Abstract
Candidemia is associated with a high mortality rate, and initial adequate antifungal therapy results in a significant decrease in the crude mortality. We herein report a rapid method that can identify eight Candida species in candidemia using imperfect match quenching probes (IM Q-probes) within three and a half hours of whole blood sample collection. Furthermore, employing the D value, which reflects the difference between the Tm signature from a clinical isolate and that registered in the database, it is possible to quickly identify samples suitable for IM Q-probe identification. We first evaluated the method using 34 Candida colonies collected from different patients, and 100% (34/34) of the identification results matched the preidentified Candida species. We then performed blind tests using eight whole blood samples artificially mixed with eight different Candida species respectively, and all identification results correctly matched the preidentified Candida species. Finally, using 16 whole blood samples collected from candidemia patients, we compared the IM Q-probe method with the culture/sequencing method. Of a total of 16 patient samples, 100% (16/16) matched the culture and sequencing results. The IM Q-probe method is expected to contribute not only to the life expectancy of candidemia patients but also to antifungal stewardship.
Similar content being viewed by others
Introduction
Candida species are the most frequent organisms involved in invasive fungal infections and major causes of not only local mucous membrane infections but also widespread propagation with multisystem organ failure1. Recently, due to advances in medicine, such as dialysis, major surgery and immunosuppressants, the rates of invasive candidiasis have been increasing. Indeed, in the United States, Candida species are the fourth-most common pathogen of nosocomial bloodstream infection2,3. Candidemia has an associated mortality that is as high as 25%, and initial adequate antifungal therapy results in a significant decrease in the crude mortality4,5,6. However, blood cultures usually take two to five days to finalize, which leads to a significant delay in the implementation of culture-driven therapy1,7,8. In addition, although most episodes of candidemia are caused by Candida albicans, opportunistic infections due to non-C. albicans species have been reported with increasing frequency9.
Pfaller et al. reported that the trends in descending order of frequency of Candida species causing candidemia were C. albicans (42.1%), C. glabrata (26.7%), C. parapsilosis (15.9%), C. tropicalis (8.7%), C. krusei (3.4%), C. lusitaniae (1.1%), C. dubliniensis (0.9%) and C. guilliermondii (0.4%)10. Because definitive therapy by selecting an appropriate anti-Candida agent should be based on the species of Candida, the rapid identification of Candida species is critical for the prompt initiation of appropriate therapy1.
Niimi et al. reported the novel “melting temperature mapping method” for rapidly identifying the dominant bacteria in a clinical sample (whole blood sample, etc.) from a sterile site11, which is our previous teamwork. This study suggested that more than 100 bacterial species can be identified by using the melting temperatures (Tm) of 7 primer sets, and these identification results can be obtained within three hours of whole blood collection. This method is useful for identifying bacteria. However, regarding the identification of Candida species, it is more difficult to design universal primers for fungi than for bacteria since Candida species are eukaryotes, like human cells. In addition, to identify many Candida species at the species level, specific primers must be generally prepared for each Candida species. As a result, the system becomes more complicated.
To address these problems, we herein report the development of a rapid and easy method for identifying Candida species in candidemia using a real-time polymerase chain reaction (PCR)-based system.
Results
Workflow of the rapid identification method for eight Candida species
The workflow of the rapid identification method for Candida species developed in our laboratory is shown in Fig. 1a. Employing Candida universal PCR (using one Candida universal primer) and a melting temperature (Tm) value analysis of three probes, eight Candida species are able to be identified within three and a half hours of whole blood collection.
Workflow of the rapid identification method for eight Candida species within three and a half hours of sample collection. (a) Using a Candida universal primer and three IM Q-probes, this method rapidly identifies eight Candida species by finding the nearest matches of the Tm values in the database. (b) We designed three IM Q-probes that can bind the position with some probe-target mismatches in the Candida universal PCR amplicon. * Candida albicans 18 S ribosomal RNA gene (Accession No. AF114470).
