have P450s. The number at the top of each bar represents the total number of similar clusters. Detailed information is presented in Table S2.
An interesting phenomenon is observed when comparing at the subfamily level where P450 subfamilies “H” and “K” are part of BGCs and the subfamily “J” P450s are not part of BGCs despite this subfamily’s dominance in the CYP107 family (Table 3), indicating subfamily level selectivity by species. P450s were found to be part of 15 BGC types (Fig. 4B). Most of the P450s are part of BGC types such as Transatpks-Nrps (Trans-AT polyketide synthase-Non-ribosomal peptide synthetase cluster) (43 P450s), followed by Nrps-Transatpks-Otherks (Nrps-Transatpks-Other types of PKS cluster) and Transatpks (each 20 P450s) and Other (19 P450s) (Fig. 4B). Analysis of most similar known clusters revealed that the majority of P450 clusters were bacillaene (41 clusters), followed by fengycin (23 clusters) and difficidin (17 clusters) (Fig. 4C). Analysis of the association between P450 families and BGCs showed that CYP107 family P450s are mostly associated with BGCs Nrps-Transatpks-Otherks, Transatpks, Transatpks-Nrps and Other, a putative gene cluster; CYP113 family P450s are associated with Transatpks-Nrps and Transatpks BGC, and CYP134 family P450s are associated with Transatpks-Nrps and Other (Table S2). The percentage identity of Firmicutes BGCs containing P450s with similar known clusters revealed that most of the clusters have 100% sequence identity, indicating that these clusters indeed produce the expected metabolites (Table S2).
Comparative analysis of P450s involved in secondary metabolism among different bacterial species revealed that more P450s from Firmicutes species (18%) are involved in secondary metabolism compared to the P450s from mycobacterial (11%) and cyanobacterial (8%) species (Table 2). However, P450s from Firmicutes species are only second to Streptomyces species P450s (23%) in terms of their involvement in secondary metabolism (Table 2). These results strongly support the recent observation that more P450s are involved in secondary metabolism in Streptomyces species compared to other bacterial species46 and it is no surprise that two thirds of all known antibiotics in the world come from these species54.
Most Firmicutes species P450s are orphans of unknown function
Among the 712 Firmicutes species P450s, only a handful of P450s are characterized for their physiological functions. The well-known and well-studied P450 CYP102A1 (P450-BM3) from B. megaterium is found to be a fatty acid hydroxylase55,56,57. CYP152A1 from B. subtilis and CYP152K6 from B. methanolicus were found to be peroxygenases that use hydrogen peroxide to drive hydroxylation and decarboxylation of fatty acids58,59,60. CYP107H1 from B. subtilis is a pimelic acid hydroxylase involved in biotin synthesis61,62. CYP134A1 from B. subtilis is involved in synthesis of pulcherriminic acid, a natural product, by three-step oxidative transformation of the diketopiperazine cyclo-l-leucyl-l-leucyl63. Based on the P450s’ location in different BGCs and their percentage identity with known similar clusters (most of them have 100% sequence identity) (Table S2), we predict 89 P450 functions in the synthesis of different secondary metabolites such as polyketides (macrolactin, bacillaene, and difficidin), lipopeptides (surfactin, lichenysin and fengycin), phosphono-oligopeptides (rhizocticin) and siderophores (Bacillibactin) (Table 4). These secondary metabolites are well known for their potential biotechnological applications such as antibacterial, antiviral and cytotoxic properties as listed in Table 1.
Analysis of association of P450s with BGCs revealed that specific P450 orthologues are involved in the production of the same secondary metabolite (Table 4), indicating horizontal gene transfer of these BGCs among Bacillus species, a well-known phenomenon of gene-cluster transfer between bacterial species33,64,65. Prominent observations include CYP107K P450s’ involvement in biosynthesis of bacillaene, CYP113L1 P450s’ involvement in the biosynthesis of difficidin and CYP107H4 P450s’ involvement in the biosynthesis of fengycin (Table 4). Considering the high conservation of ortholog P450s, it can be assumed that BGCs such as bacillaene, difficidin and fengycin populated among Bacillus species via horizontal gene-cluster transfer.
