Abstract
Basic helix–loop–helix (bHLH) proteins, one of the most important and largest transcription factor family in plants, play important roles in regulating growth and development, stress response. In recent years, many bHLH family genes have been identified and characterized in woody plants. However, a systematic analysis of the bHLH gene family has not been reported in Ginkgo biloba, the oldest relic plant species. In this study, we identifed a total of 85 GbbHLH genes from the genomic and transcriptomic databases of G. biloba, which were classified into 17 subfamilies based on the phylogenetic analysis. Gene structures analysis indicated that the number of exon–intron range in GbbHLHs from 0 to 12. The MEME analysis showed that two conserved motifs, motif 1 and motif 2, distributed in most GbbHLH protein. Subcellular localization analysis exhibited that most GbbHLHs located in nucleus and a few GbbHLHs were distributed in chloroplast, plasma membrane and peroxisome. Promoter cis-element analysis revealed that most of the GbbHLH genes contained abundant cis-elements that involved in plant growth and development, secondary metabolism biosynthesis, various abiotic stresses response. In addition, correlation analysis between gene expression and flavonoid content screened seven candidate GbbHLH genes involved in flavonoid biosynthesis, providing the targeted gene encoding transcript factor for increase the flavonoid production through genetic engineering in G. biloba.
Similar content being viewed by others
Introduction
The basic helix–loop–helix (bHLH) proteins are one of the most important and largest transcription factor families in plants. All the bHLH proteins contain a highly conserved bHLH domain comprised of HLH region and basic region. HLH region is characterized by two α-helices connected by a loop (HLH)1. Hence the name is derived from this structural motif. In addition, two α-helices constitutes dimerization motif with approximately 45 amino acids that is indispensable in the formation of bHLH homodimers or heterodimers2,3. Generally, the basic region with approximately 15 amino acids facilitates binding to DNA2. At present, a large number of bHLH gene family have been identified and characterized at genome-wide level from some plant species, such as Arabidopsis thaliana4, Phyllostachys edulis5, Daucus carota6, and Panax ginseng7.
The bHLH classifications have been improved continuously as the functions of bHLH proteins were determined. bHLH are typically classified into six major groups from A to F according to sequence similarity and evolutionary relationship and the ability to bind DNA8,9. Group A mainly binds to the E-box (CAGCTG or CACCTG), which acts as neural and mesodermal development10. Group B binds to G-box (CACGTG), which is involved in the expression of glucose-responsive genes and the sterol metabolism11. Group C contain bHLH domain and PAS domain that bind ACGTG or GCGTG sequences, which are involved in developmental signaling and environmental homeostasis12. Group D lacks a basic region and binds to group A formatting heterodimers11. Group E bind to N boxes (CACGCG or CACGAG) that function as embryonic segmentation, somitogenesis and organogenesis13. Group F contains COE domain except bHLH domain for dimerization and DNA binding, which is related to head development and formation of olfactory sensory neurons11.
Ginkgo biloba, one of relic plant species, is looked as one living fossil14, contains flavonoids and terpenoids that affect antioxidant activities, platelet-activating factors, peripheral blood vessels, and blood circulation15. Flavonoids are synthesized by the combination of the phenylpropanoid and polyketide pathways16.Transcription factor were involved in flavonoid biosynthesis by regulating expression of structural genes17. Some structural genes related to flavonoid biosynthesis were cloned and characterized from G. biloba, including phenylalanine ammonia-lyase (PAL)18, flavonol synthase (FLS)19, flavanone 3-hydroxylase (F3H)20, chalcone synthase (CHS)21, chalcone isomerase (CHI)22, isoflavone reductase-like (IFR-like)23, dihydroflavonol-4-reductase (DFR)24, anthocyanidin reductase (ANR)25, anthocyanidin synthase (ANS)26, cinnamate‑4‑hydroxylase (C4H)27. In addition, the transcription factors (bHLH, MYB, and WD40) were also reported to play important role in the biosynthetic pathway of flavonoids28. Although some literatures reported the genome-wide map and second generation and full-length transcriptome analysis related to related flavonoids biosynthesis in G. biloba29,30,31,32, little information about bHLH genes is available in G. biloba. In the present study, we used bioinformatics to identify the bHLH family gene members and analyzed the relevant characteristics of these family members based on reported genomic sequencing and full-length transcriptome databases. In addition, we screened some bHLH genes which might be involved in biosynthetic pathway of flavonoids in G. biloba. Our data provided the targeted gene resource of transcript factor involved in flavonoids biosynthesis for increase the flavonoid production through genetic engineering in G. biloba.
