Abstract
Annexins (ANN) are a multigene, evolutionarily conserved family of calcium-dependent and phospholipid-binding proteins that play important roles in plant development and stress resistance. However, a systematic comprehensive analysis of ANN genes of Brassicaceae species (Brassica rapa, Brassica oleracea, and Brassica napus) has not yet been reported. In this study, we identified 13, 12, and 26 ANN genes in B. rapa, B. oleracea, and B. napus, respectively. About half of these genes were clustered on various chromosomes. Molecular evolutionary analysis showed that the ANN genes were highly conserved in Brassicaceae species. Transcriptome analysis showed that different group ANN members exhibited varied expression patterns in different tissues and under different (abiotic stress and hormones) treatments. Meanwhile, same group members from Arabidopsis thaliana, B. rapa, B. oleracea, and B. napus demonstrated conserved expression patterns in different tissues. The weighted gene coexpression network analysis (WGCNA) showed that BnaANN genes were induced by methyl jasmonate (MeJA) treatment and played important roles in jasmonate (JA) signaling and multiple stress response in B. napus.
Similar content being viewed by others
Introduction
Annexins (ANN) are a multigene, evolutionarily conserved family of calcium (Ca2+)-dependent and phospholipid-binding proteins present in plants, animals, and microorganisms1,2. ANN contain the characteristic annexin repeat and they regulate membrane dynamics, mediate Ca2+ sensing and signaling, link Ca2+ dynamics to cytoskeletal responses, and mediate immune or stress responses and signaling during plant growth and development1,3. A typical ANN contains four annexin repeats at the C-terminal region and a highly variable N-terminal region. Each annexin repeat usually contains a characteristic type II motif for Ca2+ binding1,3. The variable N-terminal region interacts with other proteins and is responsible for the functional diversity of ANN4.
Recent studies have identified the ANN gene family in Arabidopsis thaliana (8 genes), Brassica rapa (13), Solanum lycopersicum (9), Solanum tuberosum (9), Oryza sativa (10), Triticum aestivum (25), Gossypium raimondii (14), Arachis hypogaea L. (8), Hordeum vulgare (11), Medicago truncatula (10), Populus trichocarpa (12), Vitis vinifera (14), Carica papaya (12), Glycine max (22), Cochliobolus sativus (11), Sorghum bicolor (10), Zea mays (12), Brachypodium distachyon (11), Selaginella mollendorffii (5), and Physcomitrella patens (7) via genome-wide analysis2,5,6,7,8,9,10,11.
Studies have shown that ANN gene family plays a significant role in plant development and plant protection during both abiotic and biotic stresses1,3,12,13. In Arabidopsis, two ANN genes (AtANN1 and AtANN4) were regulated by abiotic stress, negatively regulated plant tolerance to drought, salinity, and heat stress, while AtANN8 was positive regulated the plant abiotic tolerance14,15,16,17,18,19,20,21. Studies also demonstrated that AtANN1 and AtANN2 regulated root growth and development20,21,22. Downregulation of AtANN5 resulted in abnormal pollen grains and severe male sterility23,24. The rice annexin OsANN1 enhanced heat stress tolerance25, and OsANN3 positively regulated drought tolerance26. ZmANN33 and ZmANN35 enhanced chilling stress tolerance during germination of maize seeds27. Medicago truncatula annexin 1 regulated nodulation and mycorrhization in legume plants28,29. The tobacco annexin Ntann12 was induced upon Rhodoccocus fascians infection30,31. The potato annexin STANN1 promoted drought tolerance10. The cotton annexin gene GhAnn1 was induced by various phytohormones and abiotic stress, positively regulated drought and salt tolerance32. GhANN8b and phosphatase GhDsPTP3a proteins of cotton interacted with each other and regulated salt tolerance and calcium influx33. GhAnn2 was induced by IAA and GA3, and GhAnn2 downregulation inhibited cotton fiber elongation by modulating Ca2+ influx at the cell apex8. GhFAnnxA regulated fiber elongation and secondary cell wall biosynthesis34. AnxGb6 interacted with actin1 and regulated cotton fiber elongation35. Overexpression of cotton annexin gene AnnGh3 increased trichome density and length in Arabidopsis leaf36. Overexpression of Brassica juncea annexin AnnBj2 increased salt tolerance and abscisic acid (ABA) insensitivity in transgenic plants37,38. Ectopic expression of B. juncea annexin gene BjAnn1 in tobacco and cotton enhanced tolerance to various abiotic stresses and fungal pathogen39,40,41. AnnBj3 promoted oxidative stress tolerance in plants42.
Brassica napus (genome AnAnCnCn) is an important oil crop worldwide, which was formed by recent allopolyploidy between ancestors of Brassica rapa (genome ArAr) and Brassica oleracea (genome CoCo)43. The production and quality of B. napus is greatly influence by adverse environmental conditions. Therefore, it is critical to improve stress tolerance in B. napus through the identification and use of genes involved in stress response. Although there are many studies on ANN genes in various plant species, ANN genes are yet to be characterized in B. napus and B. oleracea. In this study, we investigate the potential role of ANN genes in environmental stress response in Brassicaceae plants. Therefore, we identified the ANN genes of B. napus, B. rapa and B. oleracea and compared the gene structure, chromosomal location, evolutionary relationship, and expression pattern in different tissues and under different abiotic/biotic stresses and plant hormonal treatments. The findings of this study will provide a foundation for further studies on functional characterization of ANN genes of Brassicaceae plants under adverse environmental conditions.
Results and Discussion
Identification of ANN in B. rape, B. oleracea and B. napus
A total of 13 BrANN (B. rapa ANN), 12 BoANN (B. oleracea ANN), and 26 BnaANN (B. napus ANN) proteins were identified through BLASTP using 8 Arabidopsis ANN (AtANN) proteins (Table 1). All members were verified for the presence of annexin repeats using InterPro and Conserved Domain (CD)-search in NCBI. Brassicaceae species experienced an extra whole-genome triplication (WGT) event44,45,46, based on which approximately 24 and 48 ANN genes were expected in B. rapa/B. oleracea and B. napus genomes, respectively. However, only 13, 12, and 26 ANN genes were found in B. rapa, B. oleracea, and B. napus, respectively (Table 1). In B. napus, the number of genes in the An-subgenome (12) and Cn-subgenome (14) was almost the same as that in their diploid progenitors B. rapa and B. oleracea (Table 1). These results indicate the loss of about half of ANN genes after the Brassicaceae WGT in B. rapa and B. oleracea. However, most of the duplicated ANN genes were retained after the whole-genome duplication (WGD) event in B. napus. WGD event of gene family appears to be a widespread phenomenon, such as the auxin response factor (ARF)47, Auxin/Indoleacetic acid (Aux/IAA)48, glutathione transferases (GST)49, BRI1-EMS-SUPPRESSOR1 (BES1)50, Heat stress transcription factors (Hsfs)51,52, GRAS53 family genes in diploid and allopolyploid Brassicaceae and Calcium-dependent protein kinases (CPK)54, Jasmonate ZIM-domain (JAZ)55 and Nuclear factor YB (NF-YB)56 in diploid and allopolyploid Gossypium species (G. raimondii: DD genome; G. arboretum: AA genome; G. hirsutum: AADD genome).
