Abstract
The complexity of the leaf constitution of foxtail millet (Setaria italica (L.) P. Beauv.) makes it difficult to obtain high-purity cpDNA. Here, we developed a protocol to isolate high-quality cpDNA from foxtail millet and other crops. The new protocol replaces previous tissue grinding and homogenization by enzyme digestion of tiny leaf strips to separate protoplasts from leaf tissue and protects chloroplasts from damage by undue grinding and homogenization and from contamination of cell debris and nuclear DNA. Using the new protocol, we successfully isolated high-quality cpDNAs for whole-genome sequencing from four foxtail millet cultivars, and comparative analysis revealed that they were approximately 27‰ longer than their reference genome. In addition, six cpDNAs of four other species with narrow and thin leaf blades, including wheat (Triticum aestivum L.), maize (Zea may L.), rice (Oryza sativa L.) and sorghum (Sorghum bicolor (L.) Moench), were also isolated by our new protocol, and they all exhibited high sequence identities to their corresponding reference genomes. A maximum-likelihood tree based on the chloroplast genomes we sequenced here was constructed, and the result was in agreement with previous reports, confirming that these cpDNA sequences were available for well-supported phylogenetic analysis and could provide valuable resources for future research.
Similar content being viewed by others
Introduction
Chloroplasts serve as important cytoplasmic organelles in higher plants. Multiple biochemical processes take place in chloroplasts, including photosynthesis, nitrogen metabolism, sulfate reduction, and synthesis of starch, amino acid and lipid. As semi-autonomous cellular organelles, chloroplasts possess their own independent genome and a full complement of transcription and translation machinery to express their genetic information1. The chloroplast DNA (cpDNA) contains 110–130 genes with sizes varying from 120–160 kilobases (kb) in most plants1,2,3, and only a part of the cpDNA is circular4. Most of these genes code for proteins involved in photosynthesis or gene expression, with the remainder as transfer RNA (tRNA) or ribosomal RNA (rRNA) genes2.
In green plants, the chloroplast genome is much simpler in structural complexity than the nuclear genome and generally has a highly conserved organization and gene content across species5. Most chloroplast genomes have a quadripartite structure that includes two copies of an inverted repeat (IR) that separates the genome into large and small copy regions (LSC and SSC)6,7. In addition, cpDNA is usually maternally inherited without genetic recombination8,9. Thus, the chloroplast genome sequence can provide a wealth of information for researchers on plant phylogeny, molecular ecology, comparative genomics, population genetics and evolution and also contribute to chloroplast transformation for crop improvement. Sequencing of high-quality cpDNA is the prerequisite for such studies, and cpDNA fragments amplified from total genomic DNA by polymerase chain reaction (PCR) are currently used for chloroplast genome sequencing. However, this sequencing-quality DNA preparation method is known to be time consuming and difficult to implement when gene organization differs among different plant species10,11 and the existence of promiscuous DNA sequences transferred from chloroplasts and mitochondria to the nucleus also affects the reliability of the results from related studies12,13,14. Thus, a protocol that attempts to separate chloroplasts directly before cpDNA isolation will improve the quality of cpDNA for sequencing.
Currently, cpDNA isolation from fresh plant materials uses high-salt buffers15, a sucrose density gradient or Percoll gradient to separate chloroplast first16, and then DNase to remove nuclear DNA17. All methods need to grind and homogenize leaf tissues and separate and purify chloroplasts by gradual centrifugation. However, grinding and homogenization usually destroy a portion of the chloroplasts, and cell debris and nuclear DNA can easily contaminate isolated cpDNA. Therefore, extraction of high-quality cpDNA is extremely difficult in some species, such as foxtail millet (Setaria italica (L.) P. Beauv), rice (Oryza sativa L.) and other Poaceae crops. Their leaf blades are relatively thin and fibrous with low chloroplast content in mesophyll cells, and the abundant wax, cuticle and silica on the leaf surface also make it hard to break cells to release chloroplasts18. Thus, solving these technical issues will significantly improve the quality of cpDNA isolation from foxtail millet.
In this study, we report a new protocol for the efficient isolation of high-quality cpDNA from foxtail millet. The new protocol significantly increases chloroplast and cpDNA purity and eliminates nuclear and mitochondria DNA contamination. Using this protocol, intact cpDNAs with sequencing quality were isolated from four cultivars of foxtail millet and six cultivars of wheat (Triticum aestivum L.), maize (Zea may L.), rice and sorghum (Sorghum bicolor (L.) Moench). The availability of these chloroplast genomes was proven by comparative analysis and phylogenetic analysis, and they will provide valuable information for future research.