Result
Identification and physicochemical properties of bHLH proteins from G. biloba
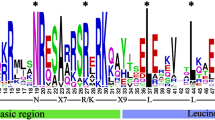
Here, a combined analysis of genome-wide and full-length transcriptome-wide was carried out to screen and identify bHLH genes in G. biloba using the publicly available genomic sequences and our recently published full-length transcriptome data32,33. A total of 85 putative bHLH proteins (GbbHLH) were obtained based on reported genomic sequencing and full-length transcriptome databases of G. biloba (Tables S1, S2). To further characterize these GbbHLHs, we analyzed the physicochemical properties of the putative proteins. These 85 GbbHLH proteins showed diversities in length, molecular weight, theoretical isoelectric points (PIs), number of negatively charged residues (Asp and Glu), and number of positively charged residues (Arg and Lys) (Table S2). Specifically, the lengths of the 85 GbbHLH proteins ranged from 98 to 1,469 amino acid residues, while their pIs were between 4.74 and 9.39 with an average of 6.78 (Table S2). The grand average of hydropathicity of the candidate GbbHLH proteins ranged from − 0.856 to 0.514. Most of GbbHLH proteins belonged to hydrophilic characteristics, except for GbbHLH042. The multiple sequence alignment of bHLH domain sequence of GbbHLH proteins showed that the basic region and two helixes were highly conserved in most of GbbHLH proteins, except the basic region was absent in GbbHLH040, GbbHLH048, GbbHLH054 and GbbHLH075, and the helix 2 region was absent in GbbHLH035 (Fig. 1A). Among amino acids of conserved bHLH domain, nineteen amino acid residues were highly conserved (> 50% consensus ratio), and eight of those were conserved with a > 75% consensus ratio. Moreover, basic region (Glu-12, Arg-13, Arg-15and Arg-16) consensus ratio were higher than 75%, helix 1 region (Leu-26, Leu-29, Val-30 and Pro-31), loop region(Asp-50 and Lys-51) and helix 2 region (Ala-52, Ser-53, Leu-55,Glu-57, Ala-58, Ile-59, Tyr-61 and Leu-65) consensus ratio beyond to 50% (Fig. 1B).
Multiple alignment of conserved domain amino acid sequences of multiple bHLH proteins from G. biloba. (A) Multiple sequence alignment of convserved bHLH domain of bHLH proteins from G. biloba. Alignment was carried out using Clustal W and represented by Adobe ExtendScript Toolkit CS6. (B) Analysis of bHLH domain motif by TBtools. Highly conserved amino acid residues in the bHLH domain across all GbbHLHs. The conservation of the sequence at that position was represents height of each stack.
Evolutionary tree analysis of bHLH gene family
To classify the G. biloba bHLH protein subfamilies and identify the evolutionary relationships among the bHLH proteins from G. biloba, Manus domestica, and A. thaliana, a phylogenetic tree were constructed using the sequences of the 85 GbbHLH proteins, 94 MdbHLH proteins, and 11 A. thaliana bHLH proteins. As shown in Fig. 2, the 85 bHLH members of G. biloba clustered into 17 subfamilies according to the topology of the tree and classification of the bHLH superfamily in A. thaliana and M. domestica. The 17 subfamilies were designated as I(a1), I(b1), I(b2), II, III(a + c), III b, III (d + e), IIIf, IVa, IVb, IVc, IVd, Vb, VII(a + b), VIII, VIIIb, VIII(c1), IX, X, XI, XII, and XV (Fig. 2). None of the G. biloba bHLH proteins were grouped into subfamilies V(a), VI, VIII(c2), XIII, and XIV possibly due to the loss of these proteins during the evolution of G. biloba. In sum, the number of G. biloba bHLHs within each subfamily varied from 1 to 10.
Phylogenetic tree constructed using the sequences of bHLH domain proteins from Arabidopsis thaliana, Manus domestica and G. biloba. The tree was generated using Clustal X2 and MEGA 6 by using neighbor-joining method with 1,000 bootstrap replicates. All bHLH genes are clustered into subclades based on the priority classification rule of Arabidopsis bHLH genes.
Gene structure and characterization of conserved bHLH motifs from G. biloba
The schematic gene structures of GbbHLH genes were analyzed using the GSDS tool (Fig. 3). Among 85 GbbHLHs, 75 were identified from the genomic database. Therefore, we analyzed the exon–intron distribution of 75 GbbHLHs of G. biloba. The 75 GbbHLH genes had a varying number of exons from 1 to 12. Among these GbbHLHs, 6 gene members, that is GbbHLH013, GbbHLH022, GbbHLH 053, GbbHLH054, GbbHLH056, and GbbHLH074, were intron-less and distributed across VIII(b) and III(d + e). Five gene members, GbbHLH003, GbbHLH011, GbbHLH038, and GbbHLH076 of subfamilies IV(b) and IV(c), were predicted to exhibit five exons and four introns, respectively. Two members (GbbHLH044 and GbbHLH012 from subfamily XI) exhibited seven exons and six introns, respectively. The members of subfamily V(b) exhibited two exons and one intron. The members of subfamilies III(a + c), III(b), I(a1), I(b1), I(b2), and XV presented two to five exons and one to four introns. The members of subfamilies III(f), VII(a + b), and XII presented six to nine exons and five to eight introns. GbbHLH043 and GbbHLH068 exhibited 12 exons and 11 introns.
Phylogenetic relationship and gene structure analysis of bHLH genes in G. biloba. (A) phylogenetic tree was constructed from the alignment of amino acid sequencing of selected bHLH proteins from G. biloba. (B) Gene structure analysis of selected bHLH genes of G. biloba, showing locations and lengths of the exons and introns. Exons and introns are presented as filled yellow round-corner rectangle and thin single lines, respectively.
MEME analysis showed that all GbbHLH proteins except GbbHLH060 contained highly conserved Gb-motif 1 and Gb-motif 2, which consists of 15 and 29 amino acids, respectively (Figs. 4 and S1). The Gb-motifs belonging to the same subfamily of bHLHs were the same or similar. GbbHLH038 and GbbHLH076 from the subfamily IV(c) contained 4 motifs, while GbbHLH003, GbbHLH011, and GbbHLH016 from subfamilies I(b2) and IV(b) all contained four motifs. Most Gb-motifs, such as subfamilies I(b2), III(a + c), IV(a), IV(c), V(b), VII(a + b), VIII(b), and XV, were located near the C-terminus. However, Some Gb-motifs, such as those found in subfamily IV(b), were located near the N-terminus.
