Abstract
Bacillus thuringiensis serovar israelensis (Bti) is used to control insect vectors of human and animal diseases. In the present study, the toxicity of four strains of Bti, named T0124, T0131, T0137, and T0139, toward Aedes aegypti and Culex quinquefasciatus larvae was analyzed. The T0131 strain showed the highest larvicidal activity against A. aegypti (LC50 = 0.015 µg/ml) and C. quinquefasciatus larvae (LC50 = 0.035 µg/ml) when compared to the other strains. Furthermore, the genomic sequences of the four strains were obtained and compared. These Bti strains had chromosomes sizes of approximately 5.4 Mb with GC contents of ~35% and 5472–5477 putative coding regions. Three small plasmids (5.4, 6.8, and 7.6 kb) and three large plasmids (127, 235, and 359 kb) were found in the extrachromosomal content of all four strains. The SNP-based phylogeny revealed close relationship among isolates from this study and other Bti isolates, and SNPs analysis of the plasmids 127 kb did not reveal any mutations in δ-endotoxins genes. This newly acquired sequence data for these Bti strains may be useful in the search for novel insecticidal toxins to improve existing ones or develop new strategies for the biological control of important insect vectors of human and animal diseases.
Similar content being viewed by others
Introduction
During sporulation, the gram-positive bacterium Bacillus thuringiensis (Bt) produces crystalline inclusions consisting of δ-endotoxins (Cry or Cyt proteins) with insecticidal activity1. Genomic analysis has contributed to the identification of new genes coding for toxins that are active against different insect species including orders such as Lepidoptera, Diptera2,3,4,5,6,7, and Coleoptera8. Proteins with nematicidal9,10,11 and molluscicidal12 activities have also been described. In addition, genome sequencing of Bt strains with diverse ecological functions has been conducted, including a endophytic strain with potential utility in the biocontrol of phytopathogens13.
Sequencing of complete Bt genomes has allowed structural and functional analysis of new plasmids that enhance our knowledge of the pathogenic properties of Bt in targeting organisms14,15,16,17. One study reported the plasmid sequence of a Bacillus thuringiensis serovar israelensis (Bti) strain, and revealed that it may produce up to seven crystal-forming toxins, named Cry4A, Cry4B, Cry10A, Cry11A, Cyt1A, Cyt2Ba, and Cyt1Ca, which are all encoded by genes found in a single 127923 bp plasmid called pBtoxis18. The average size of the complete genome sequences of Bti is 6.1 Mb, with ~35% GC content of the chromosomal DNA and an average of 6132 coding sequences19,20. Genome sequences of seven Bti isolates have been reported so far19,21,22,23,24.
In this study, we sequenced the genomes of four Bti strains, specifically T0124, T0131, T0137, and T0139 that were collected from the soil of the Tocantins state in Brazil and determined their larvicidal activity against larvae of two important mosquito species of A. aegypti and C. quinquefasciatus. Then, to better characterize these strains, we performed comparative and phylogenetic analyses among their different genomes and compared the potential insecticidal toxin genes and other virulence factors of the four Bti strains with the commercial Bti strain H14. In case we identify high anti-mosquito activity with these strains, we believe these new data are useful in the continuous search for new insecticidal toxins to improve the existing ones or develop new strategies for the biological control of important insect vectors of human and animal diseases.
Results
Larvicidal activity and features of δ-endotoxins
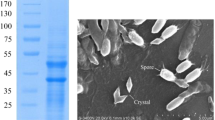
The spore-crystal mixtures of Bti strains T0124, T0131, T0137, and T0139 were tested against third instar larvae of A. aegypti and C. quinquefasciatus. The T0131 strain showed the highest larvicidal activity against A. aegypti (LC50 = 0.015 µg/ml) and C. quinquefasciatus (0.035 µg/ml) when compared to the other strains. Moreover, based on the toxicity ratios, the T0131 strain presented similar toxicity to the reference strain H14 (Toxicity ratio = 1.1 against A. aegypti and Toxicity ratio = 1.3 against C. quinquefasciatus) (Table 1). The T0124, T0137, and T0139 isolates showed lower toxicities compared to the H14 strain (Table 1). However, the SDS-PAGE analysis of crystal protein content revealed that all the strains have similar protein profiles. δ-endotoxins with molecular weights of 130, 70, and 27 kDa (Fig. 1A) and round morphology (Fig. 1B–E), characteristics of the Bti protein profile, were observed for the strains.
Crystal protein profile of Bti strains. Lane 1: molecular weight marker (Invitrogen); Lane 2: T0124; Lane 3: T0131; Lane 4: T0137; and Lane 5: T0139. Seven μg of solubilized crystals from each strain were analyzed by SDS-PAGE. The ultrastructural characterization of the spores and Cry proteins from T0124 (B), T0131 (C), T0137 (D), and T0139 (E) strains. All strains presented round crystals. Arrows indicate spores (S) and round crystals (R).