The eight Candida species causing candidemia were selected based on the trends in descending order of frequency, as described above10. This method consists of four major steps. First, fungal DNA is directly extracted from a clinical sample (e.g. 2 mL of whole blood sample) as a template for PCR. Step two involves PCR using a Candida universal primer and three imperfect match quenching probes (IM Q-probes). The Candida universal primer can detect all species of Candida. The three PCR reaction mixtures (three PCR tubes) contain the same Candida universal primer set and each IM Q-probe. We designed three IM Q-probes (IM Q-probe 1, IM Q-probe 2, IM Q-probe 3) that are able to bind to the positions with some probe-target base sequence mismatches in the same Candida universal PCR amplicon (Fig. 1b, Table 1). Allowing for some probe-target base sequence mismatches, there is substantial flexibility in these IM Q-probe designs. To obtain stable Tm values (and therefore stable identification results), we carefully selected each probe position whose target base sequence is the same among the different strains of the same Candida species reported in the DNA Data Bank of Japan (Table S1). PCR is then performed, and one PCR amplicon is obtained. In step three, the three Tm values are acquired by analyzing each IM Q-probe. Each of the Tm values varies depending on Candida species. The Tm value analysis showed various melting peaks (i.e. Tm values) among the eight Candida species with each IM Q-probe (Fig. S1). Consequently, the combination of the three Tm values was Candida species-specific (Fig. 2). In C. glabrata and C. lusitaniae, using the IM Q-probe 1, Tm values could not be obtained because of the large number of probe–target mismatches. Step four involves the identification of Candida species by comparing each of three Tm values with the database. We constructed a preliminary database of the eight Candida species showing the mean values of triplicate Tm measurements (Table S2). Candida isolates are able to be rapidly identified by comparing the Tm values to the values in the database.
In order to identify a Candida isolate, we calculate the difference (D) value using the formula shown in Fig. 3. The D value reflects the difference between the Tm signature from a clinical isolate and that registered in the database. Candida species are identified by comparing the three distances from the average of a Candida isolate to those in the database. Tm values above the average receive a “+” designation, while those below the average receive a “−” designation. The known Candida species in the database with the smallest D value is then identified as a correct match.
The assessment of the accuracy of the IM Q-probe method
In order to assess the accuracy of the IM Q-probe method, we first validated the measurement error. Using the Rotor-Gene Q instrument, whose tube-to-tube variation is within ± 0.1 °C, we tried to validate the measurement errors among 10 samples using the same C. albicans DNA template in the same trial (Table S3). The results showed that the maximum measurement error of the IM Q-probes was ± 0.25 °C. On receiving this result, we then established preliminary interpretative criteria to assess the suitability of the IM Q-probe identification. The interpretative criteria for identification based on the D value are shown in Table 2. Considering the maximum measurement error of the IM Q-probes, identification results with a D value of ≤ 0.41 are considered the Candida isolate. To determine the D threshold (= 0.41), we calculated it employing three Tm degrees (+0.25, −0.25, +0.25) and the formula (Fig. 3). If the IM Q-probe method shows identification results with D values of >0.41, the identification result with the lowest D value is highly likely to be the Candida isolate due to the high specificity of the IM Q-probe identification (Table S4). C. lusitaniae and C. glabrata are narrowed down as pathogen, but cannot be distinguished using the D value only. In this case, it is easy to distinguish them by checking the difference in their Tm values already obtained, which are apparently different (Table S4). In addition, using the current protocols, the limit of identification of the IM Q-probe method is 1.2 CFU of C. albicans per PCR tube (Table S5).
We subsequently evaluated the accuracy of the IM Q-probe method using 34 Candida colonies (8 Candida species) obtained from various type of samples (blood, sputum and urine) collected from different patients (Table 3). As a result, 100% (34/34) of the IM Q-probe results, whose D values were all ≤ 0.41, correctly matched the pre-identified Candida species by the sequencing method.
In order to assess the IM Q-probe method, we then performed blind tests using the whole blood samples artificially mixed with Candida species (200 CFU of Candida species per 2 mL of healthy whole blood). After concealing the name of Candida species, we tried to identify the Candida species in the whole blood. In all eight trials with eight different Candida species respectively, the IM Q-probe results, whose D values were all ≤ 0.41, correctly matched the pre-identified Candida species by the sequencing method (Table 4).
Finally, using 16 whole blood samples (2 mL) collected from candidemia patients, we assessed the accuracy of the IM Q-probe method compared that of the culture/sequencing method. The individual IM Q-probe results compared with the culture and sequencing results are shown in Table 5. Of the total of 16 patient samples, 100% (16/16) matched the culture and sequencing results with D values of ≤0.41. The breakdown of the identification results was, 11 cases of C. albicans, 4 cases of C. parapsilosis, and 1 case of C. tropicalis.