Results
Isolation of chloroplast DNA
To isolate cpDNA in foxtail millet, the first step is to separate chloroplasts from other components. In previous protocols, this step was achieved by grinding and homogenizing leaf samples in an isolation buffer. However, abundant wax, cuticle and silica deposited on the leaf surface make it difficult to break cells and release sufficient chloroplasts into isolation buffer. The new protocol uses several novel strategies to obtain pure chloroplasts from cells (Fig. 1). First, the tissue grinding procedure was replaced by cutting leaves into tiny pieces using a surgical blade, which minimized the contamination of cell debris and chloroplast damage. In addition, mechanical wounding from cutting can increase contact between enzymes and cells. Thus, this new method protects chloroplast intactness and facilitates the release of chloroplasts into extraction buffer. Then, cellulase and macerozyme were used to effectively separate large quantities of intact protoplast from leaf material after centrifugation at 200 g for 10 min. Eliminating the grinding and homogenizing step also minimized the contamination of cell debris and nuclear DNA from grinding tissue. Second, the protoplasts were re-suspended in Buffer 2 followed by two additional centrifugation steps at 500 g for 10 min and 3000 g for 15 min to separate chloroplasts from cell debris and mitochondrial DNA and collect crude chloroplast precipitate based on the difference in sedimentation rates between mitochondria and chloroplast. Third, an extra Percoll density gradient centrifugation step was used to purify isolated chloroplasts. The crude chloroplast pellet was dissolved in Buffer 3 and layered onto a Percoll gradient (10–50% for foxtail millet). After centrifugation at 3000 g for 30 min, intact and pure chloroplasts were recovered from the Percoll interface, and additional nuclear DNA was eliminated.
The new method was used to extract cpDNAs from ten genotypes of different species, including four foxtail millet cultivars (Gu56A, Gu572A, Datong28 and Datong29lv), three wheat cultivars (Jinqiang8, Jinnong6 and Lunxuan987), one maize hybrid cultivar (Zhengdan958), one Japonica rice cultivar (Jingeng818) and one sorghum cultivar (SbJ200), revealing that this protocol was quite valuable for the isolation of cpDNAs from not only foxtail millet but also other Poaceae species with narrow and thin leaves.
Chloroplast genome sequencing
To evaluate the quality of the cpDNAs isolated using the new protocol, we sequenced the complete chloroplast genomes of the five Poaceae crops mentioned above using Illumina next-generation sequencing technology. cpDNAs isolated using the new protocol generated a higher proportion (4.22–12.35%) of mapped reads on the reference chloroplast genomes than those (1% to 4%) from cpDNAs isolated using old protocols19, confirming that the new protocol significantly improved the purity of cpDNAs and generated enough high-quality cpDNAs to assemble the entire chloroplast genome; thus, this protocol has solved previous issues associated with the isolation of cpDNAs from plant species with narrow and thin leaves.
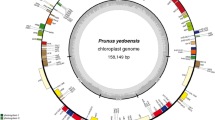
The results showed that all chloroplast genomes we sequenced had a typical quadripartite structure, including two IR regions, an LSC region and an SSC region (Fig. 2, Supplementary Figs S1–S4), with conservative genome size and gene content (Table 1, Supplementary Tables S1–S2). The sequenced chloroplast genomes are 140,454–140,659 bp long in the typical C4 plants maize and sorghum and 134,435–136,482 bp long in the C3 plants wheat and rice. The chloroplast genome of foxtail millet is 139,159–139,261 bp in length, including 129–131 genes with 90–92 genes encoding amino acids, and showed 45 Single Nucleotide Polymorphisms (SNPs) and nine Insertions and Deletions (InDels) among different cultivars, which can be used for marker development for breeding, population genetics and evolution studies.
Genome structure and mapping of genes in the foxtail millet chloroplast genome. The foxtail millet cultivars from inside to outside are Gu56A, Gu572A, Datong28 and Datong29lv, respectively. The thick lines indicate the extent of the IRA and IRB, which separate the genome into the SSC and LSC regions. Genes on the outside of the map are transcribed in the counterclockwise direction, and genes on the inside of the map are transcribed in the clockwise direction. Genes are colored according to their functions as shown in the legend.
