Abstract
Nitrogen (N) fertilizers can potentially alter spatial distribution of soil organic carbon (SOC) and total nitrogen (TN) concentrations in croplands such as switchgrass (SG: Panicum virgatum L.) and gamagrass (GG: Tripsacum dactyloides L.), but it remains unclear whether these effects are the same between crops and under different rates of fertilization. 13C and 15N are two important proxy measures of soil biogeochemistry, but they were rarely examined as to their spatial distributions in soil. Based on a three-year long fertilization experiment in Middle Tennessee, USA, the top mineral horizon soils (0–15 cm) were collected using a spatially explicit design within two 15-m2 plots under three fertilization treatments in SG and GG croplands. A total of 288 samples were collected based on 12 plots and 24 samples in each plot. The fertilization treatments were no N input (NN), low N input (LN: 84 kg N ha−1 in urea) and high N input (HN: 168 kg N ha−1 in urea). The SOC, TN, SOC/TN (C: N), δ13C and δ15N were quantified and their within-plot variations and spatial distributions were achieved via descriptive and geostatistical methods. Results showed that SG generally displayed 10~120% higher plot-level variations in all variables than GG, and the plot-level variations were 20~77% higher in NN plots than LN and HN plots in SG but they were comparable in unfertilized and fertilized plots in GG. Relative to NN, LN and HN showed more significant surface trends and spatial structures in SOC and TN in both croplands, and the fertilization effect appeared more pronounced in SG. Spatial patterns in C: N, δ13C and δ15N were comparable among different fertilization treatments in both croplands. The descending within-plot variations were also identified among variables (SOC > TN > δ15N > C: N > δ13C). This study demonstrated that N fertilizations generally reduced the plot-level variance and simultaneously re-established spatial structures of SOC and TN in bioenergy croplands, which little varied with fertilization rate but was more responsive in switchgrass cropland.
Similar content being viewed by others
Introduction
As two important bioenergy crops, perennial switchgrass (SG: Panicum virgatum L) and gamagrass (GG: Tripsacum dactyloides L) are common alternative energy sources for sustainable replacement of fossil fuels1,2,3,4,5. N fertilizers are widely amended to increase bioenergy crop yields6,7,8, but N fertilization impact on soil organic C (SOC) and total soil N (TN) contents varied in magnitude and signal9,10,11. The large variability of fertilization effects among different studies are seldom attributed to the underlying variations in space. In fact, the generally very low number of soil samples in a study (i.e., 3~5) inevitably amplified the influence of spatial variations on the detected treatment effect12. Furthermore, the crop-specific root morphology and chemistry largely regulated the spatial variation of soil biogeochemistry13,14 and may have contributed to contrasting SOC and TN stocks and their responses to N fertilization between SG and GG15. Thus, elucidating the effects of fertilization on spatial distribution of soil C and N will provide fundamental knowledge needed to develop effective strategies to improve soil quality, C sequestration, agricultural productivity, and climate change adaptation16,17,18,19.
A limited number of studies however have focused on the spatial distributions of soil C and N in bioenergy croplands. In a SG cropland, the spatial dependence of SOC and TN at soil depth of 0–30 cm is weak to moderate when the point-to-point distance is less than 200 m20. This contrasts with the observation that a strong spatial dependence of SOC and TN exists within 30 m in a loblolly pine ecosystem21. As the major driver of soil organic matter decomposition, soil microbial communities responded strongly to 3-year N fertilization such that N fertilizer inputs enhanced the spatial heterogeneity of microbial biomass C and N in bioenergy croplands22. It is expected that the generated hotspots of microbial communities in a fertilized plot may result in greater soil C and N accumulations due to fast microbial necromass incorporated into soil organic matter pool23,24,25. Besides the elevated microbial communities, higher earthworm activity, soil porosity, water holding capacity, and soil aeration as well as organic carbon storage were also revealed with decade to century long fertilizations26. Because N fertilization impacts on crop productivity and microbial activities occurred annually or over even a shorter time scale, this may likely shift spatial distribution and structure of soil C and N over month to year time scales. Such evidence is however rarely reported. Although many studies reported the effects of agricultural practices on the stocks and spatial patterns of soil C and N in forests and cultivated lands27,28,29, such studies are surprisingly scarce in bioenergy croplands. In these studies, effect of N fertilization was always masked due to its interaction with other agricultural practices (e.g., tillage and irrigation). It remains largely unknown whether the fertilization effects will depend upon the duration of fertilization.
Soil δ13C and δ15N are widely used to infer vegetation succession, SOC dynamics, and nutrient leaching in bioenergy croplands30,31,32, the spatial variability of these biogeochemical tracers has yet to be studied in SG or GG. In a subtropical woodland, the distance of dissimilar δ13C in soil appears beyond 15.6 m (i.e., spatial autocorrelation), which is far greater than individual tree canopy diameter in these lowland communities33. TN and δ15N are likely to display more spatial variations due to relative low concentrations and diverse chemical forms34, but these have not been confirmed yet. We speculate that the N fertilizer induced greater input to soil via litterfall and root exudate may have contributed to altered spatial variations of microbial biomass as observed formerly22, and this can further stimulate microbial mineralization of more 13C enriched and older soil organic matter35,36. This speculation will not only decrease δ13C values in soil under N fertilization, which resulted from greater release of 13C via heterotrophic respiration of old-aged soil37, but also change the spatial distribution of 13C. However, no studies have reported the spatial variations of soil δ13C and δ15N in different bioenergy croplands and under different N fertilization treatments.