An interesting comparison can be drawn between the role of secondary metabolites produced by Firmicutes where P450s are involved (Table 4) and the role of secondary metabolites produced by Streptomyces species45,46. In both cases, these secondary metabolites seem to be helping organisms gain the upper hand in their ecological niches, as the secondary metabolites produced by these organisms have anti-bacterial, anti-fungal and anti-viral properties, suggesting that these compounds help both the Firmicutes8 and the Streptomyces45,46 species to thrive in their ecological niches by eliminating other organisms. The structure and biological functions of bacillaene, difficidin, fengycin, lichenysin, macrolactin, rhizocticin and surfactin produced by Firmicutes were recently reviewed8.
Methods
Species and database
Nine hundred and seventy-two Firmicutes species genomes publicly available at Kyoto Encyclopedia of Genes and Genomes (KEGG)50 were used in this study. The species names, species codes, genbank accession numbers and references are presented in Table S1.
Genome data mining and annotation of P450s
Genome data mining of P450s in Firmicutes species was carried out following the standard method described elsewhere30,45,47,49. Briefly, using the information and databases presented in Table S1, the whole proteome of each Firmicutes species was downloaded and submitted to the NCBI Batch Web CD-Search Tool66. Proteins that were assigned as P450 superfamily were chosen and assessed for the presence of characteristics EXXR and CXG motifs67,68. Proteins having both motifs were selected for annotation. Annotation of P450s was accomplished following the International P450 Nomenclature Committee rules51,52,53. Proteins with an identity > 40% were classified under the same family and proteins with an identity > 55% were classified under the same subfamily. Proteins with less than 40% identity were assigned to a new P450 family. P450s and gene cluster data for Bacillus species were retrieved from a published article and used in the study30.
Phylogenetic analysis of P450s
The phylogenetic tree of Firmicutes species P450s was constructed following the method described elsewhere30,45,47,49. Briefly, the Firmicutes species P450 protein sequences were aligned using the online database, MAFFT69. Then the alignments were automatically subjected to tree inferring and optimization by the Trex web server70. Finally, the best-inferred trees were visualized, colored, and generated by iTOL71.
Generation of P450 profile heat-maps
Heat-map profiles for P450s were constructed following the method described elsewhere45,47. The heat-map profile shows the presence or absence of P450s in Firmicutes species. The data were represented as − 3 for family absence (green) and 3 for family presence (red). A tab-delimited file was loaded into MeV (Multi-experiment viewer) using a two-color array72. Hierarchical clustering using a Euclidean distance metric was used to cluster the data. Fourteen CYP families formed the vertical axis and 229 Firmicutes species formed the horizontal axis.
Secondary metabolite BGCs analysis and P450s identification
Secondary metabolite BGCs analysis and P450 identification in Firmicutes species were carried out following the method described elsewhere45,47. Briefly, each Firmicutes species genome ID presented in Table S1 was submitted to anti-SMASH73 for the identification of secondary metabolite BGCs. The results were downloaded both in the form of gene cluster sequences and Excel spreadsheets. All gene cluster sequences for each species was downloaded and captured in a separate Word file. Using these data, P450s that were part of a specific gene cluster were identified. Standard gene cluster abbreviation terminology available at the anti-SMASH database73 was maintained in this study.
Bacterial P450s and gene cluster data
P450s and their BGCs data for Streptomyces species45,46, mycobacterial species47 and cyanobacterial species49 were retrieved from published articles and used for comparative analysis.
Functional prediction of P450s
Functional prediction of P450s was carried out based on their location in a particular BGC and the percentage sequence identity of BGCs with characterized gene clusters where more than > 70% identity was set as a cut-off value. However, in the case of Firmicutes species most BGCs have 100% sequence identity with the characterized gene cluster, indicating P450s’ definite role in the production of a specific secondary metabolite.