Discussion
In relation to fungal infection, rapid identification methods of Candida species using a high-resolution melting-curve (HRM) analysis have been previously reported12,13,14. None of those reports used whole blood samples collected from candidemia patients to assess the accuracy of their rapid identification methods. HRM analyses primarily rely on differences in the melting curve shapes. The IM Q-probe method does not use melting curve shapes, instead relying only on Tm values. For this reason, although melting curve shapes are affected by various DNA concentrations15, Tm values are not. In fact, fungal concentrations in candidemia vary among individual patients.
Mylonakis et al. reported the utility of a T2 magnetic resonance (T2MR) assay for the rapid diagnosis of candidemia in whole blood16,17. Using a T2MR assay, Candida species can be detected directly from a clinical sample with high sensitivity. However, the T2MR assay detects only five kinds of Candida species (C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, and C. krusei). Furthermore, to identify the Candida species, gene sequencing analyses are additionally needed. In general, gene sequencing analyses tend to be costly and therefore have limited applications for identifying pathogens in clinical practice.
Employing only one Candida universal primer and three probes, the IM Q-probe method enabled the rapid detection and identification of the eight Candida species within three and a half hours of whole blood collection. These findings suggest that this method is rapid and easy for identifying pathogenic Candida species in cases of candidemia. If different-colored fluorescent IM Q-probes are employed, multiplexing in a single tube might be possible, which equates to even easier performance.
In order to identify various Candida species, we designed quenching probes with some probe-target mismatches. Based on the nearest neighbor thermodynamic theory18,19, the variations in melting temperature of the IM Q-probes depend on the number and position of probe-target mismatches on IM Q-probe hybridization. For this reason, the Tm values of the IM Q-probes provided adequate diversity for the IM Q-probe method to identify the eight Candida species at the species level. Chakravorty et al. reported the rapid identification of bacterial isolates using a sloppy molecular beacon (SMB) melting temperature signature technique20. SMB probes possess relatively long probe sequences, enabling them to form hybrids with PCR amplicons despite the presence of mismatched base pairs. However, we designed the imperfect-match “Q probes” so as not to form secondary structures, since Q probes tend to self-quench by forming secondary structures. To obtain stable identification results, we selected IM Q-probe positions whose target base sequence was the same among different strains of the same Candida species. If the IM Q-probe method identifies a mutant strain (D value > 0.41) with a mutation in the IM Q-probe target regions, we will register it in the database. Consequently, the IM Q-probe method would be able to identify mutant strain as well. In addition, to obtain stable identification results, the pH and salt concentration of the PCR buffer should be consistent. Therefore, preparing a reagent kit is advised.
We first set the interpretative criteria to assess the suitability of the IM Q-probe identification. Considering the maximum measurement error of the IM Q-probes (±0.25 °C) using Rotor-Gene Q instrument whose tube-to-tube variation was within ±0.1 °C, the identification results with a D value ≤ 0.41 were considered the Candida isolate. This interpretative criterion with the D value can be revised according to the maximum measurement error, and the tube-to-tube variation caused by each real-time PCR instrument is the main component of the maximum measurement error. The tube-to-tube variations in almost all commercially available real-time PCR instruments are within ±0.3 to ±0.5 °C. If the maximum measurement error is within ±0.8 °C, then the range in the theoretical D value should be within 1.31. Considering this range of D value and the high specificity of the IM Q-probe method (Table S4 shows the D value between the most similar Candida species in the database is 2.98), almost all commercially available real-time PCR instruments can feasibly be used for the IM Q-probe method to correctly identify any of eight Candida species in a clinical sample.
