Abstract
Separated river systems could create confluences via two geological processes, estuary coalescence in response to decreasing sea levels and headwater capture, allowing primary freshwater species to disperse across rivers. Squalidus multimaculatus, is an endemic and primary freshwater species restricted to the southeast coast of the Korean Peninsula. The distribution of this species is unique, given that other congeneric species, including its closely related S. gracilis majimae, as well as other cyprind species are observed throughout the peninsula except for the east coast. Phylogeographic analyses were conducted using three mitochondrial loci to identify the origin of S. multimaculatus and the historical pathways of dispersal. A strong phylogenetic affinity between S. multimaculatus and S. g. majimae and the genetic structure among populations indicated that S. multimaculatus originated from the eastward colonization of the common ancestor between S. g. majimae and S. multimaculatus via headwater capture through fault zones within successive mountain range. Following colonization, the ancestral S. multimaculatus likely migrated towards north via estuary coalescence along a well-developed continental shelf. Our study was the first empirical attempt providing insights into how freshwater organisms dispersed to the southernmost tip of East Asia, despite the potential loss of such historical imprints with anthropogenic interference.
Similar content being viewed by others
Introduction
In freshwater ecosystems, primary freshwater fish species cannot naturally move to other physically separated drainages, and landscape structures may even become barriers within a single drainage, limiting gene flow among populations1,2. Because of this fragmented nature, freshwater ecosystems can provide exceptional opportunities for analysis of the contribution of physical isolation to intraspecific genetic structure1,3,4. By combining the genetic data of intraspecific population structures with information regarding historical drainage structures and geological events, it is possible to infer how a freshwater species or its ancestor have dispersed across rivers shaping the present distribution2,4,5,6.
With time, separated river systems could create confluences via two representative geological processes, allowing freshwater species to disperse across rivers that are not currently connected7,8. First, the estuary regions of adjacent rivers were often connected in response to decreasing sea levels during the Quaternary period (estuary coalescence)9,10,11, though connections could rather rarely occur between rivers that did not share the same well-developed continental shelf12,13. This geological phenomenon seems to have occurred in regions with wide continental shelves, where rivers in geographical proximity are likely to share estuaries8,10,14. Second, the uppermost region of a river may be diverted from its own bed and merged with the headwater of other adjacent rivers (headwater capture)11,15,16. Although headwater capture is still observable with watershed erosion in a specific condition17, it is a geological phenomenon that frequently occurred via strong tectonic deformation along fault zones17,18,19.
The Korean Peninsula is a small area located at the end of the East Asian continent (Fig. 1). The Yellow Sea, located between the peninsula and western China (Fig. 1), was part of the Yellow River, a major river system flowing east through China, when the sea level was lower than at present during the last glacial maximum (LGM)6,20,21. The Yellow Sea probably created confluences with the present-day major river systems along the south and west coast on the Korean Peninsula21,22,23. Conversely, the east coast of this peninsula is geographically isolated by successive mountain ranges (Fig. 1); accordingly, the freshwater ecosystems on the east coast likely had confluences with a different large drainage system (i.e., the Paleo-Amur River)24,25. Despite the landscape isolation, the freshwater ecosystems along the southern east coast might have historically been coalesced with the Paleo-Yellow Sea9,21,25,26. However, no empirical approach has been conducted to date to reconstruct colonization processes around this area.
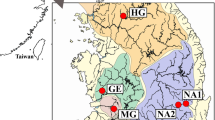
The distribution range of Squalidus multimaculatus (red-colored areas) and S. gracilis majimae (major river systems flowing the south and west coasts). The collection sites of S. multimaculatus (yellow circles) and S. g. majimae (green stars) are shown in the squares positioned on the right and middle top (see Table S1 for the detailed information). Arrows indicate the three hypothetical routes (H1, H2 and H3)16,26,28 of ancestral S. multimaculatus through successive mountain range (BD: Baekdudaegan; NJ: Nakdongjeongmaek). The map was generated by Adobe Illustrator CC 2015 using a GIS shape file retrieved from GADM database (www.gadm.org; v 2.5; freely available for non-commercial purposes) in DIVA GIS (http://www.diva-gis.org/) and modified in QGIS v 2.16.3 in accordance with the guidelines suggested in the websites.