Among the 51 Brassica ANN genes, 35 were the typical ANN, which encoded proteins ranging from 315–325 amino acids (AA) and contained four annexin repeats. All eight ANN members (315–320 AA) homologous to AtANN4 (AT2G38750) contained 2–3 annexin repeats, as same as AtANN4. While two other ANN members (157 AA) contained only a single annexin repeat and six members (183–265 AA) contained 2–3 annexin repeats (Table 1), they may were the truncated mutated duplications.
Phylogenetic and structural analysis of ANN
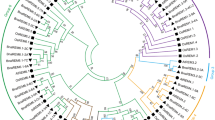
A phylogenetic tree (Fig. 1A) was generated using the sequences of 59 ANN proteins from B. rapa, B. oleracea, B. napus, and Arabidopsis (Fig. S1). These ANN proteins were divided into six groups. All eight AtANN were found to have orthologous genes in B. rapa, B. oleracea, and B. napus (Fig. 1A). Twelve pairs of BnaANN were found in the corresponding B. napus An- and Cn-homoeologous chromosomes, and ten pairs of them had homologous genes both in B. rapa and B. oleracea. Meanwhile, two pairs (BnaC03g49290D/BnaA06g23960D and BnaC09g44350D/BnaA10g20320D) only had homologous genes in B. rapa. All 12 BoANN genes were found to have homologous genes in the Cn-subgenome of B. napus, while one BrANN (Bra039578) had no homologous gene in An-subgenome of B. napus (Table 1 and Fig. 1A).
Phylogenetic tree (A), gene structure (B), and gene motifs (C) of ANN of Arabidopsis, B. rapa, B. oleracea, and B. napus. Neighbor-joining phylogenetic tree showing the relationship among 13 B. rapa (blue dots), 12 B. oleracea (yellow dots), 26 B. napus (red dots), and 8 Arabidopsis ANN proteins (A). The resulting six groups are labeled (Group I-VI). Orange boxes, black lines, and blue boxes indicate exons, introns, and untranslated regions, respectively (B). Five motifs in BnaSAP proteins were identified by MEME tools (C).
Gene structure analysis revealed that majority of the homologous ANN gene pairs had same gene structure (Fig. 1B). There were five introns in group IV/V/VI members, expect for two truncated mutant genes (Bo1g039570 and BnaC01g16910D) (Fig. 1B). This finding indicates that the ANN genes are conserved in Brassicaceae species, possibly due to their importance in plant growth and productivity.
A typical ANN protein contains four annexin repeats, each approximately 70 amino acids long1,3. Annexin repeat usually contain a characteristic type II motif for binding calcium ions with the sequence GxGT-[38 residues]-D/E3. MEME analysis showed that 42 ANN proteins contained four annexin repeats (Fig. 1C). Motif1 was the core sequence of all the four annexin repeats, and motif4 was only found in the third annexin repeat in group I–V, while motif5 was the core sequence close to the C-terminal of Motif1 in the second and fourth annexin repeats (Fig. 1C).
According to the gene structure and motif analysis, the missing parts of the truncated mutant members were readily apparent. Both the first and fourth annexin repeats were absent in Bo9g172330 and BnaC09g46400D, and the first annexin repeat was absent in BnaAnng37420D. Bo1g039570 and BnaC01g16910D had only the second annexin repeat at the C-terminal (80–159 AA), and the core sequence of annexin repeat was not detected at the N-terminal (1–79 AA). It is similar in the N-terminal of Bo6g043900, BnaC06g08410D, and AtANN4 homologues (Fig. 1B,C).
Chromosomal location and synteny analysis of ANN of B. rapa, B. oleracea, and B. napus
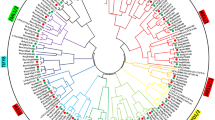
As showed in Fig. 2, the distribution of BnaANN in An- and Cn-subgenome was nearly even with 12 ANN genes from the An-subgenome and 14 from the Cn-subgenome. However, the ANN genes’ distribution was uneven on each chromosome. Three pair (2 genes/pair) of ANN genes from the An-subgenome were repeated in tandem on chromosome Bn_A03, Bn_A04, and Bn_A10 (Fig. 2); and three pair (2 genes/pair) of ANN genes from the Cn-subgenome were repeated in tandem on chromosome Bn_C03, Bn_C04, and Bn_C09 (Fig. 2C). B. napus genome analysis showed that the An- and Cn-subgenome were largely collinear to the corresponding diploid Ar and Co genomes43,57. Most of the An-Ar and Cn-Co orthologous gene pairs demonstrated similar chromosomal locations. The distribution of ANN genes in B. rapa and B. oleracea were similar to the distribution of the orthologous BnaANN genes in the B. napus An-subgenome and Cn-subgenome, respectively (Fig. 2). Two BnaANN (BnaAnng04520D and BnaAnng37420D) and one BrANN (Bra039578) genes were located on the unanchored scaffolds that were not mapped to a specific chromosome (Fig. 2). The sequence and phylogenetic analyses revealed BnaAnng04520D-Bra034402 and BnaAnng37420D-Bra031890 as two An-Ar orthologous gene pairs. Based on this, we predicate Bn_A02 and Bn_A05 as the true chromosomal locations of BnaAnng04520D and BnaAnng37420D, respectively. BnaANN (BnaC03g49290D and BnaC09g44350D) had no orthologous genes in B. oleracea (Fig. 2), though they had homologous genes in An-subgenome. These findings indicate that duplication of BnaA06g23960D and BnaA10g20320D led to the formation of BnaC03g49290D and BnaC09g44350D, respectively. Analysis of the synteny among An-subgenome and Cn-subgenome showed high collinearity between Bn_A01-Bn_C01, A02-C02, A03-C03, A04-C04, A05-C05, A06-C06, A07-C07, A08-C08, A09-C09, and A10-C09, and 83.7% orthologous gene pairs between B. rapa and B. oleracea were retained as homologous gene pairs in B. napus43,57. 90.9% ANN gene pairs (10/11 pairs) between B. rapa and B. oleracea were retained as homologous gene pairs between B. napus An-chromosomes and Cn-chromosomes (Fig. 2).
