Abstract
Chlorella is a unicellular green microalga that has been used in fields such as bioenergy production and food supplementation. In this study, two promoters of N (nitrogen) deficiency-inducible Chlorella vulgaris N Deficiency Inducible (CvNDI) genes were isolated from Chlorella vulgaris UTEX 395. These promoters were used for the production of a recombinant protein, human granulocyte-colony stimulating factor (hG-CSF) in Chlorella vulgaris UTEX 395 and Chlorella sp. ArM0029B. To efficiently secrete the hG-CSF, the protein expression vectors incorporated novel signal peptides obtained from a secretomics analysis of Chlorella spp. After a stable transformation of those vectors with a codon-optimized hG-CSF sequence, hG-CSF polypeptides were successfully produced in the spent media of the transgenic Chlorella. To our knowledge, this is the first report of recombinant protein expression using endogenous gene components of Chlorella.
Similar content being viewed by others
Introduction
Chlorella is a single-celled eukaryote belonging to the green algae. It can grow phototrophically, heterotrophically, or mixotrophically to high biomass, producing different amounts of proteins and lipids1. It grows rapidly, dividing into four cells in about 6 h. Because Chlorella synthesizes large amounts of proteins and lipids, it has been used as a raw material for biodiesel and also as a food additive2,3,4. Chlorella undergoes posttranslational modifications such as protein N-glycosylation5. The use of Chlorella for the production of valuable proteins has therefore been attempted for more than two decades6,7,8,9. Chen et al.10 showed expression of rabbit neutrophil peptide using a translational enhancer omega in Chlorella ellipsoidea. Kim et al.11 produced flounder growth hormone using a 35S cauliflower mosaic virus (35S) promoter in C. ellipsoidea and showed a 25% growth increase of flounder fry. No recombinant proteins produced from Chlorella species have yet been commercialized, although the species has been studied intensively for the potential production of biodiesel. An optimized expression system for producing proteins in Chlorella is urgently needed.
Recombinant protein production studies require a highly efficient method for the transformation of Chlorella. Various transformation techniques, including electroporation, PEG transformation, particle bombardment, and Agrobacterium cocultivation, have been used for the efficient transformation of Chlorella spp.10,11,12,13,14. Recently, Kumar et al.15 reported an effective method for enhancing transformation efficiency by more than 100-fold using electroporation combined with efficient protoplasting of Chlorella.
Besides efficient transformation, an optimized system for the endogenous expression of proteins is indispensable. An expression system involving appropriate promoters is one of the most important factors for high-efficiency production of recombinant proteins. In Chlamydomonas reinhardtii, the promoter of the heat shock protein gene HSP70a has been widely used for the expression of foreign genes in C. reinhardtii16,17. In recent years, the promoter of the nitrate reductase gene has been used for recombinant protein production in Phaeodactylum tricornutum18,19. The addition of NaNO3 to growth media caused an increase in the activity of the nitrate reductase promoter, resulting in an increase in production of recombinant protein. It has been reported that both the cauliflower mosaic virus 35S promoter and the C. reinhardtii HSP70 promoter are effective in transgenic C. vulgaris15. Recently, the Photosystem I protein D (psaD) gene homologous to C. reinhardtii psaD was identified in C. vulgaris in silico20. The 0.4 kb psaD promoter region was shown to act in Nicotiana benthamiana as well as in C. reinhardtii, but not in Chlorella, although it was not as strong as the promoter of the C. reinhardtii psaD gene20. To date, the use of the promoters of the endogenous genes of Chlorella has not been studied in Chlorella.
Under N depletion conditions, Chlorella stores carbon as triglycerides and starch, causing massive lipid production in the cell21. Recently, it has been reported that protein production under these conditions depends upon the N level and the duration of treatment22. The study showed that protein productivity in N starvation media was more than 40% higher than that in N-sufficient media over 4 days of Chlorella culture. Moreover, lipid productivity under the N starvation media was least at the fourth day of incubation, indicating that N starvation could be used for enhancing recombinant protein production.
Granulocyte-colony stimulating factor (G-CSF) is a member of CSF family: hormone-like glycoproteins produced by a variety of tissues to regulate the proliferation and differentiation of neutrophilic granulocytes23,24,25. G-CSF functions as a primary regulatory factor controlling the neutrophil response to inflammatory stimuli24. Cloning and expression of a human G-CSF (hG-CSF) gene in Escherichia coli was achieved in 1986 by Amgen26. The commercialized hG-CSF, which functions as an immunostimulator, has been used for patients with congenital or acquired neutropenia, and forms of recombinant hG-CSF being sold include filgrastim, pegfilgrastim, and lenograstim25. The nonglycosylated form was reported to have similar efficacy to that of the O-glycosylated form, indicating that the glycosylation of hG-CSF might not be important for the function27.
Recently, a new Chlorella species, ArM0029B, was isolated from drift ice in the Arctic region. The organism was characterized, and its genome was partially sequenced28,29. ArM0029B grows rapidly and accumulates high levels of lipids at a range of temperatures28. ArM0029B might be more easily handled and more productive than other Chlorella species.
In this study, we developed a novel expression system controlled by endogenous gene promoters to produce desired proteins in Chlorella. Two genes induced by N deficiency were identified using RNA-Seq of Chlorella ArM0029B grown under N deficiency. The homologous genes were isolated from C. vulgaris UTEX 395 using BLAST searching. The genes were named Chlorella vulgaris N Deficiency Inducible 1 (CvNDI1) and 2 (CvNDI2). The promoter regions of those genes were used to construct expression vectors for the hG-CSF gene. Novel signal peptides (SPs) were also used for the production of target proteins in the culture media. The vector systems were successfully used to produce hG-CSF in transgenic Chlorella grown under media lacking N.