Genome features
The average size of the chromosomal draft sequences of the T0124, T0131, T0137, and T0139 isolates was 5.4 Mb, with GC contents of ~35%. Chromosomes of these isolates contained 5477 (T0124), 5473 (T0131) and 5472 (T0137 and T0139) protein-coding genes. The number of tRNA genes was consistent over the strains (122) while small variation was seen in the number of rRNA (39–42) among them (Table 2).
Genome assembly showed high similarities in nucleotide identity with the HD-789 reference genome (Fig. 2). Regions (in blank) within positions 3 407 568 – 3 451 845 bp and 4 278 610 – 4 319 513 bp indicate two prophage sequences located on the reference chromosome which are absent in the T0124, T0131, T0137, and T0139 isolates (Fig. 2). All four strains contain six plasmids with average sizes of 5.4, 6.8, 7.6, 127, 235 and 359 kb. These replicons showed nucleotide identity greater than 99% with the extrachromosomal elements pTX14-1 (NC_002091), pTX14-2 (NC_004334), pTX14-3 (X56204), pBTHD789-2 (NC_018509), pBtoxis (NC_010076), and pHD1002-1 (NZ_CP009349), respectively (Table 3). The coverage for the plasmid assemblies was between 4000 and 14000 times for the 5.4, 6.8, and 7.6 kb plasmids and between 20 and 109 times for the 127, 235 and 359 kb plasmids.
Comparative chromosomal nucleotide analysis of Bti strains. The concentric rings represent the sequences of T0124, T0131, T0137, and T0139 against the reference CP003763 strain. The black circle represents GC content of CP003763. The blue circle represents T0124, red circle represents T0131, green circle represents T0137, and purple circle represents T0139. Regions with less than 80% identity appear as blanks on each ring. This circular map was generated using the BLAST Ring Image Generator (BRIG) tool48.
The 127 kb plasmid is the only one that encodes crystal-forming protein genes that are toxic to Diptera (cry11Aa, cry4Aa, cry4Ba, cry10Aa, cyt1Aa, cyt1Ca, and cyt2Ba) (Fig. 3) (Tables 3 and 4). No SNPs were found when the 127 kb plasmids of the different strains were compared.
Comparative sequence map of pT0124-4, pT0131-4, pT0137-4 and pT0139-4 plasmids. The circles starting with the outermost ring are as follows: circle 1 (pT0124-4) showing the position of δ-endotoxins; circle 2 (pT0139-4), circle 3 (pT0137-4), and circle 4 (pT0131-4) show regions of sequence similarity representing darker regions detected by BLASTN in the primary sequence (pT0124-4). Circle 5 shows GC content (deviation from average) and circle 6 illustrates the GC skew in green (+) and purple (−). The circle with δ-endotoxins and the map was generated using the Geneious47 and CGView56 tool.
Comparative genomic analysis
The genome drafts of the isolates T0124, T0131, T0137, and T0139 were compared to 14 other complete chromosomes of B. thuringiensis (Table 5) by phylogenetic analysis and Mauve alignment (Fig. 4). The Mauve alignment showed collinearity of genes among the isolates from this study and the Bti strains AM65-52 and HD-789, forming 32 locally collinear blocks (LCB) (Fig. 4A). The SNP-based phylogeny revealed close relationship between the isolates T0124, T0131, T0137, T0139 and the Bti strains AM65-52 and HD-789 (Fig. 4B). Although a total of 2190 SNPs positions were found in all analyzed chromosomes, no SNPs were found in the chromosomes of the isolates used in this study compared to Bti strains AM65-52 and HD-789 (Fig. 4C). Furthermore, the plasmids with 127 kb found in the four isolates (T0124, T0131, T0137, T0139) differed only by minor nucleotide changes (1 to 7 mutations) from the pBtoxis plasmid (NC_010076), and none of the nucleotide changes was related to the δ-endotoxins (Table 6).
Comparative chromosome sequences of the isolates T0124, T0131, T0137, and T0139 with 14 genomes of other strains of B. thuringiensis. (A) Reciprocal LCBs in different sequences are indicated by the same colors and are connected by lines. (B) Phylogenetic tree based on the concatenated SNPs. The SNPs were called by CSI phylogeny 1.452 using HD-789 strain as reference. The branch structure was confirmed by a bootstrap consensus tree inferred from 1,000 replicates in MEGA 1053. The scale bar indicates the evolutionary distance between the sequences determined by 0.10 substitutions per nucleotide at the variable positions. Red dots indicate the genomes of strains from the present study and blue dots indicate other genomes of Bti from the GenBank database. (C) The matrix shows the 2190 SNPs after pairwise comparison between isolates.