The synteny analysis of ANN genes in the B. rapa, B. oleracea and B. napus chromosomes. Br_A (B. rapa chromosomes): yellow trapezoid; Bo_C (B. oleracea chromosomes): blue trapezoid; Bn_A (B. napus An-subgenome chromosomes) and Bn_C (B. napus Cn-subgenome chromosomes): red trapezoid; Bn_C02_Random means genes were randomly distributed to B. napus Cn-subgenome chromosome C02. S1 and S2 are two unanchored scaffolds from B. napus; S3 is an unanchored scaffold (Scaffold000169) from B. rapa. The orthologous and paralogous ANN genes were mapped onto the chromosomes and linked by each other. Yellow lines linked two syntenic ANN genes from B. rapa and B. napus; Blue lines linked two syntenic ANN genes from B. oleracea and B. napus; Green lines linked two syntenic ANN genes from B. rapa and B. oleracea; Red lines linked two syntenic ANN genes from B. napus An- and Cn-subgenome.
There were two tandem pairs (AtANN3/4 and AtANN6/7) on chromosome 2 and chromosome 5 in Arabidopsis, respectively58. Bra009048/Bra009049, Bo9g172330/Bo9g172340, BnaA10g22010D/BnaA10g22020D, and BnaC09g46400D/BnaC09g46410D were homologous to AtANN6/7 tandem pair in B. rapa, B. olearcea, B. napus An-subgenome and Cn-subgenome, respectively. We identified two tandem pairs each homologous to AtANN3/4 in B. rapa (Br_A03 and Br_A04), B. olearcea (Bo_C03 and Bo_C04), B. napus An-subgenome (Bn_A03 and Bn_A04), and Cn-subgenome (Bn_C03 and Bn_C04) (Fig. 2). AtANN8 (AT5G12380) was located near the AtANN6/7 tandem pair on chromosome 5 in Arabidopsis58. Correspondingly, there was a gene homologous to AtANN8 located near the tandem pair homologous to AtANN6/7 in B. rapa (Br_A10), B. napus An-subgenome (Bn_A10) and Cn-subgenome (Bn_C09) (Fig. 2). There was no gene homologous to AtANN8 in B. olearcea (Bo_C09). Instead, we found a truncated mutated gene (Bo1g039570) homologous to AtANN8 in B. olearcea (Bo_C01). Meanwhile, a truncated mutated gene (BnaC01g016910D) was homologous to Bo1g039570 in B. napus Cn-subgenome (Bn_C01) (Fig. 2). Bra039578 had no homologous gene in An-subgenome of B. napus. These findings suggest that majority of the ANN genes are conserved in Brassicaceae species, only a few ANN genes are missing or duplicating in B. napus.
To better understand the evolutionary constraints acting on the ANN gene family, we estimated the number of nonsynonymous substitutions per nonsynonymous site (Ka), the number of synonymous substitutions per synonymous site (Ks), and the Ka/Ks ratio. Ka/Ks value <1 indicates that a gene pair has experienced purifying selection; Ka/Ks>1 indicates positive selection; and Ka/Ks = 1 indicates neutral selection59. The Ka/Ks ratio was <1 for majority of the ANN collinear gene pairs (209/210), except for the gene pair Bra024346/BnaA06g23960D (Ka/Ks > 1) (Table S1). These results indicate that majority of genes experienced purifying selection, whereas Bra024346 and BnaA06g23960D experienced positive selection.
Expression profile of ANN genes in different tissues
ANN genes exhibit tissue-specific expression, which is usually consistent with their substantially differentiated functions14,16,17,18,22,23,58. We investigated the expression of all ANN genes in different tissues of Arabidopsis, B. rapa, B. olearcea, and B. napus based on the Arabidopsis eFP Browser data (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) and RNA-Seq data (B. rapa: GSE43245; B. oleareaca: GSE42891; B. napus: PRJNA394926) (Table S2)57,60,61. The ANN genes were expressed across different vegetative and reproductive organs during different developmental stages of the four species (Fig. 3). In general, the ANN expression pattern was different between groups; however, expression pattern was very similar within a group in the four plant species.
Group I (ANN6/7) members showed expression in young siliques (ovules) and seeds, which indicate their importance in ovule and seed development in Brassicaceae plants. However, two truncated mutated members (Bo9g172330 and BnaC09g46400D) homologous to ANN7 were at low abundance expression levels (Fig. 3). Unlike Bo9g172330 and BnaC09g46400D, other five truncated mutated members (BnaAnng37420D, BnaA06g23960, BnaC06g08410D, Bo1g039570 and BnaC01g16910D) have a similar expression level to their homologous genes which have complete gene structure (Figs. 1 and 3). So, truncated mutated gene structures may decrease their own genes’ expression level, but not always. The expression levels of group 2 (ANN2) members were highest in roots and young siliques (ovules), while that of group 3 (ANN1) members were higher in roots, stems, and young siliques (pericarps) (Fig. 3). These expression levels are consistent with the role of AtANN1 and AtANN2 in root growth and development20,21,22. It was indicated that ANN1/2 regulates the development of young siliques and seeds. We detected low level of expression for group 4 (ANN8) members. AtANN5, which regulates pollen development23,24, showed specific expression in mature pollen. The B. napus genes homologous to AtANN5 were mainly expressed in buds and new pistils. The genes homologous to AtANN3 and AtANN4 demonstrated similar expression pattern. Both genes were expressed in flowers and young siliques (ovules), though they belong to group IV and VI, respectively (Fig. 3). All these indicated that ANN genes may be involved in various developmental processes with different functions. In Arabidopsis, AtANN3 and AtANN4 had similar expression pattern because they share a 5′ promoter region (2654 bp)58. In B. rapa, Bra000090 and Bra000091 share a 5′ promoter region (2079 bp), while in B. oleareca, Bo3g032760 and Bo3g032770 share a 5′ promoter region (2412 bp); In B. napus, BnaA03g18070D and BnaA03g18080D share a 5′ promoter region (2455 bp) and BnaC03g21590D and BnaC03g21600D share a 5′ promoter region (6199 bp). They were homologous to AtANN3/AtANN4 pair, and had similar expression pattern. But another gene pairs (Bra017102/Bra017103, Bo4g187790/Bo4g187780, BnaA04g22190D/BnaA04g22180D, and BnaC04g45920D/BnaC04g45910D) homologous to AtANN3/AtANN4 pair were at low abundance expression levels (Fig. 3B,C). All the results suggested that there were gene duplications, gene expression pattern differentiations and subsequent functional diversifications in ANN family genes in Brassicaceae species, and the functions of homologs of a given group ANN genes might be redundant as they share similar expression patterns.