Concerning the specificity of the IM Q-probe method, the method detects not only the eight Candida species (C. albicans, C. glabrata, C. parapsilosis, C. tropicalis, C. krusei, C. lusitaniae, C. dubliniensis, C. guilliermondii) but also other kinds of Candida species (C. fermentati, C. inconspicua, C. kefyr, C. nivariensis, C. norvegensis, C. orthopsilosis, C. pelliculosa, C. sake, C. zeylanoides, etc.) since the Candida universal primer can detect all species of Candida. In this regard, the problem is that C. tropicalis and C. orthopsilosis cannot be distinguished using the IM Q-probe method (Table S6). Likewise, C. guilliermondii, C. fermentati and C. zeylanoides cannot be distinguished as well. However, the frequency of other kinds of Candida species causing candidemia is only 0.8% (Table S6), so this does not appear to present an issue in identifying Candida species in clinical practice. Of note, regarding specificity, the IM Q-probe method can also identify genus Aspergillus and genus Cryptococcus at the genus level (Table S7). It is easy to distinguish genus Candida from genus Aspergillus and genus Cryptococcus due to the high specificity of the IM Q-probe identification. However, the purpose of the IM Q-probe method is to identify any of eight Candida species in a clinical sample. In order to identify various kinds of fungi at the species level, additional developments will be required.
In conclusion, the IM Q-probe method enabled the rapid identification of eight Candida species in a clinical sample within three and a half hours of whole blood collection. Furthermore, using the D value, it was possible to quickly identify samples suitable for IM Q-probe identification. The IM Q-probe method is expected to contribute not only to the life expectancy of candidemia patients but also to antifungal stewardship.
Methods
Candida isolates
Candida isolates were obtained from patient’s blood, sputum and urine culture isolates at Toyama University Hospital, then were sequenced and identified at the species level.
Clinical samples
A total of 16 whole blood samples were collected from patients with candidemia at Toyama University Hospital. All procedures were performed under a protocol approved by the Ethics Committee of the University of Toyama, and written informed consent was obtained from all patients. The methods were carried out according to the approved guidelines.
Isolation of candida genomic DNA from colonies
The Candida colonies were selected with a sterile inoculating loop and suspended in 1 mL of molecular-grade distilled water (water deionized and sterilized for molecular biology; NACALAI TESQUE, INC., Kyoto, Japan). The sample was subsequently centrifuged at 20,000 × g for 10 minutes, and 900 μL of the supernatant was carefully removed so as not to lose the pellet. DNA was isolated from the resulting pellets using a DNA extraction kit (QIAamp UCP Pathogen Mini Kit; Qiagen, Hilden, Germany) and glass beads (Pathogen Lysis Tubes S; Qiagen) in accordance with the supplier’s instructions. Finally, Candida DNA was eluted with 100 μL of elution buffer.
Isolation of candida genomic DNA from whole blood
A total of 2 mL of venous blood was collected in EDTA-2K tube (NIPRO, Osaka, Japan). The blood sample was then centrifuged at 100 × g for 5 minutes to spin down the blood cells, and the resulting supernatant fraction (1 mL) was used. The supernatant was centrifuged again at 20,000 × g for 10 minutes, and 900 μL of the supernatant fraction was carefully removed so as not to disturb the pellet. Next, 1 mL of molecular-grade distilled water (water deionized and sterilized for molecular biology; NACALAI TESQUE, INC.) was added to the pellet, and the mixture was gently turned upside down several times, and subsequently centrifuged again at 20,000 × g for 5 minutes. Finally, 1 mL of the supernatant fraction was again carefully removed unless you resuspend the pellet. DNA was isolated from the pellet employing a DNA extraction kit (QIAamp UCP Pathogen Mini Kit; Qiagen) and glass beads (Pathogen Lysis Tubes S; Qiagen) in accordance with the supplier’s instructions. Finally, Candida DNA was eluted with 100 μL of elution buffer.
PCR assays
In each of the following processes, the QIAgility system (Qiagen) provided an automated PCR setup. The Rotor-Gene Q (Qiagen) was used for the amplification, real-time detection of the target DNA and Tm value analysis of the Imperfect Match Quenching probes (IM Q-probes). All PCR assays were performed as single-tube assays (no multiplex PCR). We used 1.5 mL PCR-clean Eppendorf tubes that were RNase- and DNase-free (Eppendorf, Hamburg, Germany) and 0.2 mL PCR tubes (Qiagen) for PCR. Fungal (Candida) universal primer targeting the 18 S ribosomal RNA gene (18 S rDNA), which has been described previously21, were synthesized by Life Technologies Japan, Ltd. (Tokyo, Japan). The IM Q-probes were designed using a multiple alignment software program (Clustal X) and were synthesized by NIPPON STEEL and SUMIKIN Eco-Tech Corporation (Ibaragi, Japan). The IM Q-probes were as follows: IM Q-probe 1 (5′-CTTTCCTTCTGGGTAGCCATTT-3′, probe size: 22 bp), IM Q-probe 2 (5′-TGGAATAATAGAATAGGACGTTATGGTTC-3′, probe size: 29 bp), IM Q-probe 3 (5′-GCATCAGTAATCAGTTGTCAGAGGAGAAATTC-3′, probe size: 32 bp).