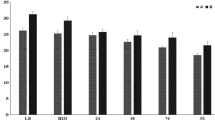
A functional gene ontology analysis was performed among the four strains (T0124, T0131, T0137, T0139) and two strains of B. thuringiensis (HS18-1, and YWC2-8), which presented toxic bioactivity to dipteran insect and not associated to the serotype H14, followed by a summary from shared OrthoVenn clusters. The comparison of the inferred proteins among the strains of this study and the two other strains revealed 4 829 proteins shared by the strains and a total of 231 orthologous clusters shared by HS18-1 and YWC2-8 (Fig. 5A). The HS18-1 and YWC2-8 strains presented specific genes with 6 and 64 single clusters, respectively (Fig. 5A). The analysis of all Gene Ontology (GO) terms assigned to 4 829 orthologous clusters shared by the species showed 1 180 for metabolic processes, 1 001 for ion binding, and 1317 for cell parts (GO-inferred terms) (Fig. 5B–D).
The Venn diagram of the strains from this study and other strains with toxicity to Diptera and summary of the functional gene ontology from shared OrthoVenn clusters. The Venn diagrams of T0124, T0131, T0137, T0139, HS18-1, and YWC2-8 (A). Summary of the functional gene ontology categories using GO slim57 for orthologous clusters in the Venn diagram overlapping regions are represented in the biological process (B), molecular function (C), and cellular component (D) categories.
The important genes of the sporulation process previously described as variable and absent in some Bacillus species25 were analyzed. ORF sequences coding for the germination gene (GerB), small acid soluble proteins genes (SspP and SspH), sensor kinase (SerK) genes, coat gene of the spore (CoatB), and sigma factor genes (SigB, SigE, SigF, and SigH) were compared to the same genes present in other species of the Bacilli group (Fig. 6). Higher sequence identity was observed for B. thuringiensis HD-789 and B. thuringiensis serovar israelensis AM65-52. The GerB and SspH genes showed the highest sequence variability when compared with the sequences acquired in this study.
Heatmap comparison of the distribution of sporulation (GerB, SspP, SspH, SerK, CoatB) and sigma factor (SigB, SigE, SigF, and SigH) gene sequences among some species from the Bacilli group. Each column and line represents a gene and a Bacillus lineage, respectively, and percentage sequence identities between these species and the strains from this study were shown as colors ranging from 70% (dark blue) to 100% (red) as shown in the bottom. Undetected gene sequences are shown in black. The comparative analysis was performed using blastx and the Heatmap was generated using version 4.9.0 of the MeV tool49.
Discussion
Here, the four new Bti strains T0124, T0131, T0137, and T0139, collected from the soil of the Tocantins state in Brazil, showed toxic activity to larvae of A. aegypti and C. quinquefasciatus. In addition to the fact that the mosquito strains were collected from locations not targeted by insecticide applications and hence presenting low risk of insecticide resistance build up. The Bti mode of action is distinct from neurotoxic or growth-regulating compounds used for mosquito control. These facts make Bti an effective alternative for controlling mosquito populations displaying or not resistance to these insecticides26.
The Bti strains analyzed in this study presented different lethal concentrations among them and when compared to a reference commercial strain of Bti (H14). However, the δ-endotoxin gene content and toxin protein profiles assessed by SDS-PAGE were very similar. The T0131 strain presented the highest toxicity for both insect vectors and, therefore, it is probably the most promising strain for biological control among the four isolated strains.
We performed whole genome sequencing of T0124, T0131, T0137, and T0139. However, differently from the toxicity results, the genomic analysis of these isolates indicated highly similar sequences. Previous genome comparison among strains of Bt revealed that 80% of the genes of this species are conserved, and the variability among Bt strains can be attributed to the acquisition of essential or non-essential genes from other microorganisms residing in the same microbial community27. In addition, Bt has an open pan-genome which is a characteristic of species that colonize different environments and have different genetic material exchange pathways28. Bt species comprise different subspecies and comparative analysis of the same subspecies may reveal genomically identical or highly related strains, even from different geographic regions. Such findings could be explained by the emergence of clonal lineages of pathogens that successfully colonized the biosphere, undergoing limited genetic exchange, thus representing homogeneous subspecies23. Similarly, studies have shown that Bti also present genomically similar strains, indicating the close relationship among them and suggesting a high degree of genomic conservation29,30 thus corroborating the results obtained in this study.
The assembled chromosomes of T0124, T0131, T0137, and T0139 did not show the presence of the two prophages present in the Bti reference HD-78921. Bolotin et al.24 also identified the sequences the two prophages in the Bti AM65-52 strain. Phage sequences, as well as plasmids, are said to be mobile genetic elements which also contribute to genetic diversity among species and are considered important tools in the divergence of strains and closely related bacterial species31.
Despite the presence of rearrangements and sequence inversions also that have been linked to the variability of genetically related species32, the strains from this study, HD-789 and AM65-52 showed collinear chromosomes. Although the polymorphism analysis indicated the presence of various SNPs in the Bti isolates, none of the mutations reported was related to the insecticidal activity33. The SNP-based phylogeny revealed close relation among the four isolates and other Bti isolates (HD-789 and AM65-52), in agreement with previous study33, reinforcing the close genetic relationship among these bacteria.