Expression pattern of ANN genes in response to abiotic stress and hormonal treatment
Accumulating evidence from various plant species has shown the regulation of ANN genes in response to abiotic stress and hormonal treatment5,6,7,9,58. To examine the expression pattern of BnaANN genes under various abiotic stress conditions and hormonal treatments, we utilized the data on transcriptional profiling (Table S3). As shown in Fig. 4, most of the expressed BnaANN genes in group II/III/V/VI/were up-regulated under salinity and PEG stress in roots and MeJA treatment in leaves. BnaA06g23960, BnaA03g18070D and BnaC03g21590D were down-regulated under cold stress, whereas BnaAnng04520D and BnaC05g27530D were up-regulated under cold stress at 12 hours point (Fig. 4).
Expression of BnaANN under abiotic stress and plant hormone treatments. Leaf: untreated leaves; Cold: leaves treated with 4 °C; Hot: leaves treated with 40 °C; ABA: leaves treated with 100 μM abscisic acid; MeJA: leaves treated with 100 μM methyl jasmonate; ETH: leaves treated with 10 μg/ml ethephon; SA: leaves treated with 1.0 mM salicylic acid. Root: untreated roots; NaCl: roots treated with 200 mM NaCl; PEG: roots treated with 20% polyethylene glycol 6000. Coloured rectangles indicate TPM values.
B. napus is a winter biennial crop with excellent tolerance to low-temperature stress during vegetative stage. The response mechanisms are different under chilling and freezing temperatures, as well as cold shock and cold acclimation in plants62,63. Based on the transcriptional profiling of early-maturing, cultivated B. napus varieties under different low-temperature treatments with or without cold acclimation (GSE129220: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129220) (Table S4)64, transcriptome analysis revealed that group III BnaANN were induced slightly by chilling stress, and were up-regulated by freezing stress strongly, regardless of cold acclimation (Fig. 5A). This finding indicates that group III BnaANN genes play important roles in freezing stress in B. napus.
Expression profile of BnaANN under different low-temperature treatments in two early-maturing semi-winter B. napus varieties HX17 and HX58 (A) and in susceptible (Westar) and tolerant (ZY821) genotypes of B. napus infected with Sclerotinia sclerotiorum (B). MA: untreated leaves of 6-weeks-old seedlings; CA: leaves of 6-weeks-old seedlings treated with cold acclimation (4 °C for two weeks) and then treated 4 °C for 12 hours; FA: leaves of 6-weeks-old seedlings treated with cold acclimation (4 °C for two weeks) and then treated −4 °C for 12 hours; MB: untreated leaves of 6-weeks-old seedlings; CB: leaves of 6-weeks-old seedlings treated with 4 °C for 12 hours; FB: leaves of 6-weeks-old seedlings treated with −4 °C for 12 hours. Coloured rectangles indicate the gene FPKM values.
Sclerotinia sclerotiorum is a hemibiotroph pathogen with a wide host range. It is the causative agent of stem rot, one of the most devastating diseases of B. napus65,66. Previous studies have shown the role of JA signaling in plant resistance to hemibiotroph pathogens67,68,69,70. The transcriptional profiling of B. napus susceptible (Westar) and tolerant (ZY821) genotypes infected with S. sclerotiorum (GSE81545: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE81545) (Table S5) showed that the group II BnaANN were induced by S. sclerotiorum infection, and the expression level in the susceptible genotype (Westar) was more than that in the tolerant (ZY821) genotype; some members from group III and group V BnaANN were induced, while some members were repressed by S. sclerotiorum infection (Fig. 5B). These findings indicate a complex response mechanism and the role of some BnaANN in B. napus response to S. sclerotiorum.
To validate the results on transcriptional profiling, we performed a qRT-PCR to detect the transcript levels of three genes (BnaC03g49290D, BnaC05g27530D, and BnaC03g21590D) from group II/III/VI in the roots challenged with salt and PEG and in the leaves treated with cold and MeJA. The expression pattern (of these BnaANN genes) was consistent with the RNA-Seq data (Figs. 4–6). All three BnaANN genes were induced under salinity and PEG stress in roots and induced by MeJA in leaves (Fig. 6A). BnaC03g49290D and BnaC03g21590D were repressed by cold treatment (Fig. 6A). BnaC05g27530D was significantly upregulated under freezing stress, with or without cold acclimation (Fig. 6B). In B. rapa, Bra034402 (gene to homologous BnaC05g27530D) was strongly induced by hormone and stress treatments11. All these results indicated the role of these three genes in multiple abiotic stress response and JA signaling response in B. napus.
qRT-PCR analysis of three BnaANN genes under abiotic stress and hormone treatments. The relative qRT-PCR expression level (blue bar) is shown on the left y-axis. The RNA-Seq TPM/FPKM values (red line) are shown on the right y-axis. BnaActin (BnaC05g34300D) was used as the endogenous reference gene. The relative transcript levels were averaged over the three technical replicates.
Weighted gene co-expression network analysis (WGCNA) of BnaANN in response to environmental stress
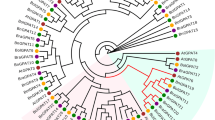
Weighted gene co-expression network analysis (WGCNA) is an effective way to identify clusters of highly correlated genes71. To reveal the divergent functions of BnaANN genes in development, abiotic stress response, and hormone signaling, coexpression networks were constructed on the basis of pairwise correlations of all B. napus gene expression across 12 tissues samples and 8 treatment (abiotic stress and hormone) samples using WGCNA. The analysis identified 56 distinct modules (labeled with different colors) as shown in the dendrogram (Fig. S2). In total, 16 of 26 BnaANN genes were identified in six different modules: light green module (5), blue module (3), green module (3), turquoise module (3), salmon module (1), and magenta module (1) (Table S6). The lightgreen module (845 genes) was positively correlated with the MeJA treatment in leaves (Fig. S2). Five BnaANN genes (BnaA03g18070D, BnaA03g18080D, BnaC03g21590D, BnaC03g21600D and BnaC05g27530D) were induced by MeJA treatment in lightgreen module (Fig. 4 and Table S6). The top two hub genes with the highest the module membership kME (k-means clustering algorithm) values were BnaA03g18070D (BnaANN4A-1) and BnaC06g31830D (BnaTIFY7) in the light green module (Fig. 7A and Table S6). The jasmonate acid (JA) signaling repressor, TIFY, was induced by JA and regulates plant development and stress response72,73,74,75. Additionally, there were some B. napus JA biosynthesis genes and JA responsive genes in the light green module, such as the Lipoxygenase (LOX), Allene oxide cyclase (AOC), Allene oxide synthase (AOS), 12-oxophytodienoate reductase (OPR), Jasmonate O-methyltransferase (JMT), and Ethylene-responsive factor (ERF) (Fig. 7A and Table S6). Transcriptional profiling and qRT-PCR analysis results showed that BnaA03g18070D/BnaANN4A-1, BnaC06g31830D/BnaTIFY7, BnaC04g38070D/BnaERF42, and BnaC02g29610D/BnaAOS were all induced by MeJA (Fig. 7B and Table S7). However, there was little research at the functions of annexins in JA signaling. ZmAnx6.1 and ZmAnx7 were induced at 12 h by JA, and the JA-responsive cis-elements exist in their promoters76. We analyzed the promotor sequences (2000 bp upstream of transcription start sites) of BnaANN, and founded that there were so many cis-elements involved in stress (drought, low-temperature, heat, anaerobic, wounding, defense and stress) response and plant hormones (MeJA, ABA and SA) response in their promotors, MeJA-responsive cis-element (CGTCA-motif, TGACG-motif and G-box) was the most numerous cis-element and all BnaANN members contain MeJA-responsive cis-elements (1 to 9) in promotors (Fig. S3). It suggested that the BnaANN genes in lightgreen module involved in JA signaling response in B. napus.