Comparative analysis of chloroplast genomes
To further examine the quality of cpDNAs isolated using the new protocol, we aligned the ten sequenced chloroplast genomes with their corresponding reference genome sequences in National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) using the web-based VISualization Tool for Alignments (mVISTA, http://genome.lbl.gov/vista/index.shtml)20. The chloroplast genome sequence of the maize hybrid Zhengdan958 is identical to its reference chloroplast genome (cultivar B73, AY928077.1) (Supplementary Fig. S5). Rice cultivar Jingeng818 and sorghum cultivar SbJ200 exhibited high sequence identity to their reference chloroplast genomes (Nipponbare, GU592207.1 for rice and BTx623, EF115542.1 for sorghum) (Supplementary Figs S6–S7). Those results showed that the new protocol produced high-quality chloroplast cpDNAs for sequencing, and the derived genome sequences should be reliable for further studies.
The sequenced chloroplast genomes of wheat varied with variety. The chloroplast genome sequence of Jinnong6 was 647 bp (4.76‰) longer, and those of Jinqiang8 and Lunxuan987 were 261 bp (1.92‰) and 398 bp (2.93‰) shorter, respectively, than the Chinese Spring chloroplast reference genome (KJ614396.1, 135,835 bp). Several large InDels in intergenic spacers (IGS) and genic regions of psbA, rps7 and rrn23S accounted for the majority of the size variation (Supplementary Fig. S8).
The complete chloroplast genome sizes of the sequenced four foxtail millet cultivars were 3,643–3,745 bp longer than that of their reference genome (NC_022850.1, 135,516 bp in length). The mVISTA alignment results showed that a total of 337 sequence variations (88 insertions and 249 deletions) were detected in NC_022850.1 when compared with the four foxtail millet chloroplast genomes sequenced in our study (Fig. 3, Supplementary Table S3). The majority (272) of InDels were located in IGS of the chloroplast genome, and a small fraction of InDels were distributed in exons (22) or introns (43) of different genes. Among the 337 variations, 219 (64.99%) were 1-10-bp-long small InDels, 89 (26.41%) were 11–40 bp in length, and the remaining 29 (8.60%) were 41–200 bp long. We also found that the chloroplast genome size difference between the sequenced four foxtail millet cultivars and NC_022850.1 foxtail millet reference (27‰) was similar to that between foxtail millet and wheat (19.6–28.2‰), slightly smaller than that between foxtail millet and rice (35.14–35.9‰), and significantly larger than those between foxtail millet and maize (8.6–9.3‰) or sorghum (10.0–10.8‰) (Table 2), suggesting that the size differences between the four foxtail millet chloroplast genomes sequenced here and NC_022850.1 reached or even exceeded the degree of differences between chloroplast genomes of some different species.
Alignment and percentage identity of complete chloroplast genome sequences of five foxtail millet cultivars using mVISTA. Measures of similarity are relative to the reference chloroplast genome NC_022850.1. Exons are shaded in dark blue, and the conserved non-coding sequences (CNS) are shaded in pink. Arrows indicate positions of annotated genes in reference sequences.
Phylogenetic analysis
To assess their phylogenetic relationships, a phylogenetic analysis was performed based on the entire chloroplast genomes from foxtail millet, wheat, maize, rice and sorghum we reported here and five other Poaceae species available in NCBI (Fig. 4). The chloroplast genome of Arabidopsis thaliana (accession number AP000423.1) was used as an out-group. As shown in Fig. 4, Pooideae species, Triticum aestivum (Jinong6, Jinqiang8 and Lunxuan987) and Branchypodium distachyon (accession number NC_011032) formed a clade with Ehrhartoideae species, Oryza sativa (Jingeng818) and Leersia tisserantii (accession number NC_016677). Five Panicoideae species and one Chloridoideae species, Cenchrus ciliaris (accession number MH286942.1), formed another separate clade. In this clade, Zea mays (Zhengdan958) and Sorghum bicolor (SbJ200) were the earliest diverged species, followed by Urochloa brizantha (accession number NC_030067), then Cenchrus americanus (accession number NC_024171) and Cenchrus ciliaris were parallel to Setaria italica (Gu56A, Gu572A, Datong28 and Datong29lv).