Motif composition and distribution of 85 bHLH proteins in G. biloba. The motifs of the GbbHLH proteins were analyzed using the MEME web server. The length of the black line indicates the length of a sequence relative to all the other sequences. The position of each block indicates the location of a motif with a matching sequence.
Analysis of GO annotation and subcellular localization
The GO annotation of GbbHLHs showed three aspects of functional classifications, namely, molecular function, cellular component, and biological process (Fig. 5). GbbHLH002, GbbHLH024, and GbbHLH069 were annotated in the molecular function, which is related to transcriptional regulation. Only GbbHLH069 was annotated in the cellular component. Among 85 bHLH members, 14 GbbHLH genes, including GbbHLH002, GbbHLH009, GbbHLH023, GbbHLH024, GbbHLH032, GbbHLH038, GbbHLH039, GbbHLH043, GbbHLH056, GbbHLH060, GbbHLH069, GbbHLH072, GbbHLH073, and GbbHLH076, were annotated in biological process and involved in DNA binding, oxidoreductase activity, and protein dimerization activity.
The remaining 71 GbbHLH genes cannot be annotated to GO databases, which accounts for 83.53% of the total GbbHLH genes. We conducted annotated 71 GbbHLH genes according to evolution of G. biloba, Manus domestica, and A. thaliana. We found that the number of bHLH genes involved in biological regulation was up to 71. Among these genes, 69 bHLH genes were involved in cell, cell part and organelle, respectively. A total of 66 bHLH genes and 68 bHLH genes were classified into binding nucleic acid and binding transcription factor activity, respectively.
The subcellular localization analysis of the 85 bHLH protein were performed online with WOLF PSORT. As shown in Table S3, a total of 74 GbbHLHs were predicted to located in the nucleus (up to 87%), 8 of which, including GbbHLH015, GbbHLH020, GbbHLH051, GbbHLH061, GbbHLH062, GbbHLH063, GbbHLH064, and GbbHLH068, were predicted to located in the chloroplast (0.09%). Only GbbHLH035 was supposed to located in the plasma membrane. GbbHLH022 and GbbHLH083 were likely to located in the peroxisome.
Promoter analysis and protein–protein interaction network prediction
Many bHLH genes play important roles in plant growth and development, as well as in response to various abiotic stresses. To further investigate the putative functions of GbbHLH genes, we identified and analyzed the potential cis-elements in the promoter regions of 2000-bp upstream of the start codon of bHLH genes using PlantCARE software. As shown in Fig. 6, three main categories were found in the cis-elements of GbbHLH genes. Category one was related to plant growth and development, such as cell differentiation, circadian control, and cell cycle regulation. This category was composed of ARE, AT-rich sequence, HD-Zip-1, RY-element, GCN4_motif, AACA_motif, circadian, and MSA-like. Category two was involved in phytohormones, such as abscisic acid (ABA), auxin, gibberellin, methyl jasmonate (MeJA), and salicylic acid (SA). This category included ABA response element (ABRE), AuxRR-core, CGTCA-motif, TATC-box, TCA-element, and TGACG-motif. Category three was associated with abiotic stresses, such as light responsiveness, drought inducibility, wound responsiveness, anaerobic induction, and low-temperature responsiveness. Category three contained 3-AF1 binding site, AAAC-motif, ACE, C-box, G-Box, GT1-motif, LTR, MBS, MRE, P-box, Sp1, TC-rich repeats, and WUN-motif.
Cis-element analysis of 74 bHLH gene promoters in G. biloba. The potential cis-regulatory elements in the promoter regions 2,000 bp upstream of the G. biloba were predicted by PlantCARE software. Different colors indicated the elements related to growth and development (circadian control), plant hormones (abscisic acid, auxin, methyl jasmonate, gibberellic acid, and salicylic acid) and stress responsiveness (anaerobic induction, light, low temperature, and drought inducibility).
The interaction of 85 GbbHLHs was predicted by STRING (Fig. 7). In the protein sequence homology to A. thaliana, GbbHLH002 and GbbHLH014 that were homologous with PHYTOCHROME-INTERACTING FACTOR 3 (PIF3) belonged to the subfamily VII(a + b) group. GbbHLH028 [homologous PIF3-LIKE 5 (PIL5)] belonged to subfamily VII(a + b) group. PIF3 and PIL5 are related to light signaling and phytohormones signals33. GbbHLH028 could interact with GbbHLH002 and GbbHLH014 that regulated light signaling and phytohormones signals pathway GbbHLH005, GbbHLH019, and GbbHLH032 (homologous MYC2) belonged to subfamily III(d + e) group; and GbbHLH004 (homologous MYC4) belonged to subfamily III(a + c) group. MYC2, MYC3 and MYC4 controls additively jasmonate-related defense responses by reducing expression of GS biosynthesis genes. MYC interact directly with GS-related MYBs to regulation of defense secondary metabolite production34. Hence, we speculate GbbHLH004 could interact with GbbHLH005, GbbHLH019, and GbbHLH032. That involved in jasmonate-related defense responses. GbbHLH001, GbbHLH041 and GbbHLH066 were homologous with FMA. GbbHLH033and GbbHLH083 were homologous with ICE1. GbbHLH030 was homologous with SCRM2. GbbHLH001, GbbHLH041 and GbbHLH066 could interact with GbbHLH033and GbbHLH083. GbbHLH030 could interact with GbbHLH033and GbbHLH083.