Conclusions
Cytochrome P450 monooxygenases (CYPs/P450s) have been well known to biologists for more than five decades. These enzymes perform enzymatic reactions in stereo- and regio-specific manner and have thus gained attention in the production of chemicals valuable to human beings. Studies indicated that P450s performing such enzymatic reactions help organisms to adapt to specific ecological niches. This includes loss of P450s if an organism adapts to a lifestyle where abundant simple carbon sources are present. In this study, we examined such a phenomenon where Firmicutes species that are pathogens and adapted to simple lifestyles lost P450s in their genomes. This strongly supports the earlier hypothesis put forward by our laboratory that the impact of lifestyle shapes P450 content in an organism. Analysis of P450s in Firmicutes species revealed the presence of the lowest number of P450s and the lowest P450 diversity in these species compared to bacterial species belonging to the genera Streptomyces and Mycobacterium and from the phylum Cyanobacterium. The lowest P450 diversity is due to the P450 family blooming/expansion especially in P450 families such as CYP107, CYP102, CYP152, CYP109 and CYP106. Firmicutes species were found to have the second highest number of BGCs in their genomes after Streptomyces. Most of the P450s belonging to the expanded families were found to be part of BGCs indicating that these P450s were favored by these species, possibly to gain some advantage, as observed in other organisms. Interestingly, the preference of certain P450 subfamilies for being part of BGCs were found in Firmicutes species despite these P450 subfamilies not being expanded. Based on the presence of ortholog P450s in Firmicutes species and the highest percentage of sequence identity with characterized BGCs, in this study we successfully predicted 89 P450s’ function in the synthesis of different secondary metabolites. Interestingly, most of these P450s are involved in the synthesis of bacillaene and difficidin (polyketides) and fengycin (lipopeptide). Based on the P450 profiles among species belonging to the same phylum and their lifestyles, one can connect P450 profiles to their lifestyles and this concept has been successfully demonstrated in eukaryotes and in some prokaryotes. It is worth mentioning that the phylogenetic age of bacteria from old to young is Firmicutes, Streptomyces, Cyanobacteria and Mycobacterium74, indicating that there is no link with the number and diversity of P450s and the age of the bacteria and thus P450 profiles may be more strongly related to the lifestyle. Studies are in progress to determine the impact of lifestyle in other bacterial species.
References
Arumugam, M. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011).
Dieterich, W., Schink, M. & Zopf, Y. Microbiota in the gastrointestinal tract. Med. Sci. 6, 116 (2018).
Thursby, E. & Juge, N. Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836 (2017).
Seong, C. N. et al. Taxonomic hierarchy of the phylum Firmicutes and novel Firmicutes species originated from various environments in Korea. J. Microbiol. 56, 1–10 (2018).
Filippidou, S. et al. A combination of extreme environmental conditions favor the prevalence of endospore-forming Firmicutes. Front. Microbiol. 7, 1707 (2016).
Bintsis, T. Lactic acid bacteria as starter cultures: an update in their metabolism and genetics. AIMS Microbiol. 4, 665 (2018).
Verma, D. K. et al. Bioprocessing Technology in Food and Health: Potential Applications and Emerging Scope 97–140 (Apple Academic Press, Burlington, 2018).
Caulier, S. et al. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 10, 302 (2019).
Lopetuso, L. R., Scaldaferri, F., Petito, V. & Gasbarrini, A. Commensal Clostridia: leading players in the maintenance of gut homeostasis. Gut Pathogens 5, 23 (2013).
Rood, J. I. General physiological and virulence properties of the pathogenic Clostridia. In Clostridial diseases of animals (eds. Uzal, F.A., Prescott, J.F., Songer, J.G. and Popoff, M.R.) 7–12 (Wiley, 2016).
Wells, C. L. & Wilkins, T. D. Medical Microbiology 4th edn. (University of Texas Medical Branch, Galveston, 1996).
Kho, Z. Y. & Lal, S. K. The human gut microbiome—a potential controller of wellness and disease. Front. Microbiol. 9, 1835 (2018).
Koliada, A. et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 17, 1–6 (2017).
Yuan, J. et al. Antibacterial compounds—macrolactin alters the soil bacterial community and abundance of the gene encoding PKS. Front. Microbiol. 7, 1904–1904. https://doi.org/10.3389/fmicb.2016.01904 (2016).