Discussion
Due to its highly conserved gene content and organization between species, cpDNA has been widely used to study genome-wide phylogenetics and ecology, transcriptomics, and complete plastid proteome characterization21,22,23. However, an effective method for cpDNA isolation from foxtail millet is currently lacking due to its complicated leaf structure, which makes it difficult to break cells to release chloroplasts18. In this study, we developed a new cpDNA extraction protocol that can be used for foxtail millet and several other crop species that have difficulty in cpDNA extraction (Fig. 1). Compared to previously reported methods15,16,17, the new protocol has been technically improved by the following steps: 1) using a surgical blade to cut leaves into tiny pieces instead of grinding tissue in a grinding machine; 2) extracting protoplasts instead of a homogenization process; 3) selecting appropriate centrifugation speeds and density gradients to separate chloroplasts from other DNA and cell components. These changes significantly increased chloroplast quality and solved several issues including broken chloroplasts from grinding the narrow and thin leaves of foxtail millet, contamination of cell debris in isolated chloroplasts due to the high content of minerals, and nuclear and mitochondrial DNA contamination. The new protocol protected the intactness of chloroplasts of foxtail millet, thus significantly improving chloroplast quality, which is critical for sequencing the chloroplast genome of foxtail millet and lays a solid foundation for further understanding the genetic basis of foxtail millet.
To determine the quality of isolated cpDNA, cpDNA of all five species with narrow and thin leaves was sequenced, and high proportions (4.22–12.35%) of reads were mapped onto their corresponding chloroplast genomes. The percentage of mapped reads was much higher than those (1% to 4%) from cpDNA isolated using other protocols19, indicating that the purity of the cpDNA isolated using the new protocol was much higher than that with existing protocols and is good enough for whole chloroplast genome assembly. We sequenced ten complete chloroplast genomes from five species (Fig. 2, Supplementary Figs S1–S4) using the new protocol and obtained high-quality sequence data, indicating that the newly developed cpDNA isolation protocol is suitable not only for foxtail millet but also other Poaceae species with narrow and thin leaves. Based on the chloroplast genomes we sequenced here and cpDNA sequences from five other Poaceae species available in NCBI, we constructed a phylogenetic tree using the maximum-likelihood method in MEGA 7.0 with 1000 replicates. The topology of this phylogenetic tree is consistent with a previous report19, indicating that the chloroplast genome sequences we reported here can be used for well-supported phylogenetic reconstruction, and these data will provide invaluable resources for future research.
We compared the chloroplast genome sequences of ten genotypes from five species to their reference genomes in NCBI (Fig. 2, Supplementary Figs S5–S8) and found that the maize cultivar Zhengdan958 had an identical chloroplast genome to its reference genome, the cultivar B73 (AY928077.1) (Supplementary Fig. S5). Moreover, the chloroplast genomes of the rice cultivar Jingeng818 and sorghum cultivar SbJ200 also showed high sequence similarity to their reference sequences (Supplementary Figs S6–S7), demonstrating the reliability of our sequence data. According to the maternal inheritance of cpDNA in plants, the completely matched chloroplast genomes of Zhengdan958 and B73 revealed that they have a common maternal parent. The pedigree analysis of Zhengdan958 reflected that Zhengdan958 was derived from the cross Zheng58 × Chang7-2, and Zheng58 was a variant from the inbred line Ye478 that was derived from U8112 × 500324,25, demonstrating that Zhengdan958, Zheng58, Ye478 and U8112 derived from the same maternal parent. Thus, we speculated that Zhengdan958, Zheng58, Ye478, and U8112 may share a common maternal parent with B73.
Compared to the nuclear genome, a chloroplast genome is much smaller with a simpler structure and more conserved gene content and arrangement5; thus, the chloroplast genome is more useful for taxonomic studies. For instance, the chloroplast genes matK and rbcL have been developed as effective molecular markers for the identification of land plant species26. Non-coding regions in the chloroplast genome, such as the trnL intron, intergenic spacer trnH-psbA and trnL-trnF, have been widely used as chloroplast barcoding markers in plant systematics and phylogeography27,28,29,30,31. Comparative analysis of complete chloroplast genomes of four foxtail millet cultivars and three wheat cultivars identified several InDels in intergenic spacer regions and genic regions (Fig. 3, Supplementary Fig. S8). These InDels could be developed as DNA markers for the identification of accessions in germplasm collections and breeding in foxtail millet and wheat.