Using a bioenergy crop field experiment in Middle Tennessee, we investigate the effects of N fertilization on the plot-level variances and spatial patterns in elemental and stable isotopic characteristics of soil C and N in two bioenergy croplands (SG and GG). N fertilization input represents the primary management practice in our research plots and no tillage, plowing, or minor mechanical disturbance was applied. We hypothesize that relative to those soils that have not been fertilized for 3 years, continued N fertilization at either low or high rates (84 and 168 kg N ha−1) will reduce the within-plot variances but restructure the spatial patterns of SOC, TN, C:N, δ13C, and δ15N; Furthermore, these aforementioned fertilization effects little depend upon fertilization rate or crop type.
Material and Methods
Site description and characteristics
Prior to the switchgrass and gamagrass croplands, the land use is the mowed grassland for decades. No fertilizers have been applied in the prior land use. Therefore, the indigenous variations are similar in both croplands. Initially established in 2011, the bioenergy crop field experiment is located at the Tennessee State University (TSU) Main Campus Agriculture Research and Education Center (AREC) in Nashville, TN, USA (Lat. 36.12° N, Long. 36.98° W, elevation 127.6 m above sea level). Climate in the region is a warm humid temperate climate with an average annual temperature of 15.1 °C, and total annual precipitation of 1200 mm38. Two crop types and three N fertilization levels were included in a randomized block design22,39. The two crop types were Alamo SG (Panicum virgatum L.) and GG (Tripsacum dactyloides L.). The three N levels included no N fertilizer input (NN), low N fertilizer input (LN: 84 kg N ha−1 yr−1 as urea), and high N fertilizer input (HN: 168 kg N ha−1 yr−1 as urea), and each treatment had four replicated plots with a dimension of 3 m × 6 m (Fig. 1). The low N fertilization rate represented the agronomically optimum nitrogen rate to maximize cellulosic ethanol production in established northern latitude grasslands40. The high N rate doubled the low rate in order to create appreciable gap and detectable effect between the two levels. The fertilizer was manually applied in June or July each year after cutting the grass. Fertilizer was applied after cutting grass during June to July each year. The soil series for the plots is Armour silt loam soil (fine-silty, mixed, thermic Ultic Hapludalfs) with acidic soil pH (i.e., 5.97) and intermediate organic matter content of 2.4%22,41.
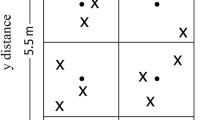
Left: the diagram of the experiment plots in the three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. Two of four blocks (R1, R2) were sampled in this study. Right: illustration of a clustered random sampling design in a plot. Filled circles represent centroids (n = 8) and each plot consists of eight centroids with one in subplot (1.375 m × 1.375 m). In each subplot, a circular was determined for soil sampling. Xs represent sample locations determined from random directions and distances from a centroid within each circular sampling area (dashed circle). The extent of an interpolation map was thus determined by the minimum and maximum values at horizontal and vertical axes, and each map can attain its extent less than or equivalent to the study area (2.75 m × 5.5 m rectangle).
Soil collection and laboratory analysis
We collected soil samples (0–15 cm) from 12 plots (2 crops × 3 N inputs × 2 replicated plots) on June 6, 2015. Within each plot, we divided a 2.75– m × 5.5– m rectangular zone into eight equal-sized square subplots. Within each subplot, a centroid was established and a circular sampling zone was created with the diameter equivalent to side length of the square (Fig. 1). Three cores were randomly collected within each circular sampling zone. Relative to the origin at the southwestern corner in each plot, the distances at the horizontal and vertical directions of each soil core were measured and assigned to each soil core as x, y coordinates. A total of 288 soil cores were obtained based on 24 cores in each of 12 plots. Similar clustered soil sampling approaches were described in formerly published work12,21,22. The soil samples were transported to the TSU lab in a cooler filled with ice packs and were then subsequently stored at 4 °C until analysis. Visible roots and rocks were removed from the samples, and soil samples were then passed through a 2 mm soil sieve. The air-dried soil subsamples were ground and homogenized to fine powder for the analysis of SOC, TN, stable carbon, and nitrogen isotopic compositions (δ13C, δ15N). The analysis was conducted at the University of North Carolina Wilmington Isotope Ratio Mass Spectrometry facility using a Costech 4010 elemental analyzer interfaced with a Thermo Delta V Plus stable isotope mass spectrometer.
Descriptive and geospatial analysis
The frequency distribution, within-plot variance, plot level coefficient of variation (CV), and sample size requirement were calculated to describe plot level variances21,22. The Pearson Moment correlation coefficients were derived among SOC, TN, C: N, microbial biomass carbon (MBC), microbial biomass N (MBN) and MBC: MBN (C: Nmb). The microbial biomass data were adapted from the previous publication22. The study derived the sample size requirement (N) in each plot given specified relative errors (γ, 0~100%) in order to evaluate how within-plot variances (i.e., sample size requirements) are altered by N fertilization or crop type at certain relative error.
Where CI, \(\overline{X,}\) s, n, N, CV, and γ denote confidence interval, plot mean, plot standard deviation, sample number (n = 24), coefficient of variation, sample size requirement and relative error, respectively. t0.975 = 1.96. The log transformed sample size requirement (N) has a negative linear relationship (i.e. slope = 2) with the log transformed relative error (γ).