Results
Screening of N deficiency-induced CvNDI1 and CvNDI2 genes from Chlorella UTEX 395
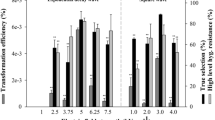
The development of an optimal promoter is a prerequisite for the production of recombinant proteins in heterologous biological systems. For the controlled expression of recombinant proteins in Chlorella, inducible promoters have been investigated as potential target promoters. N deprivation is well known to produce a dramatic increase in lipid content in C. vulgaris30, and these lipids can be used in biodiesel production. To identify the genes induced under N starvation, an arctic Chlorella sp., ArM0029B28, was subjected to RNA sequencing under N-deficient conditions. Differential expression (DE) analysis using the RNA-seq data identified 20 genes whose expression was increased more than tenfold and were thus considered significant (Supplementary Table 1). The transcript level of scaffold326G00910 was increased up to 20 times. Three genes, scaffold326G00270, 73G00080, and 253G00910, also showed significant increases in transcription levels (Supplementary Table 1). Using these four gene sequences, BLAST search was performed against the UTEX 395 sequencing data, and the four corresponding candidate homologous genes were isolated from the genome sequence of UTEX 395. To confirm the expression of those genes in the UTEX 395 under N-deficient conditions and further screen for an optimal gene among the candidates, RT-PCR was performed after 3 days cultivation of UTEX 395 in media lacking N. The numbers of transcripts of the scaffold326G00910 and 326G00270 homologs were shown to be significantly elevated under these conditions, consistent with the results of the DE analysis (Fig. 1). The expression of the scaffold326G00910 homolog was rapidly induced on day 1 in the N-deficient media, expression was maintained until day 2, and then, the expression level decreased to about half on day 3 (Fig. 1). The scaffold326G00270 homolog showed a similar expression pattern to that of the scaffold 326G00910 homolog, although the transcript of the scaffold326G00270 homolog was almost undetectable in normal growth media containing N, indicating that its expression is N starvation specific (Fig. 1). The other genes examined (scaffold37G001690 and 73G00080 homologs) were not induced by N starvation in UTEX395 (Fig. 1). To examine the rate at which the transcription of the scaffold326G00910 and 326G00270 homologs was triggered under N starvation, the expression patterns of the two genes were further checked by performing quantitative PCR (qPCR) of the genes at 0, 6, 12, and 24 h (N0, N6, N12, and N24, respectively) after N starvation treatment (Supplementary Fig. 1). Transcription of both the scaffold326G00910 and 326G00270 homologs was rapidly triggered in 6 h to about 1,500-fold and 30-fold, respectively. The transcript levels at N12 increased by up to 3,500-fold and 60-fold, respectively, compared with N0. The results indicate that the scaffold326G00910 and 326G00270 homologs are expressed quickly and strongly under N starvation conditions. Therefore, the scaffold326G00910 and 326G00270 homologs were selected for the development of a N starvation deficiency-inducible promoter gene expression system and were named CvNDI1 and CvNDI2, respectively (GenBank accession numbers MN971585 and MN971586).
Expression pattern of N starvation-induced genes screened from UTEX395 based on RNA-Seq data from ArM0029B. (A) Chlorella UTEX 395 grown in N-rich media was transferred into N-deficient media and then cultivated for 3 days. Total RNA was extracted from the cells harvested every day after the transfer, and RT-PCR was performed using scaffold-specific primers. (D0) before transfer; (D1) 1 day after transfer; (D2) 2 days after transfer; (D3) 3 days after transfer. Both CvUbi and CvAct1 were used for normalization.
The amino acid sequences of the CvNDI1 and CvNDI2 genes were deduced from the full-length DNA sequences obtained by BLAST search. BLAST searches using the amino acid sequences showed that CvNDI1 has a conserved urea carboxylase domain (Supplementary Fig. 2). CvNDI1 showed 79% homology with the urea carboxylase of Micractinium conductrix and 77% homology with that of C. sorokiniana (Supplementary Fig. 2). The biotin carboxylation domain is conserved in the N-terminal region of CvNDI1, and the carboxyltransferase domain is conserved at the C-terminal (blue and red boxes, respectively, in Supplementary Fig. 2). Urea carboxylase converts urea into ammonium, the first step for the utilization of urea as a N source31,32. Urea generated by the degradation of N compounds is known to be used as a N source in many plants, fungi, and bacteria33,34,35.
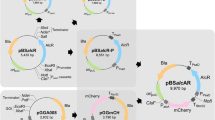
Construction of hG-CSF expression vectors controlled by CvNDI promoters. Using pJKS136 as a backbone, the RAmy3D promoter (PRAmy3D) was replaced with either CvNDI1 (pSK401 and pSK402) or CvNDI2 (pSK403 and pSK404). The hG-CSF sequence of pJKS136 (white box) was replaced with a codon-optimized hG-CSF sequence (yellow box) for Chlorella. The signal peptide of a putative cellulase (SPc), screened from our secretome data from UTEX395, was introduced into pSK401 and pSK403 for the transformation of UTEX 395. The signal peptide of a Ras-related RABF1 (SPR), screened from our secretome data of ArM0029B, was introduced into pSK402 and pSK404 for the transformation of ArM0029B. The black bar indicates a length of 100 bp.
The CvNDI2 sequence showed homologies with the ammonium transporters (AMTs) of M. conductrix and C. sorokiniana of 73% and 74%, respectively (Supplementary Fig. 3a). Hydrophobicity analysis indicated that the CvNDI2 protein contains 11 transmembrane domains, which are generally conserved among the AMT proteins of a range of organisms (Supplementary Fig. 3b). The AMT plays an important role in N metabolism by maintaining an optimal level of N in the cell36,37. Therefore, both CvNDI1 and CvNDI2 genes are expected to be involved in metabolism to support N sources for cell survival under N starvation conditions.