In some previous studies, independent deletions mediated by short direct-repeat sequences were successively found among the cpDNAs of three species of wheat, Aegilops crassa, Aegilops squarrosa and Triticum aestivum32, and four species of rice, Oryza punctata, Oryza officinalis, Oryza australiensis and Oryza sativa33. The variations in the deletions are genotype-specific and species-specific in rice and wheat respectively. In 1993, similar deletions were also found within the single species of Oryza sativa, suggesting that the occurrence of this deletion was comparatively easy between species and within a single species during evolution34. In this study, we sequenced the complete chloroplast genomes of four foxtail millet cultivars. Does this type of specific variation exist within these four cultivars or even more cultivars of foxtail millet? Where are the locations of these variations? Additionally, what are the sequences around them? These questions will be examined in our further studies and may provide a basis for cultivar classification if there are intraspecific variations of cpDNAs within foxtail millet.
Materials and Methods
Plant materials
Ten cultivars of five species were used as testing materials for protocol optimization and sequencing in this study, including a summer maize hybrid cultivar Zhengdan958, a Japonica rice cultivar Jingeng818, a sorghum cultivar SbJ200, a spring wheat cultivar Jinqiang8, two winter wheat cultivars Jinnong6 and Lunxuan987, and four foxtail millet cultivars Gu56A, Gu572A, Datong28 and Datong29lv. All plant materials were grown in the greenhouse of Tangshan Normal University, China. For each plant genotype, ~10 g of fresh leaves were harvested at the 6th to 8th leaf stage for chloroplast extraction.
Reagent preparation
Enzyme solution containing 0.5% cellulase R10 (w/v), 0.3% macerozyme R10 (w/v), 0.4 M mannitol, 20 mM KCl, and 20 mM MES (pH 5.7) was prepared by warming at 55 °C for 10 min, cooling to room temperature (25 °C), and then adding 10 mM CaCl2 and 0.1% BSA (w/v). Buffer 1 contained 0.9% NaCl (w/v), 1.4% CaCl2 (w/v), 0.04% KCl (w/v), 0.1% glucose (w/v), and 1 mM MES (pH 5.7). Buffer 2 contained 0.3 M sorbitol, 1 mM MgCl2·6H2O, 2 mM EDTA·2Na, 50 mM HEPES, and 0.04% β-mercaptoethanol (w/v), and Buffer 3 consisted of 0.3 M sorbitol, 1 mM MgCl2·6H2O, 2 mM EDTA·2Na, and 50 mM HEPES.
Chloroplast DNA isolation
Protoplast isolation
Leaves were cut into 0.5–1-mm strips by a sharp razor blade without crushing tissue at the cutting site and completely submerged into the enzyme solution (10 g of leaves in 50–150 ml) for digestion at room temperature in the dark for at least 3 h without shaking. The enzyme solution was diluted with an equal volume of Buffer 1 and filtered through a copper mesh (cell strainer, 200 meshes) to remove undigested leaf tissues. The filtered solution was centrifuged at 200 g afterwards to pellet the protoplasts in a 50-ml round-bottomed tube for 10 min at 4 °C.
Crude chloroplast extraction
The protoplasts were re-suspended in 50 ml of Buffer 2, kept on ice and inverted once every 5 min. After 15 min, the protoplasts were centrifuged at 500 g for 10 min at 4 °C, and the supernatant was harvested gently. The supernatant was then centrifuged at 3000 g for 15 min at 4 °C, and the resulting crude chloroplast precipitate was collected.
Chloroplast purification and cpDNA isolation
The crude chloroplasts were re-suspended with Buffer 3 (500–1000 µl), layered onto a Percoll gradient (10–50% for foxtail millet, 25–45% for wheat, 10–45% for maize, 15–50% for rice and 15–45% for sorghum) made with the same buffer, and centrifuged at 3000 g for 30 min at 4 °C. The resulting chloroplasts between the two gradients were collected, re-suspended in 1 ml of Buffer 3, and centrifuged at 12000 rpm for 5 min at 4 °C. The supernatant was removed, and the last step was repeated once. The high-purity chloroplast pellets were then collected, frozen with liquid nitrogen and stored at −80 °C for cpDNA isolation.
cpDNA was extracted from the intact chloroplasts using the DNAquick Plant System DP321 (Tiangen Biotech, Beijing, China) following the manufacturer’s instructions.