With regard to plasmids sequences, the high number of copies of extrachromosomal elements per chromosome can explain the high coverage of the plasmids with sizes of 5.4, 6.8, and 7.6 kb in this study24,34. Although some studies reported the ability of Bti strains to harbor up to nine plasmids, the assembly generated here revealed the presence of only six plasmids in the genomes of the isolates, which have also been reported elsewhere21,24. The 235 kb plasmids are presented in all sequenced genomes of these Bti strains. The 359 kb plasmid was described previously24 and is also found in the genomes of the T0124, T0131, T0137, and T0139 isolates.
Since the plasmids with 235 kb and 359 kb are not reported to show any crystal-forming proteins linked with toxic activity, they have not been described in this study. In fact, the plasmid with 235 kb has been described as a conjugative plasmid and the plasmid with 359 kb encodes various metabolite transporters24.
Since the 1980s, the direct relationship between plasmids and the pathogenicity of Bt was established, as they are responsible for carrying genes that express toxins active against target insects35. Plasmids of 127 kb were found in all isolates of Bti containing cry and cyt genes involved in insect toxicity. This type of plasmid, termed pBtoxis, is widely studied and described as the only plasmid capable of encoding the crystal-forming toxins of this bacterium36. In addition, pBtoxis also presents sequences with functions predicted to increase crystal formation and subsequent cell viability, acting as chaperones32,36. The additional cry4Ba coding gene in plasmid pBtoxis-like has been reported to account for the increased effectiveness in mosquito larvae killing of Bti strain33. In the present study, SNPs analysis of the plasmids carrying the genes encoding the mosquitocidal endotoxins did not reveal any mutation in pT0131-4 what could explain the high toxicity of T0131 strain.
Different strains of Bt within the same serotype may share a highly related plasmid pattern; this relationship is most evident in different strains of Bti H14 serotype, which, although isolated from different geographic origins, have the same basic plasmid pattern, sometimes even identical37. Therefore, our results, that show a high degree of genomic conservation among the strains T0124, T0131, T0137, and T0139, are consistent with previous studies.
The functional gene ontology analysis from shared clusters showed a unique set of proteins identified only in the genome of the YWC2-8 isolate associated with magnesium transport and in the HS18-1 isolate associated with vitamin B6 catabolic processes and pyridoxal 4-dehydrogenase activity (Fig. 5A). The summary of the functional gene ontology showed diversity for metabolic process category (Fig. 5B). The metabolic processes play important roles in the insecticidal activity of Bt because metabolic pathways are regulated to provide amino acid, carbon, and energy substances for sporulation and massive synthesis of crystal toxins38,39.
Heatmap analysis shows that sporulation genes and sigma factors are conserved among Bti strains, while the SspH and GerB gene showed highest variability. Although spore formation is central to the definition of Bacilli, these genes have been described as variable and absent in some species as a result of niche-specific constraints that may lead to variability in the detection of stress conditions, spore resistance, and germination25.
The comparative analysis of four new genomes of Bti carried out in the present study revealed their very high identity of nucleotide sequence. Furthermore, the results presented here are important for evolutionary studies of this species and potentially may contribute to the improvement of existing strategies or the development of new approaches in biological control that use these bacteria. Further investigations aiming to evaluate potential differences at transcriptomic/proteomic levels during specific phases (e.g., middle vegetative, early sporulation and late sporulation) of the four Bti strains will contribute to clarify the higher larvicidal activity described here for the T0131 strain.
Methods
Isolation of Bti strains
B. thuringiensis serovar israelensis (serotypes H14) T0124, T0131, T0137, and T0139 strains were isolated from a soil sample collected in Tocantins state (Brazil) according to the previously described protocol40. The bacterial strains were cultured at 28 °C for 12 h using the streak plate method on Luria-Bertani (LB) solid medium (10 gL−1 tryptone, 5 gL−1 yeast extract, 10 gL−1 NaCl, and 20 gL−1 Agar). Single bacterial colonies of each strain were inoculated in the LB liquid medium at 28 °C with shaking for 16 h.
Spore-crystal protein preparation and crystal analysis by SDS–PAGE
Spore-crystal mixtures were obtained according to the protocol described previously41. For SDS-PAGE analysis, the crystals were purified using hexane and low speed centrifugation according to the previously described method42. Proteins were suspended in a small volume of phosphate-buffered saline (136 mM NaCl, 1.4 mM KH2PO4, 2.6 mM KCl, 8 mM Na2HPO4, and 4.2 ml H2O; pH 7.4), and fractionated by electrophoresis on 12% SDS-PAGE gels43.
Scanning electron microscopy
The characterization of the spores and Cry proteins from the T0124, T0131, T0137, and T0139 strains was performed by scanning electron microscopy. The strains were cultivated in NYSM agar medium at 30 °C for 72 h, then a loop of the isolate was collected and diluted in sterile water. A volume of 100 µL of this dilution was deposited over metallic supports and dried for 24 h at 37 °C, covered with gold for 180 s using an Emitech apparatus (model K550; Quorum Technologies, Lewes, UK), and observed under a Zeiss scanning electron microscope (model DSM 962; Carl Zeiss AG, Oberkochen, Germany) at 10 or 20 Kv.