Molecular characterization of transgenic Chlorella spp. (a) Genomic DNA PCR of the transgenic Chlorella, UTEX 395 (left panel) and ArM0029B (right panel), using hG-CSF primers. NC, negative control; PC, positive control. Vertical black lines indicate the grouping of gels cropped from different parts of the same gel. (b) Expression of an hG-CSF transcript by N deficiency in the transgenic UTEX 395. (c) Expression of hG-CSF transcript by N deficiency in the transgenic ArM0029B. Transgenic Chlorella grown in standard media was transferred to N-deficient media and harvested at 0 days (D0), 2 days (D2), and 3 days (D3) after the transfer. CvUbi gene was used for normalization.
Construction of hG-CSF expression vectors driven by CvNDI promoters
To construct protein expression vectors driven by the promoters of the CvNDI genes, the sequences of the promoter region spanning a 1 kb-long region upstream of the translation start codon (ATG), including the 5′-untranslated region (UTR), were amplified by PCR, using the genomic DNA of UTEX 395 as templates (Supplementary Fig. 4). As shown in Fig. 2, the promoter sequences of the CvNDI1 and CvNDI2 genes were inserted into pJKS136, replacing the RAmy3D promoters.
Expression of hG-CSF polypeptides in the transgenic Chlorella spp. (a) Transgenic Chlorella cells grown in standard N-rich media were transferred to N-deficient media and harvested the following day. Total proteins from the cell lysates were separated on 12% NuPAGE bis–tris gel (left panel) and transferred to nitrocellulose membrane for western blotting (right panel) using a polyclonal antibody of hG-CSF diluted 1: 2000. M, protein size marker; (1) UTEX 395-pSK401; (2) UTEX 395-pSK403; (3) ArM0029B-pSK402; (4) ArM0029B-pSK404. (b) Total proteins of ArM0029B-pSK404 were extracted from either cell lysates (left panel) or spent media (right panel) for 3 days after transfer to the N-deficient media and used for western blotting. The media was concentrated 250-fold and used for the western blotting. M, protein size marker; C1, protein extracts of cell lysates from day 1; C2, protein extracts of cell lysates from day 2; C3, protein extracts of cell lysates from day 3; M1, media harvested day 1; M2, media harvested day 2; M3, media harvested day 3; C3′, C3 incubated at 4 °C overnight after denaturation at 95 °C with dithiothreitol; + , hG-CSF positive control. Numbers indicate the sizes of protein markers in kDa. (c) Expression of hG-CSF in the N-rich media. ArM0029B-pSK404 cells harvested on days 3 (C3), 5 (C5), 7 (C7), and 11 (C11) in the same media. Spent media harvested on day 11 (M11) was concentrated 250-fold. M, protein size marker; + , hG-CSF positive control. Numbers indicate the sizes of protein markers in kDa. Red and black arrows indicate hG-CSF monomers and dimers, respectively.
Besides the promoter, the use of an appropriate SP is essential for establishing an efficient protein expression and secretion system. The use of an appropriate SP facilitates the harvesting of proteins from liquid media38. To select appropriate SPs for Chlorella spp. UTEX 395 and ArM0029B, proteins secreted into media were purified and analyzed using mass spectrometry. Whole-genome sequence contigs of UTEX 395 and ArM0029B obtained from the NCBI (UTEX395-WGS Project accession: LDKB01; ArM0029B-WGS Project accession: JTEE01) were used for gene prediction using the AUGUSTUS software (https://augustus.gobics.de/) with Chlamydomonas reinhardtii parameters39. From this analysis, a putative cellulase in UTEX 395 and a Ras-related RABF1 in ArM0029B were selected as highly secreted proteins (Supplementary Fig. 5; manuscript in preparation). The sequences predicted using the SignalP program were MAGRITLLLCLCLVAGAAA for the cellulase and MKGALLLLLLALAASAAIA for the Ras-related RABF1 (bold letters in Supplementary Fig. 5). The SP sequences were fused in front of a codon-optimized hG-CSF sequence (Fig. 2). The vectors containing the SP of the cellulase were named pSK401 and pSK403 and were controlled by the promoters of CvNDI1 and CvNDI2, respectively. The vectors carrying the SP of the Ras-related RABF were named pSK402 and pSK404 and were driven by the CvNDI1 and CvNDI2 promoters, respectively (Fig. 2). Those vectors were introduced into Chlorella using the electroporation method described by Kumar et al.15.
Characterization of transgenic Chlorella expressing CvNDI::hG-CSF
Hygromycin-resistant colonies of UTEX 395 and ArM0029B were obtained within 4 weeks of transformation, using selection agar plates. Individual colonies were transferred to liquid media containing hygromycin and grown for 7 days. Genomic DNA from the liquid culture was subjected to PCR using hG-CSF primers (Supplementary Table 2). As shown in Fig. 3a, the incorporation of hG-CSF gene into the Chlorella spp. was confirmed in the UTEX 395 lines harboring pSK401 or pSK403 (Left panel in Fig. 3a) and the ArM0029B lines harboring pSK402 or pSK404 (Right panel in Fig. 3a). These transformants were successfully maintained for more than 1 year, which indicates that the transformation was stable, as reported previously15. To investigate whether the CvNDI1 and CvNDI2 promoters were able to induce hG-CSF expression under N deficiency conditions, the transgenic Chlorella grown in standard nitrogen-rich liquid media were transferred to N-deficient media and cultivated for 3 days. As shown in Fig. 3b and c, all transgenic lines of UTEX 395 and ArM0029B examined expressed hG-CSF under N starvation treatment. The amount of transcription of hG-CSF increased from day 1 after the induction (Fig. 3b and c). These results indicate that the 1 kb-long CvNDI promoters induced the downstream gene in the transgenic Chlorella responding to nitrogen deficiency.
hG-CSF protein was successfully produced in the transgenic Chlorella
We next examined whether the hG-CSF transcripts induced by the CvNDI promoters were translated into the hG-CSF polypeptides in the transgenic Chlorella. The production of hG-CSF was examined using the transgenic cell lysates of both UTEX 395 and ArM0029B after 1 day of transfer to the N-deficient media. As shown in Fig. 4a, hG-CSF polypeptides were detected in the transgenic Chlorella by western blotting (black arrow). The size of the bands was around 35 kDa in the dimer form, larger than the expected size of 19 kD for the monomer. This observation indicated that the expression system induced the production of hG-CSF polypeptides under N deficiency conditions, although the main form detected was the dimer.