Whole chloroplast genome sequencing, assembly and annotation
The cpDNAs were sequenced in an Illumina HiSeq Sequencer at Beijing Ori-Gene Science and Technology (China). Low-quality reads, reads with adaptor sequences and duplicated reads were removed, and the remaining high-quality data were used for assembly. The clean reads were first combined into contigs using SOAPdenovo35, Then, the assembled contigs were aligned to the reference genome to detect assembly errors using BLAT36. The gaps between contigs were further bridged using the GapCloser package to refine the assembly. Finally, the physical map was constructed using OrganellarGenomeDRAW37 by anchoring the scaffolds onto chloroplast genome sequences.
Sequenced genomes were annotated using CpGAVAS38. This program identified both protein-coding genes and rRNA genes by performing Blastx, Blastn, protein2genome and est2genome39 searches against a custom database of published plastid genomes. tRNAs were identified by tRNAscan40 and ARAGORN41. Inverted repeat regions (IRs) were identified using vmatch42.
Sequence alignment and phylogenetic analysis
Chloroplast genome sequences were aligned using VISualization Tool for Alignments (mVISTA) (http://genome.lbl.gov/vista/index.shtml)20. Phylogenetic analysis was performed using the maximum likelihood method, as implemented in the MEGA 7.0 program with 1000 bootstrap replicates43.
Data availability
The chloroplast genome sequences we reported in this study and corresponding annotations have been deposited to GenBank under the following accession numbers: 56 A (MK348603), 572 A (MK348609), Datong28 (MK348605), Datong29lv (MK348604), Jinnong6 (MK348601), Jinqiang8 (MK348611), Lunxuan987 (MK348610), Zhengdan958 (MK348606), Jingeng818 (MK348618), SbJ200 (MK348612).
References
Sugiura, M. The chloroplast genome. Plant Mol Biol. 19, 149–168 (1992).
Jansen, R. K. et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods in Enzymology. 395, 348 (2005).
Sugiura, M. The chloroplast genome. Essays Biochem. 30, 49–57 (1995).
Bendich, A. J. Circular chloroplast chromosomes: the grand illusion. The Plant Cell. 16, 1661–1666 (2004).
Shaffer, C. Next-generation sequencing outpaces expectations. Nat Biotechnol. 25, 149 (2007).
Raubeson, L. & Jansen, R. Chloroplast genomes of plants. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. 45–68 (2005).
Saski, C. et al. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol Biol. 59, 309–322 (2005).
Shaver, J. M., Oldenburg, D. J. & Bendich, A. J. Changes in chloroplast DNA during development in tobacco, Medicago truncatula, pea, and maize. Planta. 224, 72–82 (2006).
Ravi, V., Khurana, J. P., Tyagi, A. K. & Khurana, P. An update on chloroplast genomes. Plant Syst Evol. 271, 101–122 (2008).
Atherton, R. A. et al. Whole genome sequencing of enriched chloroplast DNA using the Illumina GAII platform. Plant Methods. 6, 1–6 (2010).
Cronn, R. et al. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 36, e122 (2008).
Ayliffe, M. A., Scott, N. S. & Timmis, J. N. Analysis of plastid DNA-like sequences within the nuclear genomes of higher plants. Mol Biol Evol. 15, 738–745 (1998).
Goremykin, V. V., Salamini, F., Velasco, R. & Viola, R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol. 26, 99–110 (2009).
Rousseau-Gueutin, M., Ayliffe, M. A. & Timmis, J. N. Conservation of plastid sequences in the plant nuclear genome for millions of years facilitates endosymbiotic evolution. Plant Physiology. 157, 2181–2193 (2011).
Bookjans, G., Stummann, B. M. & Henningsen, K. W. Preparation of chloroplast DNA from pea plastids isolated in a medium of high ionic strength. Anal Biochem. 141, 244–247 (1984).
Heinhorst, S., Gannon, G. C., Galun, E., Kenschaft, L. & Weissbach, A. Clone bank and physical and genetic map of potato chloroplast DNA. Theor Appl Genet. 75, 244–251 (1988).
Kolodner, R. & Tewari, K. K. Inverted repeats in chloroplast DNA from higher plants. Proc Natl Acad Sci USA 76, 41–45 (1979).
Shen, Z., Sun, C. & Zhu, J. Isolation and purification of rice chloroplast DNA. Chinese Biochemical Journal. 5, 7–11 (1989).