Cochran’s C test is used to test the assumption of variance homogeneity. The test statistic is a ratio that relates the largest empirical variance of a particular treatment to the sum of the variances of the remaining treatments. The theoretical distribution with the corresponding critical values can be specified42,43,44. Soil properties that exhibited non-normal distributions were log-transformed to better conform to the normality assumption of the Cochran’s C test21. The variances were compared between all plots under three fertilization treatments for each crop and among two crops. When p-value is < 0.05 for the comparison conducted in each crop, the largest variance was identified.
Three different geostatistical approaches were used to display the spatial pattern, structure and distribution of all variables within and among plots and they include trend surface analysis (TSA), Moran’s I index for spatial autocorrelation, and inverse distance weighting (IDW) for distribution map. Details can be found in former publications21,22. Brief introduction of the three geostatistical approaches were also attached. To note, the plot-level variation (i.e., CV) was calculated based on the 24 measurements of soil samples randomly collected in a plot. By accounting for their respective spatial coordinates and distance among samples, the same number of measurements was also used to derive the spatial variability (i.e., spatial heterogeneity) via the three different geostatistical methods. Therefore, there is no direct correlation between the spatial variability (i.e., spatial heterogeneity) and plot-level variation (i.e., CV).
First, the trend surface analysis (TSA) is the most common regionalized model in which all sample points fit a model that accounts for the linear and non-linear variation of an attribute. The relationships between the soil properties and the x and y coordinates of their measurement location within the sampling plots are estimated with the trend surface model:
with regression coefficients β0 to β5. The presence of a trend in the data was determined by the significance of any of the parameters β1 to β5, while the β0 term modeled the intercept45,46. Linear gradients in the x or y directions were indicated by significance of the β1 or β2 parameters. A significant β3 term indicated a significant diagonal trend across a plot. Significant β4 and β5 parameters indicated more complex, nonlinear spatial structure such as substantial humps or depressions. Trend surface regressions were estimated using S-plus (version 7.0, Insightful Inc.). Model parameters were determined to be significant at a level of P < 0.05. The number of significant parameters was counted for each plot and compared among treatments.
Second, residuals from the trend surface regressions were saved for subsequent spatial analysis using a Moran’s I index46. The Moran’s I analysis47,48,49 was used to quantify the degree of spatial autocorrelation that existed among all soil cores taken from each plot. The resulting local Moran’s I statistics are in the range from –1 to 1. Positive Moran’s I values indicate similar values (either high or low) are spatially clustered. Negative Moran’s I values indicate neighboring values are dissimilar. Moran’s I values of 0 indicate no spatial autocorrelation, or spatial randomness. A significant autocorrelation is determined if the observed Moran’s I value is beyond the projected 95% confidence interval at certain distance. Correlograms for local Moran indices were estimated for each soil variable in each plot in a range of 0–5.5 meter with 0.5 meter incremental interval. The number of distance corresponding to the significant Moran’s I was counted for each plot and compared among treatments.
Third, due to relatively small sample sizes (n = 24) per plot50, we used inverse distance weighting (IDW) interpolation rather than ordinary kriging51. The maps produced by IDW offered direct and visual assessments from which to compare the spatial distributions of the soil properties among the plots. The IDW method derived maps was able to distinguish effects of different land uses on spatial distributions of soil biogeochemical features in South Carolina, USA21. The weights for each observation are inversely proportional to a power of its distance from the location being estimated. Exponents between 1 and 3 are typically used for IDW, with 2 being the most common52. Tests with different IDW exponents indicated that 2 was optimal with these data, as estimated values generated with an exponent of 2.0 showed the best fit with actual data in cross validation tests. ArcGIS 9.0 was used to generate the IDW maps and perform cross validations.
Results
Frequency distribution, plot-level variance, CV and sample size requirement
The frequency distributions of all variables appeared concentrated in relatively narrow ranges across different fertilization treatments (Fig. S1). The concentrated ranges of SOC and TN were shifted to greater values under HN and LN relative to NN, but substantial overlaps between fertilization treatments were identified for other variables (Fig. S1). The within-plot variances generally appeared the highest in one of NN plots for all variables in SG, and in one of LN or HN plots for SOC and C: N in GG (Table 1). Only exception is δ13C in SG with the highest within-plot variance in one of HN plots meanwhile other plots of HN and LN were at least two times lower than that for NN plots (Table 1). The within-plot variances appeared insignificantly different between fertilization treatments for TN, δ13C and δ15N (Table 1).
The within-plot CVs were less than 20% for all variables in both croplands and were generally larger in SG than that in GG (Fig. 2). The within-plot CVs were less than 10% for all plots of δ13C in both croplands, C: N in GG, and most plots C: N in SG and δ15N in both croplands. The within-plot CVs varied among different variables and followed a descending order as SOC > TN > δ15N > C:N > δ13C (Fig. 2). The within-plot CV differed less pronouncedly among fertilization treatments for GG (Fig. 2). The highest CV was 19.0% for SOC in SG and the lowest 2.3% for δ13C in GG. The average CV of two plots under NN was 20~77% higher than that under LN or HN in SG (Fig. 2a), not in GG (Fig. 2b). The within-plot CV appeared the largest in one of NN plots for SG (except δ13C).
Within-plot CVs of SOC and TN, C: N, δ13C, and δ15N under three N fertilization (i.e. NN, LN and HN) in (a) SG and (b) GG in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. The line represents CV of 10% in both panels.