BnaANN involved in JA-response in B. napus. (A) Co-expression network for BnaANN genes in the lightgreen module. Red indicates candidate hub genes and light red indicates JA biosynthesis/responsive genes and BnaANN genes. (B) qRT-PCR analysis of four JA-response genes under MeJA treatments. The relative qRT-PCR expression level (blue bar) is shown on the left y-axis. The RNA-Seq TPM values (red line) are shown on the right y-axis. BnaActin (BnaC05g34300D) was used as the endogenous reference gene. The relative transcript levels were averaged over the three technical replicates.
Three BnaANN genes (BnaA04g22190D, BnaC03g49290D, and BnaC02g43450D) in the blue module were expressed with NaCl and PEG treatments in roots, while genes (BnaC08g06690D, BnaA10g20320D and BnaC09g44350D) in the green module were expressed in roots. BnaC01g16910D, BnaA06g23960D, and BnaC06g08410D in the turquoise module were positively correlated with bud, stamen, ovule, and silique (Fig. S2 and Table S6). All the results indicate the different functions of B. napus ANN genes during plant development and stress response.
Materials and Methods
Identification of ANN of B. rapa, B. oleracea, and B. napus
B. rape, B. oleracea and B. napus ANN proteins have been identified using BLASTP (E-value < 1e-5) to look for homologs of Arabidopsis ANN among B. rape, B. oleracea and B. napus genome sequences database in Ensembl gemones (http://ensemblgenomes.org/)77. The annexin motifs in ANN proteins were characterized using InterPro (http://www.ebi.ac.uk/interpro/)78 and the NCBI conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
The molecular weight (Mw), isoelectric point (pI), and subcellular localization of ANN proteins were predicted using the Compute pI/Mw tool (http://web.expasy.org/compute_pi/)79 and ProtComp 9.0 (http://linux1.softberry.com/). The exon and intron organization of the ANN genes were analyzed using the Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/)80. The conserved motifs of ANN were analyzed with MEME (http://meme.nbcr.net/meme/cgi-bin/meme.cgi)81.
Phylogenetic analysis
Multiple sequence alignment of all identified ANN proteins (Arabidopsis, B. rapa, B. oleracea, and B. napus) were performed using ClustalW and a phylogenetic tree was constructed using the neighbour-joining (NJ) phylogenetic method in MEGA782 with 1000 bootstrap replicates.
Chromosomal localization of ANN genes
The position of ANN genes on the chromosomes of B. rapa, B. oleracea, and B. napus were obtained using TBtools v0.6683183.
Nonsynonymous and sunonymous substitution rate ratio (Ka/Ks)
DnaSP (DNA Sequence Polymorphism) v684 was used to calculate the ratio of the nonsynonymous substitution rate (Ka) to the synonymous substitution rate (Ks) and the Ka/Ks value between paralogous gene pairs.
Plant materials and treatments
ZS11 (B. napus L. cv. Zhongshuang 11)57 seeds were allowed to germinate and then the seedlings were transplanted to pots containing soil or vermiculite. The growth conditions, hormone treatments, and abiotic stress conditions were as described previously85. Hormone treatments were performed by spraying leaves of 6-week-old seedlings with ABA (100 μM), MeJA (100 μM), SA (1 mM), and ETH (10 μg/ml); To simulate hot and cold stresses, seedlings were grown in chamber with 40 °C or 4 °C. To simulate salt and PEG stresses, seedlings were irrigated with NaCl (200 mM) or PEG-6000 (20%) solutions.
For chilling and freezing treatments with or without cold acclimation, the seedlings of two early-maturing semi-winter rapeseed varieties (HX17 and HX58) were used. They were treated as described previously64. Seedlings were cultured in incubators under 20 °C (14 h light: am6:00–pm8:00)/16 °C (10 h dark: pm8:00–am6:00) 4 weeks, then treated with 4 °C (14 days) → 4 °C (12 h) (CA) or −4 °C (12 h) (FA), 20 °C/16 °C (light/dark) 6 weeks → 4 °C (12 h) (CB), 20 °C (14 h light: am6:00–pm8:00)/16 °C (10 h dark: pm8:00–am6:00) 6 weeks → −4 °C (12 h) (FB). For the acclimation condition, after the 14 days at 4 °C, 4 °C/−4 °C (12 h) mean a treatment with 4 °C or −4 °C at pm8:00–am8:00 (10 h dark and 2 h light).
RNA isolation and sequencing and gene expression analysis
The collected samples were sent to the sequencing cooperations of Sangon Biotech (Shanghai) Co., Ltd. and Novogene Co., Ltd. for RNA isolation, examination, and sequencing64,85. qRT-PCR analysis was performed as described previously85. The primers used in this study were listed in Table S8.
Heat map analysis
The RPKM (Reads Per kb Per Million reads) and TPM (Transcripts Per Million) values were used to represent the expression levels of the ANN genes. A heat map of the expression profile of the ANN genes was plotted using Heatmap Illustrator, version 1.086.
Weighted gene coexpression network analysis (WGCNA)
Weighted gene coexpression network analysis was performed using WGCNA package in R71. The networks were visualized using Cytoscape v387.
Data availability
The authors declare that all the data and plant materials will be available without restrictions.