Mosquitoes and toxicity bioassays
The colonies of A. aegypti and C. quinquefasciatus were established from insects collected from the field in regions of transition between the urban and rural areas in the state of Tocantins, Brazil, (11°40′55.7″ latitude S, 49°04′3.9″ longitude W), where no insecticides have been used for the control of mosquitoes. The insects were maintained in the Entomology Laboratory of the Federal University of Tocantins, Gurupi Campus, according to the methodology described previously44. Adult mosquitoes were maintained on a 10% aqueous sucrose solution and the blood of live Wistar rats (Rattus norvegicus albinus). The larvae were reared in plastic containers (35 cm × 5 cm) and were fed a sterilized diet (80/20 mix of chick chow powder/yeast). All bioassays were conducted at 26 ± 1 °C, 60.0 ± 5% RH, with a 12 h light-dark photoperiod. All applicable international, national, and institutional guidelines for the care and use of animals were followed. Bioassays were conducted using the suspension isolated from the spore-crystal mixtures against third instar A. aegypti and C. quinquefasciatus larvae. The concentrations were determined as described previously45. Seven concentrations were used for each spore-crystal mixture from each strain. Sterile distilled water was used as a control, and the larval mortality was recorded 24 h post inoculation. Three replicates with 25 larvae for each spore-crystal mixture were performed for all tested concentrations and for the non-treated control group. The spore-crystal mixture from the H14 strain was used as a reference.
Whole genome sequencing, assembly, and annotation
Total genomic DNA was extracted and purified using a Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. DNA concentration and purity were measured using a NanoDrop™ 8000 (Thermo Fisher Scientific, Waltham, MA, USA). Whole genome sequencing was performed on the Illumina MiSeq™ platform using a paired-end application (2 × 150 bp) (Illumina, San Diego, CA, USA). The read quality of the sequenced libraries was analyzed using FastQC software v 0.11.346 and sequence reads were trimmed to yield a minimum Phred quality score > 20. The genome assembly was performed using Geneious v 10.1.347. The de novo assembly was performance using Geneious assembler with medium sensitivity settings and allowing contigs with matching ends to circularize. The linear contigs were extended. For this, the reads were mapped back to the linear contigs and the resulting contigs were used as seeds for another attempted assembly until no further extension. Finally, the extended linear contigs were aligned and reordered using as reference the genome Bti HD-789 (accession number CP003763) from the “map to reference” tool with minimum overlap identity of 85%. The circular contigs were used to investigate plasmid-like sequences, by matching them against plasmid bank with custom BLAST tool. Genome annotation was added by the NCBI Prokaryotic Annotation.
Comparative genomic and phylogenetic analysis
Comparative chromosome-sequences analysis among T0124, T0131, T0137, T0139 and reference HD-789 was performed using BRIG (BLAST Ring Image Generator)48. The comparative analysis of the gene sequence of sporulation for the strains considered in this study and other species form the Bacilli group was performed using blastx, and the heatmaps were generated using the MeV tool version 4.949. Venn diagrams generation and orthologous cluster annotation for biological process, molecular function, and cellular component categories were achieved using OrthoVenn50. The collinearity and phylogenetic analysis among T0124, T0131, T0137, and T0139 and others 14 chromosomes of Bt was performed. The collinear analysis and display of results were conducted using Mauve with the parameters reported previously51. The phylogenetic tree based on single nucleotide polymorphisms (SNPs) was performed by CSI phylogeny 1.4 web52 using the default parameters and HD-789 as reference. The SNPs were named, concatenated and aligned, and the tree was constructed using the maximum likelihood method. The phylogeny tree inferred was reviewed using MEGA X software53 with 1000 replicates. The pBtoxis (NC_010076) was used as reference for the SNPs analysis of pT0124-4, pT0131-4, pT0137-4, and pT0139-4 using Geneious v 10.1.347, “Find SNPs/InDels” tool, with minimum coverage of 10, minimum variance frequence 0.75.
Nucleotide sequence accession number
The Whole Genome Shotgun projects of PRJNA521267, PRJNA521275, PRJNA521276, and PRJNA521307 Bti strains were deposited in DDBJ/ENA/GenBank under the accession numbers CP037890, CP035735, CP035736, and CP035737.
Statistical analyses
Concentration–mortality curves were estimated via probit analysis using the PROBIT procedure in the SAS statistical software package54. The differential susceptibility among mosquito species to H14 and the T0124, T0131, T0137, and T0139 Bti strains was assessed based on the estimated LC50 (i.e., the lethal concentration capable of killing 50% of tested mosquito species) of each strain and the toxicity ratios (TR50) were estimated by dividing the LC50 value obtained for the T0124, T0131, T0137, and T0139 Bti strains by the LC50 value obtained for the H14 standard strain55. The 95% confidence intervals estimated for these toxicity rates were considered to be significantly different if they did not include the value 155.