Functional regulatory network of 85 G. biloba bHLH proteins. The protein–protein interaction of bHLH proteins was predicted using STRING software. Cyan line presents data from curated databases, purple line experimentally determined, green line gene neighborhood, red line gene fusions, blue line gene co-occurrence; yellow line presents text mining, black line co-expression and gray line protein homology.
Candidate bHLHs involved in flavonoids biosynthesis in G. biloba
Our previous work performed Illuminate sequencing of 24 independent cDNA libraries of eight organs (root, stem, immature leaf, mature leaf, microstrobilus, ovulate strobilus, immature fruit and mature fruit) from G. biloba with three biological replicates each organ31. Based on the RNA-seq data, a total of 80 GbbHLHs were expressed in eight different organs of G. biloba (Fig. 8A). No expression was observed in GbbHLH009, GbbHLH016, GbbHLH061, GbbHLH069 and GbbHLH071. The spatial expression patterns of 80 GbbHLHs were diverse. GbbHLH041, GbbHLH047, GbbHLH056, GbbHLH068 and GbbHLH081 were predominantly expressed in root. GbbHLH040 and GbbHLH076 were preferentially expressed in microstrobilus. GbbHLH084 was highly expressed in stem. GbbHLH083 was mainly expressed in mature leaves. GbbHLH045 was highly expressed in immature fruit. Based on correlation analysis between the expression level of GbbHLHs and flavonoids content using OmicShare tools, the flavonoids content was significantly correlated with expression levels of seven GbbHLHs in eight organs of G. biloba. In detail, the expression levels of GbbHLH034 (R2 = 0.536), GbbHLH029 (R2 = 0.733), GbbHLH083 (R2 = 0.762), GbbHLH066 (R2 = 0.599), GbbHLH059 (R2 = 0.610), GbbHLH080 (R2 = 0.541) and GbbHLH017 (R2 = 0.722) had significant positive correlation with flavonoids content (p < 0.05) (Fig. 8B). Therefore, we suggested that these 7 bHLH genes might be involved in flavonoids biosynthesis in G. biloba.
Expression of GbbHLH genes and correlation analysis between expression level of GbbHLH genes and the content of flavonoids in different organs of G. biloba. (A) A heatmap shows expression level of 80 GbbHLH genes with different subfamilies (left column) in different organs (bottom row) of G. biloba. Expression differences are observed in different colors. The R, S, IL, ML, M, OS, IF and MF represent root, stem, immature leaf, mature leaf, microstrobilus, ovulate strobilus, immature fruit and mature fruit, respectively. Changes in expression level are indicated by a change in color; green indicates a lower expression level, whereas red indicates a higher expression level. All data shown reflect the average mean of three biological replicates (n = 3). (B): Correlation analysis between the expression level of selected 7 GbbHLH genes and the content of flavonoids and in different organs of G. biloba.
Chromosomal distribution of GbbHLH genes
To characterize the chromosomal distribution of these GbbHLH genes, we integrated 12 scaffolds of the G. biloba genome (named Chr.1 to Chr. 12) from the genome database32. Among these GbbHLH genes, 82 members were successfully mapped to the ginkgo chromosomes (Fig. 9). The number of bHLH genes range from 4 to10 in chromosome 1 to 12. Chromosome 8 and chromosome 10 contain 10 bHLH genes. Chromosome 5 and chromosome 11 contain 4 bHLH genes. In particular, GbbHLH022 was mapped onto the hic_scaffold_9926 of ginkgo and GbbHLH041 was mapped onto the hic_scaffold_22302 of ginkgo.
Discussion
The bHLH family genes were previously divided into 21 subfamilies in A. thaliana4, 23 in M. domestica35, and 19 in peach36. The evolutionary analysis identified 85 bHLH genes in G. biloba, which were divided into 17 subfamilies. Our results on GbbHLHs showed similarities as well as differences compared to the classifications of the other plant species35. In general, the structures and functions of GbbHLH matched with those of other species. In other words, genes with the same or similar functions were clustered on the same branch. For example, AtbHLH045, AtbHLH097, and AtbHLH098 from I(a) subfamily are related to stomatal development control35,36. GbbHLH001, GbbHLH031, GbbHLH041, and GbbHLH066 were classified under the subfamily I(a). Thus, these four bHLH genes of G. biloba were deduced to participated in stomatal development control. AtbHLH037, AtbHLH040, AtbHLH043, and AtbHLH088 from subfamily VIII(b) regulate flower and fruit development37. Likewise, GbbHLH013, GbbHLH022, GbbHLH053 and GbbHLH054, being classified into VIII(b) subfamily, were projected to exhibit similar functions. Previous studies showed that AtbHLH038, AtbHLH039, AtbHLH100 and AtbHLH101 genes are involved in Fe-deficiency response38. Hence, the functions of GbbHLH050, GbbHLH051, GbbHLH052, GbbHLH070, and GbbHLH071 from I(b2) subfamily may be analogous as function in Fe-deficiency response of G. biloba. Same analogy existed in anthocyanin-related AtbHLH001, AtbHLH002, and AtbHLH042 from subfamily III(f)39 and GbbHLH034, GbbHLH042, GbbHLH065and GbbHLH084. Taken together, the evolutionary analysis results and bHLH genes with known functions can be combined to predict the GbbHLH genes related to growth and development, secondary metabolism, and environmental responses in G. biloba.