Between crops, the plotted lines of sample size requirement against relative sampling error departed from one another more pronouncedly among plots in SG than in GG (comparing the left and right panels, Fig. 3). Among different variables, a larger number of samples were required for SOC and TN than for C: N, δ13C, and δ15N given the same desired relative error (Fig. 3). Among fertilization treatments, the largest number of samples were required for one of NN plot in SG (except δ13C), and only for TN and δ15N in GG.
The plotted linear regression lines between log transformed sample size requirements (SSR) and desired relative errors (%) for SOC and TN, C: N, δ13C and δ15N in SG (left column) and GG (right column) in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA. The log scale was applied on both axes.
Although the mean changes of these variables were not the focus in the current study, the summary statistics (e.g., min, max, range, mean, SE, SD etc.,) were presented in Table S1. The current study focused on the spatial patterns and thus the summary results will be discussed in another relevant publication under preparation.
Trend surface analysis
More surface trends of SOC, TN and C:N, shown either in the x or y directions, diagonal direction, or as nonlinear spatial structure such as substantial humps or depressions, were significant in the fertilized plots (i.e., LN, HN or both) than unfertilized plots in both crops (Table 2). Surface trends of δ13C were not significant in all plots, and the number of significant surface trends of δ15N appeared comparable in all treatments in SG; However, the number of surface trends of δ13C and δ15N were comparable in NN and LN but not in HN in GG (Table 2). No difference was identified between the two crops.
Spatial autocorrelation
Spatial autocorrelations for SOC and TN were not identified at any distance in NN plots, but did show at different distances in LN and HN plots in SG (Table 3; Fig. S2). Spatial autocorrelations for SOC and TN were more frequently identified in either LN or HN than NN in GG. The comparable spatial autocorrelations for C: N, δ13C, and δ15N were identified in both SG and GG (Table 3; Fig. S2). These significant autocorrelations were either positive or negative, and the lagging distances ranged from 0.50 m to 4.75 m. Spatial autocorrelations for SOC, TN, C: N, and δ13C were generally more frequently identified in GG than SG but the opposite was true for spatial autocorrelations for δ15N (Table 3).
Distribution map via inverse distance weighting method
Based on the spatial distribution maps of SOC and TN, those in fertilized plots (particularly HN) exhibited darker color (i.e., higher concentration) and more evidence of spatial structures than NN plots in both croplands (Figs. 4 and 5). In contrast, C: N, δ13C, and δ15N exhibited substantial plot-to-plot variations under the same treatment, particularly for the two plots under NN in both croplands (Figs. 4 and 5). The plot-to-plot differences of the distribution maps were reduced under LN and HN in both croplands (Figs. 4 and 5).
Inverse distance weighted (IDW) maps of SOC, TN, C: N, δ13C, and δ15N in two plots (P1, P2) under three N fertilization treatments (i.e. NN, LN and HN) in SG cropland soils in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA.
Inverse distance weighted (IDW) maps of SOC, TN, C: N, δ13C, and δ15N in two plots (P1, P2) under three N fertilization treatments (i.e. NN, LN and HN) in GG cropland soils in a three-year long fertilization experimental site at the Tennessee State University (TSU) Agricultural Research Center in Nashville, TN, USA.
Discussion
N fertilization reduced the plot-level variations of soil variables in SG
Our results demonstrated that both low and high fertilization rates generally reduced the average variances and CVs across two plots in all variables in SG. However, these variations were little changed with N fertilization in GG. This was likely associated with the much higher variations in SG than in GG (i.e., under NN; Fig. 2), reflecting differential impact of plant species in regulating plot-level variations when N fertilizer was absent. Furthermore, this revealed that the substantial reduction in variance, i.e., greater responsiveness of SG than GG may be driven by their contrasting plant root chemistry and morphology such that the more labile and higher bioreactivity of SG root (unpublished data) is more sensitive to N fertilizer resulting in much faster root turnover and potentially homogenization of originally more varied soil features. Interestingly, the plot-level CVs of the same variable appeared similar under N fertilized plots in both croplands, e.g., 10~15% of CV for SOC, which likely represented a unique and common characteristics of soil biogeochemistry for the bioenergy crops due to their massive root system and high crop yield53,54. Meanwhile, it was also apparent that large plot to plot variations under the same treatment existed, e.g., CVs of most variables under NN in SG. In fact, relative to fertilized plots, the enormously larger plot-level variance always appeared in only one but not in another unfertilized plot. This suggested that despite a generally high variation of soil biogeochemistry in unfertilized bioenergy croplands, other factors such as sampling time and location, crop species and variable type may also regulate the plot-level variances. Nevertheless, with such variability for SOC and TN in unfertilized plots, three samples per plot will produce a relative error of the mean of about ± 20% in both bioenergy croplands, a sobering result given the level of interest in precise estimates of soil C and N sequestrations.
N fertilization restructured the spatial patterns of SOC and TN
Plowing, fertilization and irrigation are common management practices in cultivated row crops, which can homogenize a wide range of soil physiochemical characteristics21,55. The results of the current study supported that lack of other common management practices, N fertilizer input alone however reestablished spatial pattern and structure of SOC and TN, i.e., more significant surface trends in various directions and more pronounced spatial autocorrelations in the fertilized plots. This seemingly contradiction suggested that in the traditional croplands, the mechanical management practices, e.g., tillage and irrigation, acted as dominantly physical agents, which potentially overwhelmed the impacts of other chemical agents such as fertilizers. Besides N fertilizer input, minimal mechanical disturbance and no other chemical agents were amended in the research plots of the current study. Despite the indigenous plot level variations, the N fertilization input as a sole chemical agent, produced an overwhelming effect over the indigenous irregularity of soil nitrogen and likely other nutrient availability20 and reconstructed new spatial structure and pattern.