Romero-Tabarez, M. et al. 7-O-malonyl macrolactin A, a new macrolactin antibiotic from Bacillus subtilis active against methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, and a small-colony variant of Burkholderia cepacia. Antimicrob. Agents Chemother. 50, 1701–1709 (2006).
Moldenhauer, J., Chen, X. H., Borriss, R. & Piel, J. Biosynthesis of the antibiotic bacillaene, the product of a giant polyketide synthase complex of the trans-AT family. Angew. Chem. Int. Ed. Engl. 46, 8195–8197. https://doi.org/10.1002/anie.200703386 (2007).
Butcher, R. A. et al. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc. Natl. Acad. Sci. 104, 1506–1509 (2007).
Wu, L. et al. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 5, 12975 (2015).
Chen, X.-H. et al. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 188, 4024–4036 (2006).
Yang, J. et al. Genomics-inspired discovery of three antibacterial active metabolites, aurantinins B, C, and D from compost-associated Bacillus subtilis fmb60. J. Agric. Food Chem. 64, 8811–8820 (2016).
Kang, H. K., Seo, C. H. & Park, Y. Marine peptides and their anti-infective activities. Mar. Drugs 13, 618–654. https://doi.org/10.3390/md13010618 (2015).
Sur, S., Romo, T. D. & Grossfield, A. Selectivity and mechanism of fengycin, an antimicrobial lipopeptide, from molecular dynamics. J. Phys. Chem. B 122, 2219–2226. https://doi.org/10.1021/acs.jpcb.7b11889 (2018).
Knight, C. A. et al. The first report of antifungal lipopeptide production by a Bacillus subtilis subsp. inaquosorum strain. Microbiol. Res. 216, 40–46 (2018).
Peypoux, F., Bonmatin, J. M. & Wallach, J. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 51, 553–563. https://doi.org/10.1007/s002530051432 (1999).
Grangemard, I., Wallach, J., Maget-Dana, R. & Peypoux, F. Lichenysin: a more efficient cation chelator than surfactin. Appl. Biochem. Biotechnol. 90, 199–210 (2001).
Zhou, M. et al. Bacillibactin and bacillomycin analogues with cytotoxicities against human cancer cell lines from marine Bacillus sp. PKU-MA00093 and PKU-MA00092. Mar. Drugs 16, 22 (2018).
Hertlein, G. et al. Production of the catechol type siderophore bacillibactin by the honey bee pathogen Paenibacillus larvae. PLoS ONE 9, e108272 (2014).
Gu, Q. et al. Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant–pathogenic fungus Fusarium graminearum. Appl. Environ. Microbiol. 83, e01075-17 (2017).
Barsby, T., Kelly, M. T. & Andersen, R. J. Tupuseleiamides and basiliskamides, mew acyldipeptides and antifungal polyketides produced in culture by a Bacillus l aterosporus isolate obtained from a tropical marine habitat. J. Nat. Prod. 65, 1447–1451 (2002).
Mthethwa, B. C. et al. Comparative analyses of cytochrome P450s and those associated with secondary metabolism in Bacillus species. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19113623 (2018).
Podust, L. M. & Sherman, D. H. Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 29, 1251–1266. https://doi.org/10.1039/c2np20020a (2012).
Greule, A., Stok, J. E., De Voss, J. J. & Cryle, M. J. Unrivalled diversity: the many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat. Prod. Rep. 35, 757–791. https://doi.org/10.1039/c7np00063d (2018).
Medema, M. H. et al. Minimum information about a biosynthetic gene cluster. Nat. Chem. Biol. 11, 625–631. https://doi.org/10.1038/nchembio.1890 (2015).
Ngwenya, M. L. et al. Blooming of unusual cytochrome P450s by tandem duplication in the pathogenic fungus Conidiobolus coronatus. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19061711 (2018).
Matowane, R. G. et al. In silico analysis of cytochrome P450 monooxygenases in chronic granulomatous infectious fungus Sporothrix schenckii: special focus on CYP51. Biochim. Biophys. Acta Proteins Proteom. 1866, 166–177. https://doi.org/10.1016/j.bbapap.2017.10.003 (2018).
Qhanya, L. B. et al. Genome-wide annotation and comparative analysis of cytochrome P450 monooxygenases in Basidiomycete biotrophic plant pathogens. PLoS ONE 10, e0142100. https://doi.org/10.1371/journal.pone.0142100 (2015).