Ethical approval
All applicable international, national, and institutional guidelines for the care and use of animals were considered in the present investigation.
Informed consent
All the authors of this manuscript accepted that the paper is submitted for publication in the Scientific Reports journal, and report that this paper has not been published or accepted for publication in another journal, and it is not under consideration at another journal.
References
Ramírez-Lepe, M. & Ramírez-Suero, M. Biological Control of Mosquito Larvae by Bacillus thuringiensis subsp. israelensis. (Ed. Perveen, F.), Insecticides-P. Eng., https://doi.org/10.5772/29139 (2012).
Liu, G. et al. Complete genome sequence of Bacillus thuringiensis subsp. kurstaki strain HD73. Genome announc. 1, e00080–13, https://doi.org/10.1128/genomea.00080-13 (2013).
Barbosa, L. C. B. et al. Draft genome sequence of Bacillus thuringiensis 147, a Brazilian strain with high insecticidal activity. Mem. Inst. Oswaldo Cruz. 110, 822–823 (2015).
He, J. et al. Complete genome sequence of Bacillus thuringiensis subsp. chinensis strain CT-43. J. Bacteriol. 193, 3407–3408 (2011).
Palma, L., Muñoz, D., Berry, C., Murillo, J. & Caballero, P. Draft genome sequences of two Bacillus thuringiensis strains and characterization of a putative 41.9-kDa insecticidal toxin. Toxins. 6, 1490–1504, https://doi.org/10.3390/toxins6051490 (2014).
Guan, P. et al. Complete genome sequence of Bacillus thuringiensis serovar sichuansis strain MC28. J. Bacteriol. 194, 6975–6975 (2012).
Rusconi, B., Chen, Y., Koenig, S. S., El-Helow, E. R. & Eppinger, M. Sequence of Bacillus thuringiensis strain Btm27, an Egyptian isolate highly toxic to cotton leafworm. Genome announc. 3, e00446–15, https://doi.org/10.1128/genomeA.00446-15 (2015).
Zorzetti, J. et al. Genome sequence of the mosquitocidal Bacillus thuringiensis strain BR58, a biopesticide product effective against the coffee berry borer (Hypothenemus hampei). Genome announc. 3, e01232–15, https://doi.org/10.1128/genomea.01232-15 (2015).
Iatsenko, I., Corton, C., Pickard, D. J., Dougan, G. & Sommer, R. J. Draft genome sequence of highly nematicidal Bacillus thuringiensis DB27. Genome announc 2, e00101–14, https://doi.org/10.1128/genomea.00101-14 (2014).
Wang, P. et al. Complete genome sequence of Bacillus thuringiensis YBT-1518, a typical strain with high toxicity to nematodes. J. Biotechnol. 171, 1–2, https://doi.org/10.1016/j.jbiotec.2013.11.023 (2014).
Liu, X. et al. Draft genome sequence of Bacillus thuringiensis NBIN-866 with high nematocidal activity. Genome announc. 2, e00429–14, https://doi.org/10.3390/toxins6082229 (2014).
Wang, A., Pattemore, J., Ash, G., Williams, A. & Hane, J. Draft genome sequence of Bacillus thuringiensis strain DAR 81934, which exhibits molluscicidal activity. Genome announc. 1, e00175–12, https://doi.org/10.1128/genomea.00175-12 (2013).
Jeong, H., Jo, S. H., Hong, C. E. & Park, J. M. Genome sequence of the endophytic bacterium Bacillus thuringiensis strain KB1, a potential biocontrol agent against phytopathogens. Genome announc. 4, e00279–16, https://doi.org/10.1128/genomeA.00279-16 (2016).
Loeza-Lara, P. D. et al. The plasmid pBMBt1 from Bacillus thuringiensis subsp. darmstadiensis (INTA Mo14-4) replicates by the rolling-circle mechanism and encodes a novel insecticidal crystal protein-like gene. Plasmid. 54, 229–240, https://doi.org/10.1016/j.plasmid.2005.04.003 (2005).
Huang, J., Han, D., Yu, Z. & Sun, M. A novel cryptic plasmid pBMB175 from Bacillus thuringiensis subsp. tenebrionis YBT-1765. Arch. Microbiol. 188, 47–53 (2007).
Sheppard, A. E., Poehlein, A., Rosenstiel, P., Liesegang, H. & Schulenburg, H. Complete genome sequence of Bacillus thuringiensis strain 407 Cry. Genome announc. 1, e00158–12, https://doi.org/10.1128/genomea.00158-12 (2013).
Zorzetti, J. et al. Genome sequence of the mosquitocidal Bacillus thuringiensis strain BR58, a biopesticide product effective against the coffee berry borer (Hypothenemus hampei). Genome announc 3, e01232–15, https://doi.org/10.1128/genomea.01232-15 (2015).
Berry, C. et al. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 68, 5082–5095 (2002).