ArM0029B harboring pSK404 was shown to strongly induce the production of hG-CSF (Fig. 4a, Lane 4). Given this result, the production of hG-CSF was further examined using transgenic ArM0029B harboring pSK404 under N deficiency conditions. Cells grown in the N-rich media for 7 days were transferred into N-free media for another 3 days. As shown in Fig. 4b, the production of hG-CSF polypeptides was induced by N deficiency, an observation that matches the accumulation pattern of hG-CSF transcripts (Fig. 3b). The polypeptides were most strongly induced at day 3 in the transgenic line, and the hG-CSF produced was successfully secreted into the spent media. The amount of secreted hG-CSF in the pSK404 line was higher than that in the pSK402 line (right panel of Fig. 4b). Overall, the expression vector system controlled by CvNDI promoters led to successful production of hG-CSF under conditions of N deficiency, indicating that the system is effective for producing a target protein in Chlorella. Commercial hG-CSF synthesized from CHO cells used as positive controls showed a major band at 19 kDa, with a minor band at 35 kDa, identical to the hG-CSF produced in Chlorella (right panel of Fig. 4b). The 35 kDa hG-CSF harvested from the transgenic Chlorella (Lane C3 of Fig. 4b) was separated into two major bands of 19 and 35 kDa after 4 °C incubation overnight following denaturation at 95 °C with dithiothreitol (Lane C3′ of Fig. 4b), indicating that the 35 kDa molecules might result from dimerization of the 19 kDa monomers.
We further investigated whether hG-CSF expression was induced in N-rich media after additional cultivation in the same media for 11 days without transfer to the N-deficient media. As shown in Fig. 4c, hG-CSF polypeptides were detected in a dimeric form in the 7 day cultivated cell lysates and then decreased in concentration after 11 days of cultivation. In the spent media at 11 days, large amounts of hG-CSF were observed in the form of 35 kDa dimers.
Discussion
The N starvation condition used in this study was a potential concern with respect to the production of hG-CSF because N is a direct resource for amino acid synthesis. However, we demonstrated that hG-CSF polypeptides were successfully produced up to 3 days after transfer into the N-deficient media. Carbon sources not used for protein synthesis under N depletion conditions are stored as triglycerides and starch in the cells21. However, the negative effect of N deficiency on protein synthesis is dependent upon the N level and the duration of treatment22, indicating that the flow of carbon from protein synthesis into lipids under N starvation could require more time than we expected. Therefore, the duration—3 days of incubation—used for inducing hG-CSF might be optimal for the production of recombinant proteins under N starvation.
CvNDI promoters were successfully used for the production of hG-CSF polypeptides in the transgenic Chlorella spp. The use of an appropriate promoter is essential for the development of a platform for the high-yield production of recombinant proteins. The CvNDI1 and CvNDI2 ORFs were found to encode a urea carboxylase and an AMT, respectively, both of which are involved in N metabolism. Urea is converted into allophanate by CvNDI1 in the first step of urea metabolism31. AMT is known to play a vital role in ammonium uptake into a cell, and the transcription of AMT genes was found to be strongly induced under N starvation in Arabidopsis and Chlorella, suggesting that AMT might be an initial sensor of N deficiency37,40,41. CvNDI1 and CvNDI2 might be important for the survival of Chlorella under N starvation via the utilization of urea as N resource and the enhancing of ammonium transportation in the cell. Therefore, we suggest that the CvNDI promoter systems are efficient for the expression of heterologous proteins under N starvation.
Two different SPs identified from UTEX 395 and ArM0029B were used to facilitate the secretion of hG-CSF into the culture medium. The putative cellulase from UTEX 395 appeared to be a polypeptide of 8.4 kDa, and RABF1 from ArM0029B was 17.7 kDa, both rather small polypeptides. Cellulases are known to be secreted from various microorganisms, including fungi and algae42,43. RABF1, located at the endosomal membrane, regulates the secretory trafficking pathway in land plants and algae, under stress signaling and senescence44,45. Functional analysis would be necessary to characterize those genes. Proteins with small molecular weights (MWs) are secreted more efficiently than larger proteins, and the MW of a secreted protein is one of the major factors related to its secretion efficiency46. Strong bands were detected in western blots of the culture media, compared with the band from cell lysates in ArM0029B harboring pSK404. This observation suggests that our secretion system using the RABF1 SP worked efficiently. The cellulase SP from UTEX 395 secreted hG-CSF into the medium as well, when the pSK401 vector was transformed into UTEX 395 (data not shown). Some SPs could not secrete recombinant proteins as efficiently as endogenous proteins47,48. Highly expressed proteins with SP could even become cleaved and unfolded49. The SP structure could also disrupt the structure of the fused recombinant protein, and those disrupted proteins could induce protease activation, thereby decreasing secretion efficiency50,51. Further study into the SPs will be necessary for the establishment of optimal protein secretion systems.