Gene structures analysis provides important information on phylogenetic relationships. The numbers of exon/intron of the same subfamilies are the same or similar40. The exon/intron diversification of gene family members play an important role in the evolution of multiple gene families through the three main types of mechanisms, namely exon/intron gain/loss, exonization/pseudoexonization, and insertion/deletion41. The number of exons/introns ranges from 0 to 4 in rice42 and from 0 to 19 in apple35. In this study, the number of exons/introns in bHLH family member of G. biloba ranged from 0 to 12, indicating that the exons/introns of bHLH genes underwent loss or insertion during the evolution of G. biloba. In addition to Exon–intron structures, motif structure also expounds on phylogenetic relationships. The bHLH genes of one cluster contained the same or similar motifs as in P. edulis5. Similar to these results, our study also revealed that motifs 1 and 2 were located in all GbbHLH proteins. Therefore, motifs 1 and 2 are important characteristics for identifying ginkgo bHLH gene.
Subcellular localization can help to understand location of protein function. Cheng et al.5 performed bHLH protein prediction that most bHLH proteins are located in the nucleus, and some bHLH proteins are located in the mitochondria and cytoplasm in P. edulis. Similarly, GbbHLH proteins were mainly located in the nucleus. The minor number of the GbbHLH proteins were distributed in the chloroplast, plasma membrane, and peroxisome. These results indicated that GbbHLH proteins might play role in nucleus of G. biloba. A small difference in the location of bHLH proteins was also observed between P. edulis and G. biloba.
Plant promoters are important regulatory elements required for plant gene transcription and play important regulatory roles at the transcriptional level43. ABA response elements (ABRE) include ACGTG (A. thaliana), GACACGTGGC (Triticum aestivum), and CGTACGTGCA (Hordeum vulgare), are involved in ABA responsiveness44. Moreover, SA responsiveness (TCA element), light responsiveness cis-element (ACE and MRE), circadian control, LTR cis-element of low-temperature responsiveness, and GT-1 box were also reported45,46. Secondary metabolites synthesis is subjected to phytohormone regulation (MeJA, SA, ABA, and Eth) and low temperature stresses47. Consistent with above reports, our results demonstrated that the promoters of GbbHLHs contained numerous and various cis-elements that are likely related to plant growth and development regulation and response to various abiotic stresses. In general, bHLH proteins interact with other proteins to function in plant development and metabolism. For example, An et al.48 found that MdTTG1 interacts with bHLH proteins to regulate anthocyanin accumulation, while bHLH gene ICE1 interacts with SPCH, FAMA, and MUTE to regulate stomatal cell differentiation in Arabidopsis49. In addition, phytochrome-interacting factor (PIF), proteinase-activated receptor (PAR) and paclobutrazol resistance (PRE) were members of bHLH gene family. PAR1-PRE1 and PAR1-PIF4 form heterodimers regulated cell elongation and plant development in response to light and hormones50. In our study, some protein–protein interaction among bHLH members was also predicted to be associated with growth and development, abiotic stress, phytohormone, and secondary metabolite.
Flavonoids biosynthesis pathway has been extensively studied51,52. In G. biloba, several transcription factors were cloned and identified to be involved in biosynthetic pathway of flavonoids. For example, our previous work demonstrated that an R2R3-MYB gene GbMYBF2 act as negative regulators of flavonoids biosynthesis in G. biloba53. More recently, Zhang et al.54 stated that another MYB gene GbMYBFL played a positive role on flavonoids biosynthesis in G. biloba. To date, some bHLH proteins was found to play important role in the regulation of flavonoids biosynthesis. For instance, bHLH genes were involved in flavonoids biosynthesis in Nicotiana tabacum55 and Chrysanthemum morifolium Rama56. The VvMYC1 gene encoding bHLH transcription factor interacts with MYB to regulate the expression of three flavonoids biosynthetic genes, including ANR, UFGT, and CHI57. bHLH, WD40, and MYB proteins also regulate flavonoids biosynthesis by forming complexes45,50. In this study, our data revealed that 7 GbbHLH genes were significantly correlated with flavonoids content, implying GbbHLH genes that might be involved in flavonoids biosynthesis in G. biloba. However, since this conclusion was based on the correlation analysis between the expression levels and flavonoids content, additional experimental information is necessary to establish the claim. The further study could include transgenic research and transcription factor interaction with promoters of key structural genes related to biosynthetic pathway in G. biloba.
Materials and methods
Identification and classification of bHLH genes in G. biloba
A local protein database of G. biloba was created by the obtained genomic sequences and transcriptome sequences, genomic database comes from Ginkgo biloba GigaScience Database. (https://doi.org/10.5524/100.209)32, transcriptome sequences database was obtained from NCBI (https://www.ncbi.nlm.nih.gov/sra). The accession is no. SRR7948405 ~ SRR7948413 and SRP14911331. The bHLH proteins of A. thaliana, and Malus domestica were downloaded from the PlantTFDB (https://planttfdb.cbi.pku.edu.cn/prediction.php). The bHLH proteins were blasted by matching the 2 species (E value of 0.01) by Bioedit software58, and the bHLH proteins were searched against HMMER3.1 software (https://megasoftware.net/) by the hidden Markov model file of the HLH domain (PF00010) that was downloaded from Pfam database (https://pfam.xfam.org)59. The bHLH proteins that contain multiple termination signals and repeats were removed. Then, the rest of the bHLH protein were checked in the websites SMART (https://smart.embl-heidelberg.de/) and CDD-Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and showed that they remained present in the conserved bHLH domain60,61. All bHLH protein sequences were analyzed through bioinformatics analyses, including the prediction of ORFs and physico-chemical properties such as MW, pI, total number of negatively charged residues (Asp + Glu), and total number of positively charged residues (Arg + Lys) using ExPASy (https://web.expasy.org/protparam/)62.