References
Laohavisit, A. & Davies, J. M. Annexins. The New phytologist 189, 40–53, https://doi.org/10.1111/j.1469-8137.2010.03533.x (2011).
Jami, S. K., Clark, G. B., Ayele, B. T., Ashe, P. & Kirti, P. B. Genome-wide comparative analysis of annexin superfamily in plants. PloS one 7, e47801, https://doi.org/10.1371/journal.pone.0047801 (2012).
Konopka-Postupolska, D., Clark, G. & Hofmann, A. Structure, function and membrane interactions of plant annexins: an update. Plant science 181, 230–241, https://doi.org/10.1016/j.plantsci.2011.05.013 (2011).
Gerke, V. & Moss, S. E. Annexins: from structure to function. Physiological reviews 82, 331–371, https://doi.org/10.1152/physrev.00030.2001 (2002).
He, M. et al. Molecular cloning and characterization of annexin genes in peanut (Arachis hypogaea L.). Gene 568, 40–49, https://doi.org/10.1016/j.gene.2015.05.004 (2015).
Xu, L. et al. Comprehensive analyses of the annexin gene family in wheat. BMC genomics 17, 415, https://doi.org/10.1186/s12864-016-2750-y (2016).
Jami, S. K., Clark, G. B., Ayele, B. T., Roux, S. J. & Kirti, P. B. Identification and characterization of annexin gene family in rice. Plant cell reports 31, 813–825, https://doi.org/10.1007/s00299-011-1201-0 (2012).
Tang, W. et al. Down-regulating annexin gene GhAnn2 inhibits cotton fiber elongation and decreases Ca2+ influx at the cell apex. Plant molecular biology 85, 613–625, https://doi.org/10.1007/s11103-014-0208-7 (2014).
Lu, Y. et al. Genomic organization, phylogenetic comparison and expression profiles of annexin gene family in tomato (Solanum lycopersicum). Gene. 499, 14–24, https://doi.org/10.1016/j.gene.2012.03.026 (2012).
Szalonek, M. et al. Potato annexin stann1 promotes drought tolerance and mitigates light stress in transgenic Solanum tuberosum L. plants. PloS one 10, e0132683, https://doi.org/10.1371/journal.pone.0132683 (2015).
Yadav, D., Ahmed, I. & Kirti, P. B. Genome-wide identification and expression profiling of annexins in Brassica rapa and their phylogenetic sequence comparison with B. juncea and A. thaliana annexins. Plant Gene 4, 109–124, https://doi.org/10.1016/j.plgene.2015.10.001 (2015).
Davies, J. M. Annexin-mediated calcium signalling in plants. Plants 3, 128–140, https://doi.org/10.3390/plants3010128 (2014).
Mortimer, J. C. et al. Annexins: multifunctional components of growth and adaptation. Journal of experimental botany 59, 533–544, https://doi.org/10.1093/jxb/erm344 (2008).
Huh, S. M. et al. Arabidopsis annexins AnnAt1 and AnnAt4 interact with each other and regulate drought and salt stress responses. Plant & cell. physiology 51, 1499–1514, https://doi.org/10.1093/pcp/pcq111 (2010).
Wang, X. et al. Proteomic study of microsomal proteins reveals a key role for Arabidopsis annexin 1 in mediating heat stress-induced increase in intracellular calcium levels. Molecular & cellular proteomics 14, 686–694, https://doi.org/10.1074/mcp.M114.042697 (2015).
Richards, S. L. et al. Annexin 1 regulates the H2O2-induced calcium signature in Arabidopsis thaliana roots. The Plant journal 77, 136–145, https://doi.org/10.1111/tpj.12372 (2014).
Yadav, D., Ahmed, I., Shukla, P., Boyidi, P. & Kirti, P. B. Overexpression of Arabidopsis AnnAt8 alleviates abiotic stress in transgenic Arabidopsis and tobacco. Plants 5, https://doi.org/10.3390/plants5020018 (2016).
Konopka-Postupolska, D. et al. The role of annexin 1 in drought stress in Arabidopsis. Plant physiology 150, 1394–1410, https://doi.org/10.1104/pp.109.135228 (2009).
Liao, C., Zheng, Y. & Guo, Y. MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. The New phytologist 216, 163–177, https://doi.org/10.1111/nph.14679 (2017).
Laohavisit, A. et al. Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1. Plant physiology 163, 253–262, https://doi.org/10.1104/pp.113.217810 (2013).
Laohavisit, A. et al. Arabidopsis annexin1 mediates the radical-activated plasma membrane Ca2+- and K+-permeable conductance in root cells. The Plant cell. 24, 1522–1533, https://doi.org/10.1105/tpc.112.097881 (2012).
Wang, J., Song, J., Clark, G. & Roux, S. J. ANN1 and ANN2 function in post-phloem sugar transport in root tips to affect primary root growth. Plant physiology 178, 390–401, https://doi.org/10.1104/pp.18.00713 (2018).
Lichocka, M. et al. Nucleus- and plastid-targeted annexin 5 promotes reproductive development in Arabidopsis and is essential for pollen and embryo formation. BMC plant biology 18, 183, https://doi.org/10.1186/s12870-018-1405-3 (2018).
Zhu, J. et al. Annexin5 plays a vital role in Arabidopsis pollen development via Ca2+-dependent membrane trafficking. PloS one 9, e102407, https://doi.org/10.1371/journal.pone.0102407 (2014).
Qiao, B. et al. A calcium-binding protein, rice annexin OsANN1, enhances heat stress tolerance by modulating the production of H2O2. Journal of experimental botany 66, 5853–5866, https://doi.org/10.1093/jxb/erv294 (2015).
Li, X., Zhang, Q., Yang, X., Han, J. & Zhu, Z. OsANN3, a calcium-dependent lipid binding annexin is a positive regulator of ABA-dependent stress tolerance in rice. Plant science 284, 212–220, https://doi.org/10.1016/j.plantsci.2019.04.019 (2019).
He, F. et al. Two annexin genes ZmANN33 and ZmANN35 encode proteins that function in cell membrane recovery during the germination of maize (Zea mays. L.) seeds. Journal of experimental botany. https://doi.org/10.1093/jxb/ery452 (2019).
De Carvalho-Niebel, F., Timmers, A. C., Chabaud, M., Defaux-Petras, A. & Barker, D. G. The Nod factor-elicited annexin MtAnn1 is preferentially localised at the nuclear periphery in symbiotically activated root tissues of Medicago truncatula. The Plant journal 32, 343–352 (2002).
Niebel Fde, C., Lescure, N., Cullimore, J. V. & Gamas, P. The Medicago truncatula MtAnn1 gene encoding an annexin is induced by Nod factors and during the symbiotic interaction with Rhizobium meliloti. Molecular plant-microbe interactions 11, 504–513, https://doi.org/10.1094/MPMI.1998.11.6.504 (1998).