During the PCR procedure, the PCR reaction mixture (50 μL) contained 7 μL of DNA template in KOD FX Neo reagents (1 × PCR Buffer with 2.0 mM of Mg2+, 0.4 mM of dNTPs and 0.02 U of KOD FX Neo → 1.0 U of KOD FX Neo U of KOD FX Neo; TOYOBO LIFE SCIENCE, Osaka, Japan), 0.2 μM of Candida universal forward primer, 0.6 μM of Candida universal reverse primer and 0.02 μM of each IM Q-probe.
Each sample was incubated for 5 minutes at 95 °C, then was denatured for 10 seconds at 98 °C, annealed for 30 seconds at 57 °C, extended for 20 seconds at 72 °C and subjected to fluorescence acquisition for 2 seconds at 82 °C for 50 cycles.
The Tm value analysis
For the Tm value analysis, the resulting amplicons with each IM Q-probe were heated at 95 °C for 10 seconds and then cooled at 40 °C for 90 seconds. A post-PCR Tm value analysis of the 3 probes was performed from 40 °C to 95 °C, increasing at 1 °C/step. The data profile was subsequently analyzed using the Rotor-Gene Q software program, and the Tm values were identified.
Analytical sensitivity tests
Prior to performing the sensitivity tests, C. albicans was cultivated in Sabouraud dextrose broth at 37 °C for 48 h, and C. albicans suspensions (1 mL) in phosphate-buffered saline (PBS) were prepared. A total of 50 µL of the diluted suspensions were inoculated on Sabouraud dextrose agar. After incubation at 37 °C for 24 h, the number of colony-forming units (CFU) was determined by counting the colonies grown on the agar plates in triplicate12.
The limits of identification and detection were determined by serially diluting cultures with known counts (CFU) of C. albicans in PBS and subjecting the samples to IM Q-probe identification using the IM Q-probes. The limit of identification was determined to be the final dilution of the template in which the IM Q-probe identification result was correct (Table S5) with a D value of ≤0.41.
A nucleotide sequence-based analysis of fungal genomic DNA
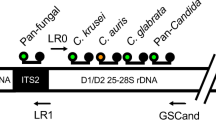
PCR amplicons amplified the D1-D2 region and the internal transcribed spacer (ITS) regions using fungal universal primers were purified (QIAquick PCR Purification Kit; QIAGEN) and then sequenced (3500 Genetic Analyzer; Thermo Fisher Scientific, Waltham, Massachusetts, USA)22. An online homology search was performed for strain identification using the BLAST nucleotide database tool of the DNA Data Bank of Japan (http://www.ddbj.nig.ac.jp/index-j.html).
Culture-based biochemical identification of Candida species
The whole blood samples (one aerobic blood culture bottle and one anaerobic blood culture bottle) were collected simultaneously with the blood sample for the IM Q-probe analysis from the same puncture site. The whole blood samples were then analyzed according to the standard methods used by the Clinical Laboratory Center (certified ISO15189) at Toyama University Hospital. The blood culture procedures were performed using the BacT/ALERT 3D system (bioMérieux, Inc., Marcy-l'Étoile, France). Identification of fungi was presumptively performed by CHROMagerTM Candida (Becton Dickinson and Company, Franklin Lakes, New Jersey, USA) and definitely performed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Bruker, Billerica, Massachusetts, USA).
References
Pappas, P. G. et al. Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin. Infect. Dis. 62, e1–50 (2016).
Blumberg, H. M. et al. Risk factors for candidal bloodstream infections in surgical intensive care unit patients: the NEMIS prospective multicenter study. Clin. Infect. Dis. 33, 177–86 (2001).
Wisplinghoff, H. et al. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39, 309–17 (2004).
Andes, D. R. et al. Impact of treatment strategy on outcomes in patients with candidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin. Infect. Dis. 54, 1110–22 (2012).
Takesue, Y. et al. Management bundles for candidaemia: the impact of compliance on clinical outcomes. J. Antimicrob. Chemother. 70, 587–93 (2015).