Phylogenetic analysis
Phylogenetic tree was constructed by Clustal X2 and MEGA 6 using neighbor-joining method with bootstrap test (1,000 replicates), Poisson model, and partial deletion63,64.
Gene structure analysis and conserved motif characterization
The exon–intron structures of GbbHLH genes was displayed by GSDS (https://gsds.cbi.pku.edu.cn/index.php)65. The conserved motifs of the bHLH proteins were searched in MEME 5.0.5 (https://meme.sdsc.edu/meme/) with a maximum of 20 motifs and analyzed by TB tools66,67.
Gene ontology (GO) annotation and subcellular localization prediction
The translated bHLH protein sequences from the full-length transcriptome of G. biloba31 were annotated using the Blast2GO program to assign the GO terms (https://amigo.geneontology.org/amigo/term/)68. The GO analysis showed that the E-value was 1.0E-6, and GO terms were provided under three main categories, namely, biological process, cellular component, and molecular function. The bHLH proteins were uploaded to WOLF PSORT69 (https://www.genscript.com/psort.html) to predict subcellular localization.
Promoter analysis and protein–protein interaction network prediction
The upstream 2,000 bp genomic DNA sequences of the bHLH gene start code were downloaded and submitted to PlantCARE to predict putative cis-elements68,70. The protein–protein interaction of bHLH proteins was predicted using STRING (https://string-db.org/) under the following parameters: A. thaliana was selected to perform the comparison analysis, and then the minimum required interaction score was set to middle confidence, that is, 0.40071.
Determination of flavonoids in G. biloba
The flavonoid contents in roots, stems, immature leaves, mature leaves, microstrobilus, ovulate strobilus, immature fruits, and mature fruits were determined according to the method of Ye et al.29. The flavonoid contents were calculated by multiplying the total content of quercetin, kaempferol, and isorhamnetin with a factor of 2.51, and were expressed as percentage (m/m)72.
Correlation analysis between flavonoid content and gene expression level
Our previous work constructed 24 independent cDNA libraries of eight organs (root, stem, immature leaf, mature leaf, microstrobilus, ovulate strobilus, immature fruit and mature fruit) from G. biloba with three biological replicates each organ31. The 24 cDNA libraries were sequenced using an Illumina Hiseq X Ten Platform by Biomarker Biotechnology (Beijing, China). The SRA accession of these sequencing raw data is nos. SRR7948405–SRR7948413 in NCBI (https://www.ncbi.nlm.nih.gov/sra). The gene expression levels were estimated by fragments per kilobase of transcript per million fragments mapped with the following equation: FPKM = cDNA Fragments/[Mapped Fragments (Millions) × Transcript Length (kb)]. Flavonoids content and expression levels of GbbHLH genes were performed to correlation analysis by applying OmicShare tools (https://www.omicshare.com/tools) to identify genes involved in flavonoids metabolism with correlation coeffcients of ≥ 0.6. Thus, r > 0.6 and P < 0.05 meant significant correlation were considered to have an expression that was significantly correlated with the expression of genes in the biosynthetic pathways of flavonoids.
The location of bHLH genes on chromosomes
The position of each GbbHLH gene on the twelve chromosomes was obtained from the GigaDB site (https://gigadb.org/dataset/100613) and was visualized using TBtools67.
Conclusion
In this study, we identified 85 GbbHLHs through HMMER and BLAST from G. biloba. These GbbHLH genes were classified into 17 subfamilies by comparative phylogenetic analysis with A. thaliana and M. domestica bHLH proteins. Meanwhile, exon/intron and motif analyses supported the results of phylogenetic analysis. A total 74 GbbHLHs were predicted to locate in the nucleus, while other 11 GbbHLHs were located in the chloroplast, plasma membrane, and peroxisome, respectively. The cis-elements in the G. biloba bHLH gene promoters were identified to be related to phytohormone and abiotic stresses. The protein–protein interaction prediction results indicated that GbbHLH proteins are involved in phytohormone. Finally, the correlation analysis between gene expression and flavonoid content revealed seven candidate GbbHLH genes involved in flavonoids biosynthesis. The results of our study provide a foundation for understanding molecular mechanism of bHLH regulating flavonoids biosynthesis in G. biloba.
References
Mao, K., Dong, Q., Li, C., Liu, C. & Ma, F. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front Plant Sci.8, 480 (2017).
Kavas, M. et al. Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol. Genet. Genomics.291, 129–143 (2016).
Xiang, L. L. et al. A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in chrysanthemums (Chrysanthemum morifolium ramat). PLoS ONE10, 1–7 (2015).
Toledo-Ortiz, G., Huq, E. & Quail, P. H. The Arabidopsis basic/helix–loop–helix transcription factor family. Plant Cell15, 1749–1770 (2003).
Cheng, X. R. et al. Basic helix-loop-helix gene family: Genome wide identification, phylogeny, and expression in Moso bamboo. Plant Physiol. Biochem.132, 104–119 (2018).
Chen, Y. Y. et al. Genome-wide analysis of basic helix–loop–helix family transcription factors and their role in responses to abiotic stress in carrot. Mol. Breed.35, 125 (2015).
Chu, Y. et al. Genome-wide characterization and analysis of bHLH transcription factors in Panax ginseng. Acta Pharm. Sin. B8, 666–677 (2018).
Atchley, W. R. & Fitch, W. M. A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl. Acad. Sci. USA94, 5172–5176 (1997).
Simionato, E. et al. Origin and diversification of the basic helix-loop-helix gene family in metazoans: Insights from comparative genomics. BMC Evol. Biol.7, 1–18 (2007).