Baucher, M. et al. Ntann12 annexin expression is induced by auxin in tobacco roots. Journal of experimental botany 62, 4055–4065, https://doi.org/10.1093/jxb/err112 (2011).
Vandeputte, O. et al. The tobacco Ntann12 gene, encoding an annexin, is induced upon Rhodoccocus fascians infection and during leafy gall development. Molecular plant pathology 8, 185–194, https://doi.org/10.1111/j.1364-3703.2007.00385.x (2007).
Zhang, F., Li, S., Yang, S., Wang, L. & Guo, W. Overexpression of a cotton annexin gene, GhAnn1, enhances drought and salt stress tolerance in transgenic cotton. Plant molecular biology 87, 47–67, https://doi.org/10.1007/s11103-014-0260-3 (2015).
Mu, C., Zhou, L., Shan, L., Li, F. & Li, Z. Phosphatase GhDsPTP3a interacts with annexin protein GhANN8b to reversely regulate salt tolerance in cotton (Gossypium spp.). The New phytologist, https://doi.org/10.1111/nph.15850 (2019).
Zhang, F. et al. A cotton annexin affects fiber elongation and secondary cell wall biosynthesis associated with Ca2+ influx, ROS homeostasis, and actin filament reorganization. Plant physiology 171, 1750–1770, https://doi.org/10.1104/pp.16.00597 (2016).
Huang, Y., Wang, J., Zhang, L. & Zuo, K. A cotton annexin protein AnxGb6 regulates fiber elongation through its interaction with actin 1. PloS one 8, e66160, https://doi.org/10.1371/journal.pone.0066160 (2013).
Li, B. et al. Cotton AnnGh3 encoding an annexin protein is preferentially expressed in fibers and promotes initiation and elongation of leaf trichomes in transgenic Arabidopsis. Journal of integrative plant biology 55, 902–916, https://doi.org/10.1111/jipb.12063 (2013).
Ahmed, I., Yadav, D., Shukla, P. & Kirti, P. B. Heterologous expression of Brassica juncea annexin, AnnBj2 confers salt tolerance and ABA insensitivity in transgenic tobacco seedlings. Functional & integrative genomics 18, 569–579, https://doi.org/10.1007/s10142-018-0614-z (2018).
Ahmed, I. et al. Constitutive expression of Brassica juncea annexin, AnnBj2 confers salt tolerance and glucose and ABA insensitivity in mustard transgenic plants. Plant science 265, 12–28, https://doi.org/10.1016/j.plantsci.2017.09.010 (2017).
Divya, K., Jami, S. K. & Kirti, P. B. Constitutive expression of mustard annexin, AnnBj1 enhances abiotic stress tolerance and fiber quality in cotton under stress. Plant molecular biology 73, 293–308, https://doi.org/10.1007/s11103-010-9615-6 (2010).
Jami, S. K., Hill, R. D. & Kirti, P. B. Transcriptional regulation of annexins in Indian mustard, Brassica juncea and detoxification of ROS in transgenic tobacco plants constitutively expressing AnnBj1. Plant signaling & behavior 5, 618–621, https://doi.org/10.4161/psb.11506 (2010).
Jami, S. K. et al. Ectopic expression of an annexin from Brassica juncea confers tolerance to abiotic and biotic stress treatments in transgenic tobacco. Plant physiology and biochemistry 46, 1019–1030, https://doi.org/10.1016/j.plaphy.2008.07.006 (2008).
Dalal, A. et al. Alleviation of methyl viologen-mediated oxidative stress by Brassica juncea annexin-3 in transgenic Arabidopsis. Plant science 219–220, 9–18, https://doi.org/10.1016/j.plantsci.2013.12.016 (2014).
Chalhoub, B. et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953, https://doi.org/10.1126/science.1253435 (2014).
Lysak, M. A., Koch, M. A., Pecinka, A. & Schubert, I. Chromosome triplication found across the tribe Brassiceae. Genome research 15, 516–525, https://doi.org/10.1101/gr.3531105 (2005).
Wang, X. et al. The genome of the mesopolyploid crop species Brassica rapa. Nature genetics 43, 1035–1039, https://doi.org/10.1038/ng.919 (2011).
Cheng, F., Wu, J. & Wang, X. Genome triplication drove the diversification of Brassica plants. Horticulture research 1, 14024, https://doi.org/10.1038/hortres.2014.24 (2014).
Wen, J. et al. The auxin response factor gene family in allopolyploid Brassica napus. PloS one 14, e0214885, https://doi.org/10.1371/journal.pone.0214885 (2019).
Li, H. et al. Genome-wide analysis of the auxin/indoleacetic acid (Aux/IAA) gene family in allotetraploid rapeseed (Brassica napus L.). BMC plant biology 17, 204, https://doi.org/10.1186/s12870-017-1165-5 (2017).
Wei, L. et al. Genome wide identification and comparative analysis of glutathione transferases (GST) family genes in Brassica napus. Scientific reports 9, 9196, https://doi.org/10.1038/s41598-019-45744-5 (2019).
Song, X. et al. Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC genomics 19, 346, https://doi.org/10.1186/s12864-018-4744-4 (2018).
Lohani, N., Golicz, A. A., Singh, M. B. & Bhalla, P. L. Genome-wide analysis of the Hsf gene family in Brassica oleracea and a comparative analysis of the Hsf gene family in B. oleracea, B. rapa and B. napus. Functional & integrative genomics 19, 515–531, https://doi.org/10.1007/s10142-018-0649-1 (2019).
Zhu, X. et al. Systematic analysis of Hsf family genes in the Brassica napus genome reveals novel responses to heat, drought and high CO2 stresses. Frontiers in plant science 8, 1174, https://doi.org/10.3389/fpls.2017.01174 (2017).
Guo, P. et al. Genome-wide survey and expression analyses of the GRAS gene family in Brassica napus reveals their roles in root development and stress response. Planta. https://doi.org/10.1007/s00425-019-03199-y (2019).
Gao, W. et al. Calcium-dependent protein kinases in cotton: insights into early plant responses to salt stress. BMC plant biology 18, 15, https://doi.org/10.1186/s12870-018-1230-8 (2018).
Sun, H. et al. The Jasmonate ZIM-domain gene family mediates ja signaling and stress response in cotton. Plant & cell. physiology 58, 2139–2154, https://doi.org/10.1093/pcp/pcx148 (2017).