In this study, hG-CSF polypeptides produced in transgenic Chlorella were detected with an MW of 35 kDa in both cell lysates and culture medium. This value is higher than the expected size of 19 kDa. This gel shifting in the western blots might be due to either protein aggregation via disulfide bonding or posttranslational modifications such as glycosylation52,53. hG-CSF has two disulfide bonds at positions 36–42 and 64–74 and a free cysteine at position 1754. The free cysteine was reported to cause dimerization of hG-CSF monomers during protein purification55,56. Moreover, recombinant hG-CSF tends to aggregate readily at pH values above 5.0 at elevated temperatures54,57,58,59. Therefore, the pH homogeneity of the culture media during cultivation of Chlorella would be important in our culture system. To avoid protein aggregation, further investigation aimed at optimizing protein purification, as well as growth conditions, should be performed. Purified hG-CSF will be used for functional efficacy tests such as growth tests in granulocyte macrophage colonies. Moreover, O-glycosylation occurs only at Thr133 in native hG-CSF60. The glycosylation of recombinant hG-CSF might depend upon the host cells used for hG-CSF production. Lenograstim produced in mammalian cells was found to be glycosylated, whereas filgrastim is produced in E. coli in its nonglycosylated form. Thus, hG-CSF synthesized from Chlorella might be in the glycosylated form, although O-glycosylation patterns were not examined in this study. Protein modification might increase the size of hG-CSF only slightly, because of the small mass of O-glycan.
Microalgae have been considered to be an attractive platform for the production of valuable proteins, because of their short life cycle, high biomass, potential for scaling up, and eukaryotic N-glycosylation45. Chlamydomonas has been an organism of choice for the production of various pharmaceuticals using either constitutive or inducible promoters of 35S, Ubi1, and HSP70-RBCS245,61,62,63,64. However, its protein production is overall 0.1–5% total soluble protein, which is relatively low compared with the productivity of other eukaryotic organisms45. To achieve optimal production of recombinant proteins in Chlorella, it will be necessary to identify the copy number and location of transgenes.
In summary, a novel system, including N starvation-inducible promoters, was developed for recombinant protein expression in Chlorella spp. hG-CSF was successfully produced under N starvation conditions. Functional assays of the protein will be conducted after purification of the protein, and optimization of production will be the next step to produce high yields of hG-CSF from Chlorella.
Materials and methods
Strain and growth conditions
Chlorella vulgaris UTEX 395 and arctic Chlorella sp. ArM0029B28 were grown in BG11 medium65, including 3% glucose at 25 °C under constant illumination (50–60 μmol photons m−2 s−1) on a rotary shaker (150 rpm) in a multi-spin shaker (Vision Sciences, Korea)15. For the transformation of Chlorella, we used electroporation, as previously described15. Transgenic Chlorella colonies were formed on agar plates containing 40 mg/L of hygromycin and were then transferred into BG11 liquid medium containing 3% glucose for further growth. To induce hG-CSF expression, the Chlorella transgenic lines were inoculated at 5% (v/v) in 100 ml of new BG11 media and cultivated for 7 days until growth approached stationary phase (OD at 680 nm of 14.0). The cells were harvested by centrifugation at 4,000 rpm for 10 min and then resuspended and cultivated in BG11 media lacking NaNO3 for 1 to 3 days. The cells and spent media were harvested for RNA or protein extraction.
cDNA library construction and massively parallel sequencing
Transcript sequencing of ArM0029B and D3 cultured for 3 days under N-deficient conditions was performed using the Illumina Hiseq2000 platform (Illumina, USA). The detailed procedure was as follows. RNA-Seq paired-end libraries were prepared using Illumina TruSeq RNA Sample Preparation Kits v2 (Illumina, USA). Starting with total RNA, mRNA was purified using poly (A) selection, and then, the RNA was chemically fragmented and converted into single-stranded cDNA using random hexamer priming. Next, the second strand was generated to create double-stranded cDNA. Library construction involved the generation of blunt-end cDNA fragments from ds-cDNA. Then, A bases were added to the blunt ends to prepare them for the ligation of sequencing adapters. After size selection of ligates, the ligated cDNA fragments that contained adapter sequences were enhanced via PCR using adapter-specific primers. The library was quantified using KAPA library quantification kits (Kapa biosystems KK4854) following the manufacturer’s instructions. Each library was loaded onto the Illumina Hiseq2000 platform, and we performed high-throughput sequencing to ensure that each sample met the desired average sequencing depth. Sequence data of quality Q greater than 20 were extracted by SolexaQA66. Trimming resulted in reads with a mean length of 69 bp across all samples and a minimum length of 25 bp.
Short read mapping and identification of differentially expressed genes (DEG) and functional annotation
Trimmed reads were mapped to reference transcripts using the Bowtie2 (v2.1.0) software67, allowing for alignments with a maximum of two mismatches. The number of mapped clean reads for each transcript was calculated and then normalized using the DESeq package in R68. The fold change and number of reads mapped onto reference transcripts were used to identify DEG between each sample. The false discovery rate calculated via DESeq was used to identify the threshold of the p-value in multiple tests and analyses. All correlation analyses and hierarchical clustering were performed using the AMAP library in R69. Using Gene Ontology (GO) information and KEGG information (https://www.genome.jp/kegg/) provided by the customer, GO and KEGG analysis of DEG was conducted. The number of genes assigned to each GO term was counted using in-house scripts produced by SEEDERS Co.