Massri, M. E. & Murre, C. Helix–loop–helix protein regualations: Regulators of transcription in eukaryotic organisms. Mol. Cell Biol.20, 429–440 (2000).
Song, X. M. et al. Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassicarapa ssp. pekinensis). Mol. Genet. Gnomics289, 77–91 (2014).
Fortunato, S. A. V., Vervoort, M., Adamski, M. & Adamska, M. Conservation and divergence of bHLH genes in the calcisponge Sycon Ciliatum. EvoDevo7, 1–12 (2016).
Gao, M. et al. Identification of the grape basic helix-loop-helix transcription factor family and characterization of expression patterns in response to different stress. Plant Growth Regul88, 19–39 (2019).
Lin, X. et al. Functional genomics of a living fossil tree, Ginkgo, based on next-generation sequencing technology. Physiol. Plant.143, 207–218 (2011).
Meng, X. X., Song, Q. L., Ye, J. B., Wang, L. L. & Xu, F. Characterization, function, and transcriptional profiling analysis of 3-hydroxy-3-methylglutaryl-CoA synthase gene (GbHMGS1) towards stresses and exogenous hormone treatments in Ginkgo biloba. Molecules22, 1706 (2017).
Saito, K. et al. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem.72, 21–34 (2013).
Zhu, J. et al. Comprehensive co-expression analysis provides novel insights into temporal variation of flavonoids in fresh leaves of the tea plant (Camellia sinensis). Plant Sci290, 110306 (2020).
Xu, F. et al. Molecular cloning, characterization and expression of phenylalanine ammonia-lyase gene from Ginkgo biloba. Afr. J. Biotechnol.7, 721–729 (2008).
Xu, F. et al. Isolation, characterization, and function analysis of a flavonol synthase gene from Ginkgo biloba. Mol. Biol. Rep.39, 2285–2296 (2017).
Shen, G. et al. Cloning and characterization of a flavanone 3-hydroxylase gene from Ginkgo biloba. Biosci. Rep.26, 19–29 (2006).
Xu, F., Cheng, S. Y., Cheng, S. H., Wang, Y. & Du, H. W. Time course of expression of chalcone synthase gene in Ginkgo biloba. J. Plant Physiol. Mol. Biol.33, 309–317 (2007).
Cheng, H. et al. Molecular cloning and function assay of a chalcone isomerase gene (GbCHI) from Ginkgo biloba. Plant Cell Rep.30, 49–62 (2011).
Cheng, H. et al. Expression patterns of an isoflavone reductase-like gene and its possible roles in secondary metabolism in Ginkgo biloba. Plant Cell Rep.32, 637–650 (2013).
Hua, C. et al. Molecular cloning and characterization of three genes encoding dihydroflavonol-4-reductase from Ginkgo biloba in anthocyanin biosynthetic pathway. PLoS ONE8, e72017 (2013).
Shen, G. A. et al. Isolation and characterization of a putative anthocyanidin reductase gene from Ginkgo biloba. J. Plant Physiol.163, 227 (2006).
Xu, F. et al. Molecular cloning and function analysis of an anthocyanidin synthase gene from Ginkgo biloba, and its expression in abiotic stress responses. Mol. Cells26, 536–547 (2008).
Cheng, S. Y. et al. Characterization and expression patterns of a cinnamate-4-hydroxylase gene involved in lignin biosynthesis and in response to various stresses and hormonal treatments in Ginkgo biloba. Acta Physiol. Plant.40, 7 (2018).
Zhao, L. et al. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct. Integr. Genomics13, 75–98 (2013).
Wu, Y. et al. De novo transcriptome analysis revealed genes involved in flavonoid biosynthesis, transport and regulation in Ginkgo biloba. Ind. Crop. Prod.124, 226–235 (2018).
Meng, J. et al. Metabolomics integrated with transcriptomics reveals redirection of the phenylpropanoids metabolic flux in Ginkgo biloba. J. Agric. Food Chem.67, 3284–3291 (2019).
Ye, J. B. et al. A global survey of full-length transcriptome of Ginkgo biloba reveals transcript variants involved in flavonoid biosynthesis. Ind. Crop. Prod.139, 11547 (2019).
Guan, R. et al. Draft genome of the living fossil Ginkgo biloba. Gigascience5, 49 (2016).
Lau, O. S. & Deng, X. W. Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol.13, 571–577 (2010).
Schweizer, F. et al. Arabidopsis basic helix–loop–helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell.25, 3117–3132 (2013).
Yang, J. et al. Identification and expression analysis of the apple (Malus × domestica) basic helix–loop–helix transcription factor family. Sci. Rep.7, 28 (2017).
Zhang, C. et al. Genome-wide analysis of basic helix–loop–helix superfamily members in peach. PLoS ONE13(4), e0195974 (2018).
Carretero-Paulet, L. et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, Poplar, Rice, Moss, and Algae. Plant Physiol.153, 1398–1412 (2010).
Wang, N. et al. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant6, 503–513 (2013).
Zhao, Y., Zhou, L. M., Chen, Y. Y., Yang, S. G. & Tian, W. M. MYC genes with differential responses to tapping, mechanical wounding, ethrel and methyl jasmonate in laticifers of rubber tree (Hevea brasiliensis Muell. Arg.). J. Plant Physiol.168, 1649–1658 (2011).
Li, X. et al. Genome-wide identification, evolution and functional divergence of MYB transcription factors in Chinese white pear (Pyrus bretschneideri). Plant Cell Physiol.57, 824–847 (2016).
Xu, G., Guo, C., Shan, H. & Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA109, 1187–1192 (2012).