Pattel, G. P. et al. The effect of time to antifungal therapy on mortality in candidemia associated septic shock. Am. J. Ther. 16, 508–11 (2009).
Taur, Y., Cohen, N., Dubnow, S., Paskovaty, A. & Seo, S. K. Effect of antifungal therapy timing on mortality in cancer patients with candidemia. Antimicrob. Agents Chemother. 54, 184–90 (2010).
Arvanitis, M., Anagnostou, T., Fuchs, B. B., Caliendo, A. M. & Mylonakis, E. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin. Microbiol. Rev. 27, 490–526 (2014).
Hachem, R., Hanna, H., Kontoyiannis, D., Jiang, Y. & Raad, I. The changing epidemiology of invasive candidiasis: Candida glabrata and Candida krusei as the leading causes of candidemia in hematologic malignancy. Cancer 112, 2493–9 (2008).
Pfaller, M. et al. Epidemiology and outcomes of candidemia in 3648 patients: data from the prospective antifungal therapy (PATH Alliance®) registry, 2004–2008. Diagn. Microbiol. Infect. Dis. 74, 323–31 (2012).
Niimi, H. et al. Melting temperature mapping method: a novel method for rapid identification of unknown pathogenic microoganisms within three hours of sample collection. Sci. Rep. 5, 12543 (2015).
Goldschmidt, P. et al. New strategy for rapid diagnosis and characterization of fungal infections: the example of corneal scrapings. PLoS One 7, e37660 (2012).
Decat, E. et al. Rapid and accurate identification of isolates of Candida species by melting peak and melting curve analysis of the internally transcribed spacer region 2 fragment (ITS-MCA). Res. Microbiol. 164, 110–7 (2013).
Nemcova, E. et al. Rapid identification of medically important Candida isolates using high resolution melting analysis. PLoS One 10, e0116940 (2015).
Ozbak, H., Dark, P., Maddi, S., Chadwick, P. & Warhurst, G. Combined molecular gram typing and high-resolution melting analysis for rapid identification of a syndromic panel of bacteria responsible for sepsis-associated bloodstream infection. J. Mol. Diagn. 14, 176–184 (2012).
Neely, L. A. et al. T2 magnetic resonance enables nanoparticle-mediated rapid detection of candidemia in whole blood. Sci. Transi Med. 24, 182ra54 (2013).
Mylonakis, E. et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: a clinical traial. Clin. Infect. Dis. 60, 892–9 (2015).
Lerman, L. S., Fischer, S. G., Hurley, I., Silverstein, K. & Lumelsky, N. Sequence-determined DNA separations. Annu. Rev. Biophys. Bioeng. 13, 399–423 (1984).
Dumousseau, M., Rodriguez, N., Juty, N. & Novère, N. L. MELTING, a flexible platform to predict the melting temperatures of nucleic acids. BMC Bioinforma. 13, 101 (2012).
Chakravorty, S. et al. Rapid universal identification of bacterial pathogens from clinical cultures by using a novel sloppy molecular beacon melting temperature signature technique. J. Clin. Microbiol. 48, 258–267 (2010).
Ueno, T. et al. Eukaryote-made thermostable DNA polymerase enable rapid PCR-based detection of mycoplasma, ureaplasma and other bacteria in the amniotic fluid of preterm labor cases. PLoS One 10, e0129032 (2015).
Leaw, S. N. et al. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J. Clini Microbiol. 44, 693–699 (2006).
Acknowledgements
This work was supported by AMED under Grant Number 18im0210217 to H.N., and partially supported by JSPS KAKENHI Grant-in-Aid for Scientific Research (B) under Grant Number JP18H02728 to I.K.
Author information
Authors and Affiliations
Contributions
Y.H. and H.N. developed the rapid identification method of Candida species (IM Q-probe method). H.N. wrote the manuscript. Y.H. designed and performed the experiments. I.S., Y.Y. and I.K. supervised the overall project. All authors read and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Higashi, Y., Niimi, H., Sakamaki, I. et al. Rapid Identification of Candida Species in Candidemia Directly from Blood Samples Using Imperfect Match Probes. Sci Rep 10, 5828 (2020). https://doi.org/10.1038/s41598-020-62276-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-62276-5
- Springer Nature Limited