Kgosiemang, I. K. R., Syed, K. & Mashele, S. S. Comparative genomics and evolutionary analysis of cytochrome P450 monooxygenases in fungal subphylum Saccharomycotina. J. Pure Appl. Microbiol. 8, 12 (2014).
Jawallapersand, P. et al. Cytochrome P450 monooxygenase CYP53 family in fungi: comparative structural and evolutionary analysis and its role as a common alternative anti-fungal drug target. PLoS ONE 9, e107209. https://doi.org/10.1371/journal.pone.0107209 (2014).
Syed, K., Shale, K., Pagadala, N. S. & Tuszynski, J. Systematic identification and evolutionary analysis of catalytically versatile cytochrome P450 monooxygenase families enriched in model basidiomycete fungi. PLoS ONE 9, e86683. https://doi.org/10.1371/journal.pone.0086683 (2014).
Suzuki, H. et al. Comparative genomics of the white-rot fungi, Phanerochaete carnosa and P. chrysosporium, to elucidate the genetic basis of the distinct wood types they colonize. BMC Genom. 13, 444. https://doi.org/10.1186/1471-2164-13-444 (2012).
Akapo, O. O. et al. Distribution and diversity of cytochrome P450 monooxygenases in the fungal class Tremellomycetes. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20122889 (2019).
Sello, M. M. et al. Diversity and evolution of cytochrome P450 monooxygenases in Oomycetes. Sci. Rep. 5, 11572. https://doi.org/10.1038/srep11572 (2015).
Hamberger, B. & Bak, S. Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos. Trans. R. Soc. B Biol. Sci. 368, 20120426 (2013).
Feyereisen, R. Insect Molecular Biology and Biochemistry 236–316 (Elsevier, Amsterdam, 2012).
Senate, L. M. et al. Similarities, variations, and evolution of cytochrome P450s in Streptomyces versus Mycobacterium. Sci. Rep. 9, 3962. https://doi.org/10.1038/s41598-019-40646-y (2019).
Mnguni, F. C. et al. More P450s are involved in secondary metabolite biosynthesis in Streptomyces compared to Bacillus, Cyanobacteria and Mycobacterium. Int. J. Mol. Sci. 21, 4814. https://doi.org/10.3390/ijms19113623 (2020).
Parvez, M. et al. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: special focus on mycobacterial P450s. Sci. Rep. 6, 33099. https://doi.org/10.1038/srep33099 (2016).
Syed, P. R. et al. Cytochrome P450 monooxygenase CYP139 family involved in the synthesis of secondary metabolites in 824 mycobacterial species. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20112690 (2019).
Khumalo, M. J. et al. Comprehensive analyses of cytochrome P450 monooxygenases and secondary metabolite biosynthetic gene clusters in Cyanobacteria. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21020656 (2020).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. https://doi.org/10.1093/nar/gkv1070 (2015).
Nelson, D. R. et al. The P450 superfamily: update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 12, 1–51. https://doi.org/10.1089/dna.1993.12.1 (1993).
Nelson, D. R. Cytochrome P450 nomenclature. Methods Mol. Biol. (Clifton, N.J.) 107, 15–24. https://doi.org/10.1385/0-89603-519-0:15 (1998).
Nelson, D. R. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. (Clifton, N.J.) 320, 1–10. https://doi.org/10.1385/1-59259-998-2:1 (2006).
de Lima Procópio, R. E., da Silva, I. R., Martins, M. K., de Azevedo, J. L. & de Araújo, J. M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 16, 466–471 (2012).
Munro, A. W. et al. P450 BM3: the very model of a modern flavocytochrome. Trends Biochem. Sci. 27, 250–257 (2002).
Li, H. & Poulos, T. L. The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat. Struct. Biol. 4, 140–146 (1997).
Noble, M., Miles, C., Reid, G., Chapman, S. & Munro, A. Catalytic properties of key active-site mutants of flavocytochrome P-450 BM3. Biochem. Soc. Trans. 27, 44–44 (1999).