Extraction of nucleic acids and PCR
To extract nucleic acids, Chlorella cells were harvested by centrifugation, frozen quickly using liquid N, and then ground using a mixer mill MM 300 (Qiagen, Germany) as described by Kumar et al.15. Genomic DNA was extracted from the powder using the cetyl trimethylammonium bromide method70. To investigate the presence of transgenes in the hygromycin-resistant cell lines, PCR was performed on genomic DNA using hG-CSF primers, which were designed to propagate 351 bp-long transcripts, from 31 to 381 nt of the codon-optimized hG-CSF gene. Total RNA was isolated using Trizol (Invitrogen, USA). After DNase I treatment, the RNA was reverse transcribed at 50 °C for 1 h using TOPscript cDNA Synthesis Kits (Enzynomics, Korea) and subjected to RT-PCR and qPCR using primers for detecting hG-CSF transcripts. For RT-PCR, the normalization for quantification was performed by PCR using Chlorella ubiquitin (CvUbi) and/or Actin1 (CvAct1) gene primers. The number of cycles for RT-PCR was 25 cycles for CvUBI and 32 cycles for hG-CSF. For qPCR, the comparative threshold cycle method (ΔΔCt) was used (LightCycler 96; Roche, USA). The CvUbi gene was used as an internal reference. The primers used in this study are summarized in Supplementary Table 2.
Vector construction
hG-CSF expression vectors were constructed using pJKS136 as a backbone (Fig. 2). The sequences of the promoter regions of the CvNDI1 and CvANDI2 genes, spanning the 1 kb-long region upstream of the translation start codon (ATG), including the 5′-UTR, were amplified by PCR, using the genomic DNA of ArM0029B as templates (Supplementary Fig. 4 and Supplementary Table 2). The CvNDI1 and CvNDI2 promoter sequences obtained were inserted into pJKS136 using the restriction enzyme sites HindIII and BamHI, thereby replacing the RAmy3D promoter. Using secretomic data analysis (manuscript in preparation), SPs for protein secretion were extracted from the sequence of the putative cellulase genes from UTEX 395 and Ras-related RABF1 from ArM0029B (Supplementary Fig. 5). SP sequences were fused with the codon-optimized hG-CSF sequence (GenScript, USA) by PCR using forward primers containing the SP nucleotide sequence with the 5′ end of the hG-CSF (red letters in Supplementary Table 2) and a reverse primer of the 3′ region of the hG-CSF sequence. The hG-CSF sequence fused with SP was introduced into the region between the CvNDI promoters and the RAmy3D terminator using the restriction enzymes Kpn I and BamHI.
Immunoblotting
Chlorella cultivated in N-deficient media were harvested by centrifugation at 8,000 rpm for 20 min, and then, the spent medium and Chlorella were sampled. For extraction of total protein from Chlorella cells, 50 mg of cells frozen in liquid N were homogenized in a mixer mill MM300 (Qiagen, Germany) and then resuspended in 1 M PBS buffer (pH 7.4) including 1 × cOmplete™ (Roche, Germany) at 4 °C. The solution was centrifuged at 14,000 rpm for 10 min, and then, the supernatant was transferred into a new tube. To determine the concentration of total proteins in the medium, the spent medium was filtered through a 0.2 μm membrane and was concentrated up to 250 times via 5 kDa size cut-off Viva Flow 200 and Vivaspin 20 in turn (Sartorius, USA). After the cell lysates and total proteins from the medium were separated in 12% NuPAGE gel (Invitrogen, USA), they were transferred onto nitrocellulose membranes using the TurboTransfer system (Bio-Rad, USA). After the membrane was incubated with the polyclonal antibody of hG-CSF (Abcam, USA) for 1 h (1:2000 dilution), it was exposed to anti-rabbit IgG-HRP, which was diluted to 1:5,000 (Abcam, USA). Recombinant hG-CSF synthesized in CHO cells (Abcam, USA) was used as the positive control.
In silico analysis
BLAST searching using the amino acid sequences of CvNDI1 and CvNDI2 was performed using the NCBI BLAST program (https://blast.ncbi.nlm.nih.gov/). Sequence alignment analysis was conducted using the Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/). The hydrophobicity of CvNDI2 was predicted using TMHMM Server v. 2.0 (https://www.cbs.dtu.dk/services/TMHMM-2.0/). The presence of SPs and the location of cleavage sites in proteins were predicted using the SignalP-5.0 program (https://www.cbs.dtu.dk/services/SignalP/).
References
Zhang, Y. M., Chen, H., He, C. L. & Wang, Q. Nitrogen starvation induced oxidative stress in an oil-producing green alga Chlorella sorokiniana C3. PLoS ONE 8, e69225 (2013).
Kothari, R., Pathak, V. V., Kumar, V. & Singh, D. P. Experimental study for growth potential of unicellular alga Chlorella pyrenoidosa on dairy waste water: an integrated approach for treatment and biofuel production. Bioresour. Technol. 116, 466–470 (2012).
Xue, J. et al. The pivotal role ofmalic enzyme in enhancing oil accumulation in green microalga Chlorella pyrenoidosa. Microb. Cell Fact. 15, 120 (2016).
Wang, L. et al. Chlorella vulgaris cultivation in sludge extracts from 2, 4, 6-TCP wastewater treatment for toxicity removal and utilization. J. Environ. Manag. 187, 146–153 (2017).
Mócsai, R. et al. N-glycans of the microalga Chlorella vulgaris are of the oligomannosidic type but highly methylated. Sci. Rep. 9, 331 (2019).
Hawkins, R. L. & Nakamura, M. Expression of human growth hormone by the eukaryotic alga, Chlorella. Curr. Microbiol. 38, 335–341 (1999).
Ren, X. Y., Wang, H. Q., Zhu, J. B. & Kong, Q. J. Selection of Chlorella transformed with rotavirus VP4-ST fusion gene. Vet. Sci. China 40, 41–44 (2010).
Koo, J., Park, D. & Kim, H. Expression of bovine lactoferrin N-lobe by the green alga, Chlorella vulgaris. Algae 28, 379–387 (2013).