Chen, Y. et al. Genome-wide analysis of the NF-YB gene family in Gossypium hirsutum L. and characterization of the role of GhDNF-YB22 in embryogenesis. International journal of molecular sciences 19, https://doi.org/10.3390/ijms19020483 (2018).
Sun, F. et al. The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. The Plant journal 92, 452–468, https://doi.org/10.1111/tpj.13669 (2017).
Cantero, A. et al. Expression profiling of the Arabidopsis annexin gene family during germination, de-etiolation and abiotic stress. Plant physiology and biochemistry 44, 13–24, https://doi.org/10.1016/j.plaphy.2006.02.002 (2006).
Wagner, A. Selection and gene duplication: a view from the genome. Genome biology 3, reviews1012, https://doi.org/10.1186/gb-2002-3-5-reviews1012 (2002).
Tong, C. et al. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC genomics 14, 689, https://doi.org/10.1186/1471-2164-14-689 (2013).
Liu, S. et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nature communications 5, 3930, https://doi.org/10.1038/ncomms4930 (2014).
Ruelland, E., Vaultier, M. N., Zachowski, A. & Hurry, V. Cold signalling and cold acclimation in plants. Adv Bot Res 49, 35–150 (2009).
Guy, C. Molecular responses of plants to cold shock and cold acclimation. Journal of molecular microbiology and biotechnology 1, 231–242 (1999).
He, X. et al. Comparative transcriptome analyses revealed conserved and novel responses to cold and freezing stress in Brassica napus L. G3: Genes|Genomes|Genetics 9, 2723–2737, https://doi.org/10.1534/g3.119.400229 (2019).
Kabbage, M., Yarden, O. & Dickman, M. B. Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant science 233, 53–60, https://doi.org/10.1016/j.plantsci.2014.12.018 (2015).
Bolton, M. D., Thomma, B. P. & Nelson, B. D. Sclerotinia sclerotiorum (Lib.) de Bary: biology and molecular traits of a cosmopolitan pathogen. Molecular plant pathology 7, 1–16, https://doi.org/10.1111/j.1364-3703.2005.00316.x (2006).
Guo, X. M. & Stotz, H. U. Defense against Sclerotinia sclerotiorum in Arabidopsis is dependent on jasmonic acid, salicylic acid, and ethylene signaling. Mol Plant Microbe In 20, 1384–1395 (2007).
Wang, Z. et al. Defense to Sclerotinia sclerotiorum in oilseed rape is associated with the sequential activations of salicylic acid signaling and jasmonic acid signaling. Plant Science 184, 75–82 (2012).
He, X. et al. GhJAZ2 attenuates cotton resistance to biotic stresses via the inhibition of the transcriptional activity of GhbHLH171. Molecular plant pathology 19, 896–908 (2018).
Li, C. et al. Cotton WRKY1 mediates the plant defense-to-development transition during infection of cotton by Verticillium dahliae by activating JASMONATE ZIM-DOMAIN 1 expression. Plant physiology 166, 2179–2194 (2014).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559, https://doi.org/10.1186/1471-2105-9-559 (2008).
Thines, B. et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665, https://doi.org/10.1038/nature05960 (2007).
Chini, A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007).
Hu, H. Y. et al. GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant Journal 88, 921–935 (2016).
Kazan, K. & Manners, J. M. JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci 17, 22–31 (2012).
Zhou, M. L. et al. Induction of annexin by heavy metals and jasmonic acid inzea mays. Functional & Integrative Genomics 13(2), 241–251 (2013).
Kersey, P. J. et al. Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Research 46, D802–D808, https://doi.org/10.1093/nar/gkx1011 (2017).
Mitchell, A. L. et al. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47, D351–D360, https://doi.org/10.1093/nar/gky1100 (2019).
Wilkins, M. R. et al. Protein identification and analysis tools in the ExPASy server. Methods in molecular biology 112, 531–552, https://doi.org/10.1385/1-59259-584-7:531 (1999).
Hu, B. et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297, https://doi.org/10.1093/bioinformatics/btu817 (2014).
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37, W202–208, https://doi.org/10.1093/nar/gkp335 (2009).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular biology and evolution 33, 1870–1874, https://doi.org/10.1093/molbev/msw054 (2016).
Chen, C. J., Xia, R., Chen, H. & He, Y. H. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 289660, https://doi.org/10.1101/289660 (2018).
Rozas, J. et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular biology and evolution 34, 3299–3302, https://doi.org/10.1093/molbev/msx248 (2017).
He, X. et al. Genome-wide identification of stress-associated proteins (SAP) with A20/AN1 zinc finger domains associated with abiotic stresses responses in Brassica napus. Environmental and Experimental Botany 165, 108–119, https://doi.org/10.1016/j.envexpbot.2019.05.007 (2019).
Deng, W., Wang, Y., Liu, Z., Cheng, H. & Xue, Y. HemI: a toolkit for illustrating heatmaps. PloS one 9, e111988, https://doi.org/10.1371/journal.pone.0111988 (2014).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 13, 2498–2504, https://doi.org/10.1101/gr.1239303 (2003).
Acknowledgements
This work was supported by “2011 Plan”: Institutions of Higher Learning Innovation Ability Enhancement (30552-520190100004), National Key Basic Research Program of China (2015CB150200) and National Key Research and Development Project (2017YFD0101703). In addition, He Xin wants to thank, in particular, the patience, care and support from Miss Liu Xuanzhi over those passed years. Will you marry me?
Author information
Authors and Affiliations
Contributions
Xin He designed the overall study, performed and analyzed most of the experiments, and wrote the manuscript. Li Liao, Sai Xie, Min Yao, Pan Xie, Wei Liu, Yu Kang, Luyao Huang, Mei Wang, Lunwen Qian assisted with experimental data analysis and graph draw. Chunyun Guan and Zhongsong Liu made a significant contribution to the manuscript. Wei Hua and Mei Guan revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
He, X., Liao, L., Xie, S. et al. Comprehensive analyses of the annexin (ANN) gene family in Brassica rapa, Brassica oleracea and Brassica napus reveals their roles in stress response. Sci Rep 10, 4295 (2020). https://doi.org/10.1038/s41598-020-59953-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-59953-w
- Springer Nature Limited
This article is cited by
-
Identification of WRKY transcription factors responding to abiotic stresses in Brassica napus L.
Planta (2022)
-
Genome-wide identification and expression pattern analysis of lipoxygenase gene family in banana
Scientific Reports (2021)
-
Characterization of Annexin gene family and functional analysis of RsANN1a involved in heat tolerance in radish (Raphanus sativus L.)
Physiology and Molecular Biology of Plants (2021)
-
Novel approaches to mitigate heat stress impacts on crop growth and development
Plant Physiology Reports (2020)