Zghal, R. Z., Ghedira, K., Elleuch, J., Kharrat, M. & Tounsi, S. Genome sequence analysis of a novel Bacillus thuringiensis strain BLB406 active against Aedes aegypti larvae, a novel potential bioinsecticide. Int. J. Biol. Macromol. 116, 1153–1162 (2018).
Anderson, I. et al. Comparative genome analysis of Bacillus cereus group genomes with Bacillus subtilis. FEMS Microbiol. Lett. 250, 175–184 (2005).
Doggett, N. A. et al. Complete genome sequence of Bacillus thuringiensis serovar israelensis strain HD-789. Genome announc. 1, e01023–13, https://doi.org/10.1128/genomea.01023-13 (2013).
Jeong, H., Seung-Hwan, P. & Soo-Keun, C. Genome sequence of the acrystalliferous Bacillus thuringiensis serovar israelensis strain 4Q7, widely used as a recombination host. Genome announc 2, e00231–14, https://doi.org/10.1128/genomea.00231-14 (2014).
Ankarloo, J. et al. Genome stability of Bacillus thuringiensis subsp. israelensis isolates. Curr. Microbiol. 40, 51–56 (2000).
Bolotin, A. et al. Comparative genomics of extrachromosomal elements in Bacillus thuringiensis subsp. israelensis. Microbiol. Res. 168, 331–344, https://doi.org/10.1016/j.resmic.2016.10.008 (2016).
Alcaraz, L. D. et al. Understanding the evolutionary relationships and major traits of Bacillus through comparative genomics. BMC genomics. 11, 332, https://doi.org/10.1186/1471-2164-11-332 (2010).
Araújo, A. P. et al. The susceptibility of Aedes aegypti populations displaying temephos resistance to Bacillus thuringiensis israelensis: a basis for management. Parasit vectors. 6, 297, https://doi.org/10.1186/1756-3305-6-297 (2013).
Fang, Y. et al. A pangenomic study of Bacillus thuringiensis. J. Genet. Genomics. 38, 567–57 (2011).
Medini, D., Donati, C., Tettelin, H., Masignani, V. & Rappuoli, R. The microbial pan-genome. Curr. Opin. Genetics Dev. 15, 589–594 (2005).
Rivera, A. M. G. & Priest, F. G. Molecular typing of Bacillus thuringiensis serovars by RAPD-PCR. Syst. Appl. Microbiol. 26, 254–261 (2003).
Sorokin, A. et al. Multiple-locus sequence typing analysis of Bacillus cereus and Bacillus thuringiensis reveals separate clustering and a distinct population structure of psychrotrophic strains. Appl. Environ. Microbiol. 72, 1569–1578 (2006).
Frost, L. S., Leplae, R., Summers, A. O. & Toussaint, A. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732, https://doi.org/10.1038/nrmicro1235 (2005).
Rasko, D. A., Altherr, M. R., Han, C. S. & Ravel, J. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Lett. 29, 303–329 (2005).
Fayad, N. et al. Characterization and Whole Genome Sequencing of AR23, a Highly Toxic Bacillus thuringiensis Strain Isolated from Lebanese Soil. Curr. Microbiol. 76, 1503–1511 (2019).
Galvão, P., Gitahy, P. D. M., Vidal, M., Araujo, J. L. S. & Baldani, J. Plasmídios em Bacillus thuringiensis: tamanho, mecanismo de replicação e papel na atividade entomopatogênica. Embrapa Agrobiologia- (Infoteca-e). 262, 1–32 (2009).
González, J. M., Dulmage, H. T. & Carlton, B. C. Correlation between specific plasmids and δ-endotoxin production in Bacillus thuringiensis. Plasmid. 5, 351–365, https://doi.org/10.1016/0147-619X(81)90010-X (1981).
Stein, C., Jones, G. W., Chalmers, T. & Berry, C. Transcriptional analysis of the toxin-coding plasmid pBtoxis from Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 72, 1771–1776 (2006).
Reyes-Ramírez, A. & Ibarra, J. E. Plasmid patterns of Bacillus thuringiensis type strains. Appl. Environ. Microbiol. 74, 125–129 (2008).
Deng, C., Peng, Q., Song, F. & Lereclus, D. Regulation of cry gene expression in Bacillus thuringiensis. Toxins. 6, 2194–2209, https://doi.org/10.3390/toxins6072194 (2014).
Wang, J. et al. The metabolic regulation of sporulation and parasporal crystal formation in Bacillus thuringiensis revealed by transcriptomics and proteomics. Mol. Cell Proteomics. 12, 1363–1376 (2013).
Monnerat, R. G., Silva, S. F. & Silva-werneck, J. O. Catálogo do banco de germoplasma de bactérias entomopatogênica do gênero Bacillus. Documentos, Brasília: Embrapa-Cenargen. 60, 65, https://doi.org/10.1016/j.biocontrol.2006.11.008 (2001).
Lecadet, M. M., Chaufaux, J., Ribier, J. & Lereclus, D. Construction of novel Bacillus thuringiensis isolates with different insecticidal activities by transduction and transformation. Appl. Environ. Microbiol. 58, 840–849 (1992).