Yang, B., Liu, J., Jiang, Y. & Chen, F. Chlorella species as hosts for genetic engineering and expression of heterologous proteins: progress, challenge and perspective. Biotechnol. J. 11, 1244–1261 (2016).
Chen, Y., Wang, Y., Sun, Y., Zhang, L. & Li, W. Highly efficient expression of rabbit neutrophil peptide-1 gene in Chlorella ellipsoidea cells. Curr. Genet. 39, 365–370 (2001).
Kim, D. et al. Stable integration and functional expression of flounder growth hormone gene in transformed microalga, Chlorella ellipsoidea. Mar. Biotechnol. 4, 63–73 (2002).
Niu, Y. F. et al. A new inducible expression system in a transformed green alga, Chlorella vulgaris. Genet. Mol. Res. 10, 3427–3434 (2011).
Cha, T. S., Yee, W. & Aziz, A. Assessment of factors affecting Agrobacterium-mediated genetic transformation of the unicellular green alga, Chlorella vulgaris. World J. Microbiol. Biotechnol. 28, 1771–1779 (2012).
Yang, B. et al. Development of a stable genetic system for Chlorella vulgaris: a promising green alga for CO2 biomitigation. Algal Res. 12, 134–141 (2015).
Kumar, M., Jeon, J., Choi, J. & Kim, S. R. Rapid and efficient genetic transformation of the green microalga Chlorella vulgaris. J. Appl. Phycol. 30, 1735–1745 (2018).
Lumbreras, V., Stevens, D. R. & Purton, S. Efficient foreign gene expression in chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14, 441–447 (1998).
Schroda, M., Blöcker, D. & Beck, C. F. The hsp70a promoter as a tool for the improved expression of transgenes in chlamydomonas. Plant J. 21, 121–131 (2000).
Hempel, F., Lau, J., Klingl, A. & Maier, U. G. Algae as protein factories: expression of a human antibody and the respective antigen in the diatom Phaeodactylum tricornutum. PLoS ONE 6, e28424 (2011).
Hempel, F. & Maier, U. G. An engineered diatom acting like a plasma cell secreting human IgG antibodies with high efficiency. Microb. Cell Fact. 11, 126 (2012).
Kim, J., Liu, L., Hu, Z. & Jin, E. Identification and functional analysis of the psaD promoter of Chlorella vulgaris using heterologous model strains. Int. J. Mol. Sci. 19, 1969 (2019).
Scott, S. A. et al. Biodiesel from algae: challenges and prospects. Curr. Opin. Biotechnol. 21, 277–286 (2010).
Kiran, B., Pathak, K., Kumar, R., Deshmukh, D. & Rani, N. Influence of varying nitrogen levels on lipid accumulation in Chlorella sp. Int. J. Environ. Sci. Technol. 13, 1823–1832 (2016).
Welte, K. et al. Purification and biochemical characterization of human pluripotent hematopoietic colony-stimulating factor. Proc. Natl. Acad. Sci. USA 82, 1526–1530 (1985).
Demetri, G. D. & Griffin, J. D. Granulocyte colony-stimulating factor and its receptor. Blood 78, 2791–2808 (1991).
Mehta, H. M., Malandra, M. & Corey, S. J. G-CSF and GM-CSF in neutropenia. J. Immunol. 195, 1341–1349 (2015).
Souza, L. M. et al. Recombinant human granulocyte colony-stimulating factor: effects on normal and leukemic myeloid cells. Science 232, 61–65 (1986).
Lefrère, F. et al. Comparison of Lenograstim vs Filgrastim administration following chemotherapy for peripheral blood stem cell (PBSC) collection: a retrospective study of 126 Patients. Leukemia Lymphoma 35, 501–505 (1999).
Ahn, J. et al. A new Arctic Chlorella species for biodiesel production. Bioresour. Technol. 125, 340–343 (2012).
Jeong, H. et al. Plastid and mitochondrion genomic sequences from Arctic Chlorella sp. ArM0029B. BMC Genomics. 16, 286 (2014).
Stephenson, A. L., Dennis, J. S., Howe, C. J., Scott, S. A. & Smith, A. G. Influence of nitrogen-limitation regime on the production by Chlorella vulgaris of lipids for biodiesel feedstocks. Biofuels 1, 47–58 (2010).
Roon, R. J. & Levenberg, B. An adenosine triphosphate-dependent, avidin-sensitive enzymatic cleavage of urea in yeast and green algae. J. Biol. Chem. 243, 5213–5215 (1968).
Zhao, J., Zhu, L., Fan, C., Wu, Y. & Xiang, S. Structure and function of urea amidolyase. Biosci. Rep. 38, BSR20171617 (2018).
Mobley, H. L. & Hausinger, R. P. Microbial ureases: significance, regulation, and molecular characterization. Microbiol. Rev. 53, 85–108 (1989).
Mobley, H. L., Island, M. D. & Hausinger, R. P. Molecular biology of microbial ureases. Microbiol. Rev. 59, 451–480 (1995).
Sirko, A. & Brodzik, R. Plant ureases: roles and regulation. Acta Biochim. Pol. 47, 1189–1195 (2000).
von Wirén, N. & Merrick, M. Regulation and function of ammonium carriers in bacteria, fungi, and plants. Top Curr. Genet. 9, 95–120 (2004).
Yan, D., Dai, J. & Wu, Q. Characterization of an ammonium transporter in the oleaginous alga Chlorella protothecoides. Appl. Microbiol. Biotechnol. 97, 919–928 (2013).
Huang, L. F. et al. Efficient secretion of recombinant proteins from rice suspension-cultured cells modulated by the choice of signal peptide. PLoS ONE 10, e0140812 (2015).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, 435–439 (2006).
Gazzarrini, S. et al. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11, 937–948 (1999).