Another possible explanation is that N fertilizer enhanced the number of hot spots by stimulating microbial activities and turnover56. Consequently, soil fungal hyphae might migrate from root zone to the emerging hot spots57. The emerging hot spots and associated root and microbial activities together may have restructured spatial variations beyond the root zone, leading to overall greater spatial heterogeneity in microbial biomass22, and consequently on SOC and TN. Indeed, our analyses demonstrated significantly positive correlations between SOC, TN and microbial biomass (Table S2). Our data corroborated the restructuring of spatial patterns in both soil bulk C and nutrients storage at the rhizosphere and beyond58.
N fertilization rate little impacted plot-level variation and spatial heterogeneity
Though significantly different from the control treatment, the low and high fertilization treatments appeared little different in either the plot-level variation or spatial heterogeneity in SOC and TN. We speculated that the experimental duration may have contributed to this phenomenon. The current study relied upon a three-year long experiment, but a longer N fertilization history (e.g., > 10 years) may allow us to detect the appreciable changes in spatial heterogeneity under N fertilizations. The reason lied in the fact that the changes in soil C storage were more likely detectable over decades in long-term soil experiments59,60. On the other hand, our former synthesis study demonstrated that multiple soil extracellular enzyme activities were enhanced with a longer fertilization history when compared between the experiments with <1 year and 1~10 years, and those with >10 years61. As soil exoenzymes are the proximate agent of soil decomposition, the more pronounced changes in soil exoenzyme activities over more than 10 years may inform the potentially appreciable change in SOC and TN over longer time. Nevertheless, the magnitude of fertilizer input may also play a role because the fertilization rate in the current study (i.e., 84–168 kg N ha−1) is below the high fertilization rate (e.g., >200 kg N ha−1) applied in other croplands worldwide62. Long-term soil studies will need to specifically include the spatial variability in their research theme in the future.
Mixed effects of N fertilization on the spatial patterns of C: N, δ13C, and δ15N
Relative to SOC and TN, the C: N, δ13C, and δ15N displayed relatively lower plot-level variations under all fertilization treatments in both croplands. This suggested that these specific variables, i.e., derived from other measured variables or proxy of integrated ecosystem processes, likely masked the inherently high variation of each individual process. Nevertheless, the fertilization effects on their spatial patterns depend on the type of geostatistical analysis, crop type and variable. Almost no surface trend was identified for C: N and δ13C in any plot in SG, which contrasted substantially with their counterparts in GG. Both surface trends and spatial autocorrelations for δ15N were much abundant in both croplands. However, no fertilization effects were significant in these analyses. In particular, the higher N fertilizer input resulted in no any significant surface trends of either δ13C or δ15N in all plots in GG. Meanwhile, great plot to plot variations were identified between the distribution maps of two unfertilized plots for C: N and δ13C, and these variations were reduced in fertilized plots, a fertilization effect consistent with those found in SOC and TN. Despite low plot-level variation, the high plot-to-plot variations between the two unfertilized plots for C: N and δ13C may result from a suite of interactions along the air-plant-soil-microbe continuum involved in photosynthate, root exudate, microbe-root associations, and other edaphic features. These multiple drivers themselves entailed high spatial heterogeneities in soils over various spatial scales21,22,63,64,65,66. Therefore, the complexity of biogeochemistry in soils likely has masked the effect of N fertilization on C: N, 13C, and 15N in at least some of their spatial patterns. Despite this uncertainty, the distribution maps produced in this study enabled us to detect fertilization effects that were similar across all these variables.
Conclusions
Our study demonstrated that N fertilizer input generally diminished the plot-level variances of all five variables in SG cropland but re-established the spatial structures of SOC and TN in both croplands. Although N fertilization apparently reduced the plot to plot difference in distribution maps of C: N, δ13C, and δ15N, little fertilization effect was identified on their surface trends and spatial autocorrelations due to either weak or strong patterns simultaneously identified in unfertilized and fertilized plots. Effects of low and high rates of fertilization were found identical in this study. The descending within-plot variations were also identified among variables (SOC > TN > δ15N > C: N > δ13C). This study demonstrated that distinct from the homogenization of spatial structures caused by multiple practices in traditional croplands (e.g., plowing, irrigation and fertilization), N fertilizations alone generally reduced the plot-level variance and simultaneously re-established spatial structures of SOC and TN in bioenergy croplands, which little varied with fertilization rate but was more responsive in switchgrass cropland.
References
Ontl, T. A., Hofmockel, K. S., Cambardella, C. A., Schulte, L. A. & Kolka, R. K. Topographic and soil influences on root productivity of three bioenergy cropping systems. New Phytologist 199, 727–737, https://doi.org/10.1111/nph.12302 (2013).
Gelfand, I. et al. Sustainable bioenergy production from marginal lands in the US Midwest. Nature 493, 514–517, http://www.nature.com/nature/journal/v493/n7433/abs/nature11811.html#supplementary-information (2013).
Kering, M. K., Butler, T. J., Biermacher, J. T., Mosali, J. & Guretzky, J. A. Effect of Potassium and Nitrogen Fertilizer on Switchgrass Productivity and Nutrient Removal Rates under Two Harvest Systems on a Low Potassium Soil. Bioenerg Res. 6, 329–335, https://doi.org/10.1007/s12155-012-9261-8 (2012).