Lee, D.-S. et al. Crystallization and preliminary X-ray diffraction analysis of fatty-acid hydroxylase cytochrome P450BSβ from Bacillus subtilis. Acta Crystallogr. Sect. D Biol. Crystallogr. 58, 687–689 (2002).
Lee, D.-S. et al. Substrate recognition and molecular mechanism of fatty acid hydroxylation by cytochrome P450 from Bacillus subtilis crystallographic, spectroscopic, and mutational studies. J. Biol. Chem. 278, 9761–9767 (2003).
Girvan, H. M. et al. Structural and catalytic properties of the peroxygenase P450 enzyme CYP152K6 from Bacillus methanolicus. J. Inorg. Biochem. 188, 18–28 (2018).
Bower, S. et al. Cloning, sequencing, and characterization of the Bacillus subtilis biotin biosynthetic operon. J. Bacteriol. 178, 4122–4130 (1996).
Cryle, M. J. & Schlichting, I. Structural insights from a P450 carrier protein complex reveal how specificity is achieved in the P450BioI ACP complex. Proc. Natl. Acad. Sci. 105, 15696–15701 (2008).
Cryle, M. J., Bell, S. G. & Schlichting, I. Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: a cyclo-L-leucyl-L-leucyl dipeptide oxidase. Biochemistry 49, 7282–7296 (2010).
Cimermancic, P. et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421 (2014).
Tran, P. N., Yen, M. R., Chiang, C. Y., Lin, H. C. & Chen, P. Y. Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi. Appl. Microbiol. Biotechnol. 103, 3277–3287. https://doi.org/10.1007/s00253-019-09708-z (2019).
Marchler-Bauer, A. et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200-d203. https://doi.org/10.1093/nar/gkw1129 (2017).
Syed, K. & Mashele, S. S. Comparative analysis of P450 signature motifs EXXR and CXG in the large and diverse kingdom of fungi: identification of evolutionarily conserved amino acid patterns characteristic of P450 family. PLoS ONE 9, e95616. https://doi.org/10.1371/journal.pone.0095616 (2014).
Graham, S. E. & Peterson, J. A. How similar are P450s and what can their differences teach us?. Arch. Biochem. Biophys. 369, 24–29 (1999).
Katoh, K., Kuma, K., Toh, H. & Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. https://doi.org/10.1093/nar/gki198 (2005).
Boc, A., Diallo, A. B. & Makarenkov, V. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 40, W573–W579. https://doi.org/10.1093/nar/gks485 (2012).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259 (2019).
Saeed, A. I. et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. https://doi.org/10.2144/03342mt01 (2003).
Weber, T. et al. antismash 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, 237–243. https://doi.org/10.1093/nar/gkv437 (2015).
Battistuzzi, F. U., Feijao, A. & Hedges, S. B. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 4, 44 (2004).
Acknowledgements
Tiara Padayachee and Nomfundo Nzuza thank the Department of Science and Technology—National Research Foundation (DST-NRF), South Africa for Master’s scholarships (Grant numbers MND190619448759 and MND190626451135, respectively). Khajamohiddin Syed expresses sincere gratitude to the NRF, South Africa for a research grant (Grant number 114159) and University of Zululand (Grant number C686). The authors want to thank Barbara Bradley, Pretoria, South Africa for English language editing.
Author information
Authors and Affiliations
Contributions
K.S. designed, conceptualized and provided funding for the study. All authors were involved in generation, analysis and interpretation of data. All authors reviewed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Padayachee, T., Nzuza, N., Chen, W. et al. Impact of lifestyle on cytochrome P450 monooxygenase repertoire is clearly evident in the bacterial phylum Firmicutes. Sci Rep 10, 13982 (2020). https://doi.org/10.1038/s41598-020-70686-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-70686-8
- Springer Nature Limited
This article is cited by
-
A novel alkane monooxygenase evolved from a broken piece of ribonucleotide reductase in Geobacillus kaustophilus HTA426 isolated from Mariana Trench
Extremophiles (2024)
-
The gut microbiome and metabolome of Himalayan Griffons (Gyps himalayensis): insights into the adaptation to carrion-feeding habits in avian scavengers
Avian Research (2021)