Ludewig, U., Neuhauser, B. & Dynowski, M. Molecular mechanisms of ammonium transport and accumulation in plants. FEBS Lett. 581, 2301–2308 (2007).
Bisaria, V. S. & Mishra, S. Regulatory aspects of cellulase biosynthesis and secretion. Crit. Rev. Biotechnol. 9, 61–103 (1989).
Blifernez-Klassen, O. et al. Cellulose degradation and assimilation by the unicellular phototrophic eukaryote Chlamydomonas reinhardtii. Nat. Commun. 3, 1214 (2012).
Ebine, K. et al. Endosomal trafficking pathway regulated by ARA6, a RAB5 GTPase unique to plants. Small GTPases 3, 23–27 (2012).
Yan, N., Fan, C., Chen, Y. & Hu, Z. The potential for microalgae as bioreactors to produce pharmaceuticals. Int. J. Mol. Sci. 17, 962 (2016).
Chen, A. et al. Downsizing a pullulanase to a small molecule with improved soluble expression and secretion efficiency in Escherichia coli. Microb. Cell Fact. 15, 9 (2016).
Kurys, G., Tagaya, Y., Bamford, R., Hanover, J. A. & Waldmann, T. A. The long signal peptide isoform and its alternative processing direct the intracellular trafficking of interleukin-15. J. Biol. Chem. 275, 30653–30659 (2000).
Dultz, E. et al. The signal peptide of the mouse mammary tumor virus Rem protein is released from the endoplasmic reticulum membrane and accumulates in nucleoli. J. Biol. Chem. 283, 9966–9976 (2008).
Hyyryläinen, H. L. et al. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41, 1159–1172 (2001).
Darmon, E. et al. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacterial. 184, 5661–5671 (2002).
Zhou, Y. et al. Enhancing full-length antibody production by signal peptide engineering. Microb. Cell Fact. 15, 47 (2016).
Crow, M. K., Karasavvas, N. & Sarris, A. H. Protein aggregation mediated by cysteine oxidation during the stacking phase of discontinuous buffer SDS-PAGE. Biotechniques 30, 311–316 (2001).
Magnelli, P. E., Bielik, A. M. & Guthrie, E. P. Identification and characterization of protein glycosylation using specific endo- and exoglycosidases. J. Vis. Exp. 58, e3749 (2011).
Lu, S., Boone, T., Souza, L. & Lai, P. Disulfide and secondary structure of recombinant human granulocyte colony-stimulating factor. Arch. Biochem. Biophys. 1, 81–92 (1989).
Raso, S. W. et al. Aggregation of granulocyte-colony stimulating factor in vitro involves a conformationally altered monomeric state. Protein Sci. 14, 2246–2257 (2005).
Fidler, K. et al. The characterization and potential use of G-CSF dimmers and their pegylated conjugates. Acta Chim. Slov. 58, 1–8 (2011).
Herman, A. C., Boone, T. C. & Lu, H. S. Characterization, formulation, and stability of Neupogen (Filgrastim), a recombinant human granulocyte-colony stimulating factor. In Formulation, characterization and stability of protein drugs (eds Pearlman, R. & Wang, Y. J.) 303–328 (Plenum Press, New York, 1996).
Krishnan, S. et al. Aggregation of granulocyte colony stimulating factor under physiological conditions: characterization and thermodynamic inhibition. Biochemistry 41, 6422–6431 (2002).
Chi, E. Y. et al. Roles of conformational stability and colloidal stability in the aggregation of recombinant human granulocyte colony stimulating factor. Protein Sci. 12, 903–913 (2003).
Oh-eda, M. et al. O-linked sugar chain of human granulocyte colony-stimulating factor protects it against polymerization and denaturation allowing it to retain its biological activity. J. Biol. Chem. 265, 11432–11435 (1990).
Mayfield, S. P., Franklin, S. E. & Lerner, R. A. Expression and assembly of a fully active antibody in algae. Proc. Natl. Acad. Sci. USA 100, 438–442 (2003).
Eichler-Stahlberg, A., Weisheit, W., Ruecker, O. & Heitzer, M. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 229, 873–883 (2009).
Tran, M. et al. Production of anti-cancer immunotoxins in algae: Ribosome inactivating proteins as fusion partners. Biotechnol. Bioeng. 110, 2826–2835 (2013).
Rasala, B. A. et al. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant Biotechnol. J. 8, 719–733 (2010).
Stanier, R. Y., Kunisawa, R., Mandel, M. & Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bac. Rev. 35, 171–205 (1971).
Cox, M. P., Peterson, D. A. & Biggs, P. J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 11, 485 (2010).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. Ultrafast and memory efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Antoine L. amap: Another Multidimensional Analysis Package. R package version 0.8-12. https://CRAN.R-project.org/package=amap (2014).
Murray, H. G. & Thompson, W. F. Rapid isolation of high molecular weight DNA. Nucleic Acids Res. 8, 4321–4325 (1980).
Acknowledgements
This research was supported by grants from the PAP Program (PE18900) of Korea Polar Research Institute and the Next-Generation BioGreen 21 Program (PJ01365901), Rural Development Administration.
Author information
Authors and Affiliations
Contributions
S.R.K. conceived and designed study. J.H.S. performed protein works and wrote manuscript and S.R.K. finalized the manuscript. J.C. conducted vector construction, transformation and writing manuscript. J.J. conducted transformation and analyzed transformants. M.K. confirmed RNA-seq data using RT-PCR. J.L. participated in protein works. W.J.J. conducted and analyzed RNA-seq, and wrote manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shin, JH., Choi, J., Jeon, J. et al. The establishment of new protein expression system using N starvation inducible promoters in Chlorella. Sci Rep 10, 12713 (2020). https://doi.org/10.1038/s41598-020-69620-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-69620-9
- Springer Nature Limited