Monti, A., Barbanti, L., Zatta, A. & Zegada-Lizarazu, W. The contribution of switchgrass in reducing GHG emissions. Global Change Biology Bioenergy 4, 420–434, https://doi.org/10.1111/j.1757-1707.2011.01142.x (2012).
Wright, L. & Turhollow, A. Switchgrass selection as a “model” bioenergy crop: A history of the process. Biomass and Bioenergy 34, 851–868, https://doi.org/10.1016/j.biombioe.2010.01.030 (2010).
Lee, M. S., Wycislo, A., Guo, J., Lee, D. K. & Voigt, T. Nitrogen Fertilization Effects on Biomass Production and Yield Components of Miscanthus x giganteus. Front Plant Sci 8, Artn 544 https://doi.org/10.3389/Fpls.2017.00544 (2017).
Duran, B. E. L., Duncan, D. S., Oates, L. G., Kucharik, C. J. & Jackson, R. D. Nitrogen Fertilization Effects on Productivity and Nitrogen Loss in Three Grass-Based Perennial Bioenergy Cropping Systems. PLoS ONE 11, e0151919, https://doi.org/10.1371/journal.pone.0151919 (2016).
Lemus, R. & Lal, R. Bioenergy crops and carbon sequestration. Critical Reviews in Plant Sciences 24, 1–21, https://doi.org/10.1080/07352680590910393 (2005).
Aula, L., Macnack, N., Omara, P., Mullock, J. & Raun, W. Effect of Fertilizer Nitrogen (N) on Soil Organic Carbon, Total N, and Soil pH in Long-Term Continuous Winter Wheat (Triticum Aestivum L.). Communications in Soil Science and Plant Analysis 47, 863–874, https://doi.org/10.1080/00103624.2016.1147047 (2016).
Lee, D. K., Owens, V. N. & Doolittle, J. J. Switchgrass and soil carbon sequestration response to ammonium nitrate, manure, and harvest frequency on conservation reserve program land. Agronomy Journal 99, 462–468, https://doi.org/10.2134/agronj2006.0152 (2007).
Stewart, C. E. et al. N fertilizer and harvest impacts on bioenergy crop contributions to SOC. Global Change Biology Bioenergy 8, 1201–1211, https://doi.org/10.1111/gcbb.12326 (2016).
Li, J. Sampling Soils in a Heterogeneous Research Plot. J. Vis. Exp. e58519, https://doi.org/10.3791/58519 (2018).
Stewart, C. E. et al. Nitrogen and harvest effects on soil properties under rainfed switchgrass and no-till corn over 9 years: implications for soil quality. Global Change Biology Bioenergy 7, 288–301, https://doi.org/10.1111/gcbb.12142 (2015).
Angst, G., Kogel-Knabner, I., Kirfel, K., Hertel, D. & Mueller, C. W. Spatial distribution and chemical composition of soil organic matter fractions in rhizosphere and non-rhizosphere soil under European beech (Fagus sylvatica L.). Geoderma 264, 179–187, https://doi.org/10.1016/j.geoderma.2015.10.016 (2016).
Li, J. et al. Nitrogen Fertilization Escalated Soil Carbon and Nitrogen Contents Are Likely Moderated by Plant Root Chemistry and Morphology in Switchgrass and Gamagrass Croplands. Under Review. (2019).
Ruan, L. L., Bhardwaj, A. K., Hamilton, S. K. & Robertson, G. P. Nitrogen fertilization challenges the climate benefit of cellulosic biofuels. Environmental Research Letters 11, Artn 064007 https://doi.org/10.1088/1748-9326/11/6/064007 (2016).
Tiemann, L. K. & Grandy, A. S. Mechanisms of soil carbon accrual and storage in bioenergy cropping systems. Global Change Biology Bioenergy 7, 161–174, https://doi.org/10.1111/gcbb.12126 (2015).
Turner, M. G. Landscape ecology: What is the state of the science? Annu. Rev. Ecol. Evol. Syst. 36, 319–344 (2005).
Groffman, P. M., Hardy, J. P., Fisk, M. C., Fahey, T. J. & Driscoll, C. T. Climate Variation and Soil Carbon and Nitrogen Cycling Processes in a Northern Hardwood Forest. Ecosystems 12, 927–943, https://doi.org/10.1007/s10021-009-9268-y (2009).
Di Virgilio, N., Monti, A. & Venturi, G. Spatial variability of switchgrass (Panicum virgatum L.) yield as related to soil parameters in a small field. Field Crop Res. 101, 232–239, https://doi.org/10.1016/j.fcr.2006.11.009 (2007).
Li, J. W., Richter, D. D., Mendoza, A. & Heine, P. Effects of land-use history on soil spatial heterogeneity of macro- and trace elements in the Southern Piedmont USA. Geoderma 156, 60–73, https://doi.org/10.1016/j.geoderma.2010.01.008 (2010).
Li, J. et al. Nitrogen Fertilization Elevated Spatial Heterogeneity of Soil Microbial Biomass Carbon and Nitrogen in Switchgrass and Gamagrass Croplands. Sci Rep-Uk 8, 1734, https://doi.org/10.1038/s41598-017-18486-5 (2018).
Liang, C. & Balser, T. C. Microbial production of recalcitrant organic matter in global soils: implications for productivity and climate policy. Nature Reviews Microbiology 9, https://doi.org/10.1038/nrmicro2386-c1 (2011).
Ma, T. et al. Divergent accumulation of microbial necromass and plant lignin components in grassland soils. Nature communications 9, 3480–3480, https://doi.org/10.1038/s41467-018-05891-1 (2018).