Mounsef, J. R. et al. A simple method for the separation of Bacillus thuringiensis spores and crystals. J. Microbiol. Methods. 107, 147–149 (2014).
Sambrook, J. & Russel, D. W. Molecular cloning: A laboratory manual, (Ed 3 Lab) Cold Springs Harbour Press. 2344, https://doi.org/10.1016/0307-4412(83)90068-7 (2001).
Aguiar, R. W. S., Martins, E. S., Ribeiro, B. M. & Monnerat, R. G. Cry10Aa protein is highly toxic to Anthonomus grandis Bheman (Coleoptera: Curculionidae), an important insect pest in Brazilian cotton crop fields. Bt Research. 3, 20–28, https://doi.org/10.5376/bt.2012.03.0004 (2012).
McLaughlin, R. E. et al. US standard bioassay for the potency assessment of Bacillus thuringiensis serotype H-14 against mosquito larvae. Bulletin of the ESA. 30, 26–29, https://doi.org/10.1093/besa/30.1.26 (1984).
Andrews S. FastQC: a quality-control tool for high-throughput sequence (2015).
Kearse, M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28, 1647–1649 (2012).
Alikhan, N. F., Petty, N. K., Zakour, N. L. B. & Beatson, S. A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC genomics 12, 402, https://doi.org/10.1186/1471-2164-12-402 (2011).
Saed, A. I. et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 34, 374–378 (2003).
Wang, Y., Coleman-Derr, D., Chen, G. & Gu, Y. Q. OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 43, W78–W84 (2015).
Darling, A. C., Mau, B., Blattner, F. R. & Perna, N. T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004).
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M. & Lund, O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. Plos One. 9, e104984, https://doi.org/10.1371/journal.pone.0104984 (2014).
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35, 1547–1549 (2018).
SAS Institute. SAS/STAT User’s Guide., Cary, NC, USA, (2008).
Robertson, J. L., Jones, M. M., Olguin, E., & Alberts, B. Pesticide Bioassays with Arthopods. Boca Raton: CRC Press. 224 p. (2007).
Stothard, P. & Wishart, D. S. Circular genome visualization and exploration using CGView. Bioinformatics. 21, 537–539 (2004).
Gene ontology consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–D261 (2004).
Li, Q. et al. Complete genome sequence of Bacillus thuringiensis HS18-1. J. Biotechnol. 214, 61–62 (2015).
Cheng, F. et al. Complete genome sequence of Bacillus thuringiensis YC-10, a novel active strain against plant-parasitic nematodes. J. Biotechnol. 210, 17–18 (2015).
Zhu, J. et al. The complete genome sequence of Bacillus thuringiensis serovar hailuosis YWC2-8. J. Biotechnol. 219, 38–39 (2016).
Jia, N., Ding, M. Z., Gao, F. & Yuan, Y. J. Comparative genomics analysis of the companion mechanisms of Bacillus thuringiensis Bc601 and Bacillus endophyticus Hbe603 in bacterial consortium. Sci. Rep. 6, 28794, https://doi.org/10.1038/srep28794 (2016).
Li, Y. et al. Complete genome sequence of Bacillus thuringiensis Bt185, a potential soil insect biocontrol agent. J. Integr. Agric. 16, 749–751 (2017).
Johnson, S. L. et al. Complete genome sequences for 35 biothreat assay-relevant Bacillus species. Genome announc. 3, e00151–15, https://doi.org/10.1128/genomeA.00151-15 (2015).
Dong, Z. et al. Complete genome sequence of Bacillus thuringiensis CTC—A typical strain with high production of S-layer proteins. J. Biotechnol. 220, 100–101 (2016).
Acknowledgements
This work was supported by grants from the Secretaria do Desenvolvimento Econômico, Ciência, Tecnologia e Inovação do Estado do Tocantins; the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) (Finance Code 001); the National Council of Scientific and Technological Development (CNPq); the Minas Gerais State Foundation for Research Aid (FAPEMIG); and Fundaçao de Amparo a Pesquisa do Estado de Tocantins (FAPT-SESAU/TO-DECIT/SCTIE/MS_CNPQ/No 01/2017).
Author information
Authors and Affiliations
Contributions
G.B.A., L.T.M.C. and M.L.D. performed the experiments, data analysis. F.L.M. and F.S.C. helped with the data analysis and performed the experiments (Bioinformatics); R.F.T.C., K.H., R.W.S.A., E.E.O., S.A.D, G.S. and E.J.G.P. helped with the data analysis and review the manuscript and Statistic analysis. G.R.S. and B.M.R. helped with manuscript revision, laboratory infrastructure and reagents.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Alves, G.B., Melo, F.L., Oliveira, E.E. et al. Comparative genomic analysis and mosquito larvicidal activity of four Bacillus thuringiensis serovar israelensis strains. Sci Rep 10, 5518 (2020). https://doi.org/10.1038/s41598-020-60670-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-60670-7
- Springer Nature Limited