Pessoa, M., Martins, A. & Elias Ferreira, M. Molecular dating of phylogenetic divergence between Urochloa species based on complete chloroplast genomes. BMC Genomics. 18, 516 (2017).
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M. & Dubchak, I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–279 (2004).
Gargano, D., Maple-Grødem, J., Reisinger, V., Eichacker, L. A. & Møller, S. G. Analysis of the chloroplast proteome in arc mutants and identification of novel protein components associated with FtsZ2. Plant Mol Biol. 81, 235–244 (2013).
He, Z., Li, H., Shen, Y., Li, Z. & Mi, H. Comparative analysis of the chloroplast proteomes of a wheat (Triticum aestivum L.) single seed descent line and its parents. J Plant Physiol. 170, 1139–1147 (2013).
Lee, J., Kang, Y., Shin, S. C., Park, H. & Lee, H. Combined analysis of the chloroplast genome and transcriptome of the Antarctic vascular plant Deschampsia antarctica Desv. PLos One. 9, e92501 (2014).
Du, C. et al. The breeding and application of maize hybrid Zhengdan958. Journal of Maize Sciences. 14, 43–45 (2006).
Zheng, Y., Zhang, F., Meng, C., Hou, D. & Sun, Z. Studies on characteristic differentiation of corn inbred line Ye478 and its related lines. Seed. 28, 74–75 (2009).
Group, C. P. W. A. DNA barcode for land plants. Proc Natl Acad Sci USA 106, 12794–12797 (2009).
Kress, W. J. et al. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc Natl Acad Sci USA 106, 18621–18626 (2009).
Kress, W. J. et al. Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a Puerto Rican forest dynamics plot. PLoS One. 5, e15409 (2010).
Taberlet, P. et al. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35, e14 (2007).
Schmickl, R., Kiefer, C., Dobeš, C. & Koch, M. A. Evolution of trnF (GAA) pseudogenes in cruciferous plants. Plant Syst Evol. 282, 229–240 (2009).
Ansell, S. W. et al. Recombination diversifies chloroplast trnF pseudogenes in Arabidopsis lyrata. J Evolution Biol. 20, 2400–2411 (2010).
Ogihara, Y., Terachi, T. & Sasakuma, T. Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc Natl Acad Sci. 85 (1988).
Kanno, A. & Hirai, A. Comparative studies of the structure of chloroplast DNA from four species of Oryza: cloning and physical maps. Theor Appl Genet. 83, 791–798 (1992).
Kanno, A., Watanabe, N., Nakamura, I. & Hirai, A. Variations in chloroplast DNA from rice (Oryza sativa): differences between deletions mediated by short direct-repeat sequences within a single species. Theor Appl Genet. 86, 579–584 (1993).
Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 1, 18 (2012).
Kent, W. J. BLAT-the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002).
Lohse, M., Drechsel, O., Kahlau, S. & Bock, R. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41, W575–W581 (2013).
Chang, L. et al. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13, 715–715 (2012).
Mott, R. EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA. Comput Appl Biosci. 13, 477–478 (1997).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Laslett, D. & Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16 (2004).
Abouelhoda, M. I., Kurtz, S. & Ohlebusch, E. Replacing suffix trees with enhanced suffix arrays. Journal of Discrete Algorithms. 2, 53–86 (2004).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33, 1870–1874 (2016).
Acknowledgements
This work was supported by National Natural Science Foundation of China [grant number 31560094, 31471563].
Author information
Authors and Affiliations
Contributions
Z.L. conceived and designed the research, supervised the experiments, and compiled and finalized the article together with G.F. and C.C.; D. Liu developed the new protocol and performed most of the isolation of high quality chloroplast DNAs; Y.C. performed the phylogenetic analysis, and drafted the article; S.L. performed the comparative analysis of chloroplast genomes; G.B. assisted with the comparative analysis and preparing the manuscript; Q.L., Z.Z., D. Liang, C.W., J.W. and X.S. helped with the isolation of chloroplast DNAs.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, D., Cui, Y., Li, S. et al. A New Chloroplast DNA Extraction Protocol Significantly Improves the Chloroplast Genome Sequence Quality of Foxtail Millet (Setaria italica (L.) P. Beauv.). Sci Rep 9, 16227 (2019). https://doi.org/10.1038/s41598-019-52786-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-52786-2
- Springer Nature Limited