Kallenbach, C. M., Grandy, A. S., Frey, S. D. & Diefendorf, A. F. Microbial physiology and necromass regulate agricultural soil carbon accumulation. Soil Biology & Biochemistry 91, 279–290, https://doi.org/10.1016/j.soilbio.2015.09.005 (2015).
Naveed, M. et al. Impact of long-term fertilization practice on soil structure evolution. Geoderma 217-218, 181–189, https://doi.org/10.1016/j.geoderma.2013.12.001 (2014).
Guo, L. B. B., Wang, M. B. & Gifford, R. M. The change of soil carbon stocks and fine root dynamics after land use change from a native pasture to a pine plantation. Plant and Soil 299, 251–262, https://doi.org/10.1007/s11104-007-9381-7 (2007).
Fraterrigo, J. M., Turner, M. G., Pearson, S. M. & Dixon, P. Effects of past land use on spatial heterogeneity of soil nutrients in southern appalachian forests. Ecological Monographs 75, 215–230 (2005).
Nunan, N., Wu, K., Young, I. M., Crawford, J. W. & Ritz, K. In situ Spatial Patterns of Soil Bacterial Populations, Mapped at Multiple Scales, in an Arable Soil. Microbial Ecology 44, 296–305 (2002).
Follett, R. F., Vogel, K. P., Varvel, G. E., Mitchell, R. B. & Kimble, J. Soil Carbon Sequestration by Switchgrass and No-Till Maize Grown for Bioenergy. Bioenerg Res. 5, 866–875, https://doi.org/10.1007/s12155-012-9198-y (2012).
Collins, H. P. et al. Carbon Sequestration under Irrigated Switchgrass (Panicum virgatum L.) Production. Soil Science Society of America Journal 74, 2049–2058, https://doi.org/10.2136/sssaj2010.0020 (2010).
Huang, Y., Rickerl, D. H. & Kephart, K. D. Recovery of Deep-Point Injected Soil Nitrogen-15 by Switchgrass, Alfalfa, Ineffective Alfalfa, and Corn. Journal of Environmental Quality 25, 1394–1400, https://doi.org/10.2134/jeq.1996.00472425002500060033x (1996).
Bai, E. et al. Spatial variation of soil delta C-13 and its relation to carbon input and soil texture in a subtropical lowland woodland. Soil Biology & Biochemistry 44, 102–112, https://doi.org/10.1016/j.soilbio.2011.09.013 (2012).
Robertson, G. P. & Groffman, P. M. Nitrogen transformations. Pages 421–446 in E. A. Paul, editor. Soil microbiology, ecology and biochemistry. Fourth edition. Academic Press, Burlington, Massachusetts, USA., (2015).
Li, J., Ziegler, S., Lane, C. S. & Billings, S. A. Warming-enhanced preferential microbial mineralization of humified boreal forest soil organic matter: Interpretation of soil profiles along a climate transect using laboratory incubations. Journal of Geophysical Research: Biogeosciences 117, G02008, https://doi.org/10.1029/2011jg001769 (2012).
Kuzyakov, Y. Review: Factors affecting rhizosphere priming effects. Journal of Plant Nutrition and Soil Science-Zeitschrift Fur Pflanzenernahrung Und Bodenkunde 165, 382–396 (2002).
Schlesinger, W. H. & Bernhardt, E. S. in Biogeochemistry (Third Edition) (ed William, H. & SchlesingerEmily S. Bernhardt) 173–231 (Academic Press, 2013).
Deng, Q. et al. Effects of precipitation changes on aboveground net primary production and soil respiration in a switchgrass field. Agriculture Ecosystems & Environment 248, 29–37, https://doi.org/10.1016/j.agee.2017.07.023 (2017).
Dzantor, E. K., Adeleke, E., Kankarla, V., Ogunmayowa, O. & Hui, D. Using Coal Fly Ash Agriculture: Combination of Fly Ash and Poultry Litter as Soil Amendments for Bioenergy Feedstock Production. Coal Combustion and Gasification Products. 7, 33–39, https://doi.org/10.4177/CCGP-D-15-00002.1. (2015).
Jungers, J. M., Sheaffer, C. C. & Lamb, J. A. The Effect of Nitrogen, Phosphorus, and Potassium Fertilizers on Prairie Biomass Yield, Ethanol Yield, and Nutrient Harvest. Bioenerg Res. 8, 279–291, https://doi.org/10.1007/s12155-014-9525-6 (2015).
Yu, C. L. et al. Responses of corn physiology and yield to six agricultural practices over three years in middle Tennessee. Sci. Rep-Uk 6, Artn 27504 https://doi.org/10.1038/Srep27504 (2016).
Cochran, W. G. Testing a Linear Relation among Variances. Biometrics 7, 17–32 (1951).
Underwood, A. J. Experiments in Ecology: Their Logical Design and Interpretation Using Analysis of Variance. Cambridge University Press (1997).
Cochran, W. G. The distribution of the largest of a set of estimated variances as a fraction of their total. Annals of Eugenics 11, 47–52 (1941).
Gittins, R. Trend-Surface Analysis of Ecological Data. Journal of Ecology 56, 845–& (1968).
Legendre, P. & Legendre, L. Numerical ecology. Elserier Science, Amsterdam, the Netherlands. (1998).
Legendre, P. & Fortin, M. J. Spatial Pattern and Ecological Analysis. Vegetatio 80, 107–138 (1989).