Li, X. et al. Genome-wide analysis of basic/helix–loop–helix transcription factor family in rice and Arabidopsis. Plant Physiol.141, 1167–1184 (2006).
Danino, Y. M., Even, D., Ideses, D. & Juven-Gershon, T. The core promoter: At the heart of gene expression. Biochim. Biophys. Acta1849, 1116–1131 (2015).
Yang, X. et al. Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep.34, 943–958 (2015).
Piechulla, B., Merforth, N. & Rudolph, B. Identification of tomato Lhc promoter regions necessary for circadian expression. Plant Mol. Biol.38, 655–662 (1998).
Arguello-Astorga, G. & Herrera-Estrella, L. Evolution of light-regulated plant promoters. Annu. Rev. Plant Biol.49, 525–555 (1998).
Meng, X. et al. Isolation, characterization and functional analysis of a novel 3-hydroxy-3-methylglutaryl-coenzyme A synthase gene (GbHMGS2) from Ginkgo biloba. Acta Physiol. Plant.40, 72 (2018).
An, X. H., Tian, Y., Chen, K. Q., Wang, X. F. & Hao, Y. J. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol.169, 710–717 (2012).
MacAlister, C. A., Ohashi-Ito, K. & Bergmann, D. C. Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature445, 537 (2007).
Hao, Y., Oh, E., Choi, G., Liang, Z. & Wang, Z. Y. Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol. Plant5, 688–697 (2012).
Xu, W., Dubos, C. & Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci.20, 176–185 (2015).
Lei, Z., Sumner, B. W., Bhatia, A., Sarma, S. J. & Sumner, L. W. UHPLC-MS analyses of plant flavonoids. Curr. Protoc. Plant Biol.4, e20085 (2019).
Xu, F. et al. An R2R3-MYB transcription factor as a negative regulator of the flavonoid biosynthesis pathway in Ginkgo biloba. Funct. Integr. Genom.14, 177–189 (2014).
Zhang, W. W., Xu, F., Cheng, S. Y. & Liao, Y. L. Characterization and functional analysis of a MYB gene (GbMYBFL) related to flavonoid accumulation in Ginkgo biloba. Genes Genom.40, 49–61 (2018).
Bai, Y. et al. Flavonoid-related basic helix–loop–helix regulators, NtAn1a and NtAn1b, of tobacco have originated from two ancestors and are functionally active. Planta234, 363 (2011).
Xiang, L. L. et al. A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in Chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS ONE10, e0143892 (2015).
Hichri, I. et al. The basic helix–loop–helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant3, 509–523 (2010).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res.25, 3389–3402 (1997).
Finn, R. D. The Pfam protein family’s database: Towards a more sustainable future. Nucleic Acids Res.44, D279–D285 (2016).
Letunic, I. & Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res.46, D493–D496 (2017).
Marchler-Bauer, A. et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res.43, 222–226 (2015).
Islam, S., Rahman, I. A., Islam, T. & Ghosh, A. Genome-wide identification and expression analysis of glutathione S-transferase gene family in tomato: Gaining an insight to their physiological and stress-specific roles. PLoS ONE12, e0187504 (2017).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics23, 2947–2948 (2007).
Tamura, K., Stecher, G., Peterson, D., Filipsk, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol30, 2725–2729 (2013).
Hu, B. et al. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics31, 1296–1297 (2015).
Bailey, T. L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol.2, 28–36 (1994).
Chen, C. et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant.13, 1194–1202 (2020).
Ashburner, M. et al. Gene ontology: Tool for the unification of biology. Nat Genet.25, 25 (2000).
Horton, P. et al. WoLF PSORT: Protein localization predictor. Nucleic Acids Res.35, W585–W587 (2007).
Lescot, M. et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res.30, 325–327 (2002).
Szklarczyk, D. et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res.43, 447–452 (2015).
Zhu, X. Y., Mang, Y. L., Xie, J., Wang, P. & Su, W. K. Response surface optimization of mechanochemical-assisted extraction of flavonoids and terpene trilactones from gingkgo leaves. Ind. Crop. Prod.34, 1041–1052 (2011).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 31901344 and 31370680).
Author information
Authors and Affiliations
Contributions
F.X., S.K., S.C, and X.Z. designed the whole experiment and drafted the manuscript. X.Z. Z.C and G.N. performed bioinformatic analysis. X.Z., Y.L, and F.X. wrote the manuscript. J. J.Y. contributed in bioinformatic analysis and determined the flavonoid contents. All authors have reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, X., Liao, Y., Kim, SU. et al. Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba. Sci Rep 10, 13723 (2020). https://doi.org/10.1038/s41598-020-69305-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-69305-3
- Springer Nature Limited
This article is cited by
-
Integrated metabolomic and transcriptomic analysis reveals the regulatory mechanisms of flavonoid and alkaloid biosynthesis in the new and old leaves of Murraya tetramera Huang
BMC Plant Biology (2024)
-
Multilayered regulation of secondary metabolism in medicinal plants
Molecular Horticulture (2023)
-
Genome-wide identification and characterization of PdbHLH transcription factors related to anthocyanin biosynthesis in colored-leaf poplar (Populus deltoids)
BMC Genomics (2022)
-
Genome-wide identification and characterization of the bHLH gene family and analysis of their potential relevance to chlorophyll metabolism in Raphanus sativus L
BMC Genomics (2022)
-
Genome-wide analysis of basic helix–loop–helix (bHLH) transcription factors in Aquilaria sinensis
Scientific Reports (2022)