Cressie, N. Statistics for Spatial Data. John Wiley & Sons, New York (1993).
Moran, P. A. P. Notes on continuous stochastic phenomena. Biometrika 37, 17–23 (1950).
Webster, R. & Oliver, M. A. Sample adequately to estimate variograms of soil properties. Journal of Soil Science 43, 177–192, https://doi.org/10.1111/j.1365-2389.1992.tb00128.x (1992).
Isaaks, E. H. & Srivastava., R. M. Introduction to applied geostatistics. Oxford University Press, New York. (1989).
Gotway, C. A., Ferguson, R. B., Hergert, G. W. & Peterson, T. A. Comparison of kriging and inverse-distance methods for mapping soil parameters. Soil Science Society of America Journal 60, 1237–1247 (1996).
Gregory, A. S. et al. Species and Genotype Effects of Bioenergy Crops on Root Production, Carbon and Nitrogen in Temperate Agricultural Soil. Bioenerg Res 11, 382–397, https://doi.org/10.1007/s12155-018-9903-6 (2018).
Clark, R. B. et al. Eastern gamagrass (Tripsacum dactyloides) root penetration into and chemical properties of claypan soils. Plant and Soil 200, 33–45 (1998).
Robertson, G. P., Crum, J. R. & Ellis, B. G. The Spatial Variability of Soil Resources Following Long-Term Disturbance. Oecologia 96, 451–456 (1993).
Groffman, P. M. et al. Challenges to incorporating spatially and temporally explicit phenomena (hotspots and hot moments) in denitrification models. Biogeochemistry 93, 49–77, https://doi.org/10.1007/s10533-008-9277-5 (2009).
Ghimire, S. R. & Craven, K. D. Enhancement of Switchgrass (Panicum virgatum L.) Biomass Production under Drought Conditions by the Ectomycorrhizal Fungus Sebacina vermifera. Applied and Environmental Microbiology 77, 7063–7067, https://doi.org/10.1128/aem.05225-11 (2011).
Kuzyakov, Y. & Blagodatskaya, E. Microbial hotspots and hot moments in soil: Concept & review. Soil Biology and Biochemistry 83, 184–199, https://doi.org/10.1016/j.soilbio.2015.01.025 (2015).
Richter, Dd, Hofmockel, M., Callaham, M. A., Powlson, D. S. & Smith, P. Long-Term Soil Experiments: Keys to Managing Earth’s Rapidly Changing Ecosystems. Soil Science Society of America Journal 71, 266–279, https://doi.org/10.2136/sssaj2006.0181 (2007).
Smith, P. et al. How to measure, report and verify soil carbon change to realize the potential of soil carbon sequestration for atmospheric greenhouse gas removal. Glob Chang Biol, https://doi.org/10.1111/gcb.14815 (2019).
Jian, S. et al. Soil extracellular enzyme activities, soil carbon and nitrogen storage under nitrogen fertilization: A meta-analysis. Soil Biology and Biochemistry 101, 32–43, https://doi.org/10.1016/j.soilbio.2016.07.003 (2016).
Zhang, F. S., Chen, X. P. & Vitousek, P. An experiment for the world. Nature 497, 33–35, https://doi.org/10.1038/497033a (2013).
Rhoades, C. C. Single-tree influences on soil properties in agroforestry: Lessons from natural forest and savanna ecosystems. Agroforestry Systems 35, 71–94 (1997).
Dockersmith, I. C., Giardina, C. P. & Sanford, R. L. Persistence of tree related patterns in soil nutrients following slash-and-burn disturbance in the tropics. Plant and Soil 209, 137–156 (1999).
Gross, K. L., Pregitzer, K. S. & Burton, A. J. Spatial Variation in Nitrogen Availability in 3 Successional Plant-Communities. Journal of Ecology 83, 357–367 (1995).
Wang, L. X., Okin, G. S., Caylor, K. K. & Macko, S. A. Spatial heterogeneity and sources of soil carbon in southern African savannas. Geoderma 149, 402–408, https://doi.org/10.1016/j.geoderma.2008.12.014 (2009).
Acknowledgements
This study was supported by funding awarded to JL from a US National Science Foundation (NSF) DEB HBCU UP EiR Grant (No. 1900885), US Department of Agriculture 1890 Faculty Research Sabbatical Program (58-3098-9-005) and Evans-Allen Grant (No. 1017802). Funding for MAM was provided by the US Department of Energy (DOE) Office of Biological and Environmental Research through the Terrestrial Ecosystem Science Scientific Focus Area at Oak Ridge National Laboratory (ORNL). ORNL is managed by UT-Battelle, LLC, under contract DE-AC05-00OR22725 with the US DOE. We thank staff members at the TSU’s Main Campus AREC in Nashville, Tennessee for their assistance. We appreciate the anonymous reviewers for their constructive comments and suggestions.
Author information
Authors and Affiliations
Contributions
J.L. and K.D. designed the study; J.L., S.J., C.G., Q.D., C.L., Y.L. and D.H. conducted the field and laboratory analyses; S.J. conducted the data analysis and J.L. wrote the paper; M.M. analyzed the result and revised the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, J., Jian, S., Lane, C.S. et al. Nitrogen Fertilization Restructured Spatial Patterns of Soil Organic Carbon and Total Nitrogen in Switchgrass and Gamagrass Croplands in Tennessee USA. Sci Rep 10, 1211 (2020). https://doi.org/10.1038/s41598-020-58217-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-58217-x
- Springer Nature Limited