The spotted-barbel gudgeon, Squalidus multimaculatus, is an endemic freshwater species (Cyprinidae) restricted to the southern east coast of the Korean Peninsula25,27. The distribution of this species is somewhat unique, given that other congeneric species (S. gracilis majimae, S. chankaensis tsuchigae and S. japonicus coreanus) are observed in most freshwater systems throughout the peninsula except for the east coast25,27. Among these congeneric species, S. g. majimae is morphologically and genetically close to S. multimaculatus28. Thus, it is conceivable that S. multimaculatus originated from the migration of the common ancestor of S. multimaculatus and S. g. majimae individuals inhabiting the drainage areas close to the east coast such as the Nakdong River, the longest river flowing to the south coast in South Korea28. Three hypothetical routes for this geodispersal process could be envisaged based on the geographical features on the east coast and previous studies (Fig. 1)16,26. There might have been headwater confluences from the tributaries of the Han (second longest river flowing to the west coast) and Nakdong River to the Samcheok-Osip River (H1)16 or directly to rivers on the southern east coast (H2; Fig. 1)26,28. The second hypothesis (H2) was established based on the existence of Yangsan Fault Zone between the Nakdong River and the southern east coast and its tectonic history. Considering the geographic proximity, the mouths of the Nakdong River (or adjacent small streams) and southernmost east coastal rivers likely coalesced following the Quaternary climate changes (H3; Fig. 1), though this hypothesis has yet to be validated empirically.
In this study, three mitochondrial loci were used to analyze the phylogenetic relationship among the populations of S. multimaculatus and S. g. majimae (Supplementary Table S1). In addition, divergence ages among phylogenetic groups of S. multimaculatus and S. g. majimae were estimated for comparison with the history of geological changes in the landscape structures isolating the east coast. These analyses could allow us to identify the most valid one among the three biogeographic routes presented above for the colonization of S. multimaculatus. These data were also used to reconstruct the historical migration pattern of S. multimaculatus following its colonization within the east coast by analyzing the demographic histories of the phylogenetic groups. Although this study is a biogeographic investigation of a single species, our results are anticipated to provide critical insight into the historical pattern of drainage formation and freshwater dispersal on the tip of East Asian continent.
Results
The primers used in this study successfully amplified all individuals. A total of 16, 61 and 26 haplotypes were detected from the sequences of COI (670 bp), cyt b (1,128 bp) and 12S (949 bp), respectively. Among these, Squalidus gracilis majimae contained 5, 23 and 9 haplotypes in COI, cyt b and 12S, respectively. Except for two populations, JA and YO, no haplotypes were shared between these two species (Supplementary Tables S2–S4). COI, cyt b and 12S contained 41 (6.1%), 137 (12.1%) and 34 (3.5%) polymorphic sites, and 29 (4.3%), 104 (9.2%) and 21 (2.2%) parsimonious informative sites, respectively.
All three algorithms (NJ, ML and BI) generated highly concordant tree topologies in the phylogenetic analysis reconstructed based on cyt b sequences (Supplementary Fig. S2). S. g. majimae haplotypes (SG) were resolved as the most likely sister group of S. multimaculatus, and Japanese S. gracilis was placed as the sister to the cluster of S. g. majimae and S. multimaculatus (Supplementary Fig. S2). Two other congeneric species inhabiting the Korean Peninsula, S. japonicus coreanus and S. chankaensis tsuchige, were somewhat separated from S. g. majimae and S. multimaculatus in the tree (Supplementary Fig. S2). Haplotypes of S. multimaculatus were likely to be allocated into three haplogroups, though the distinction of ME and NE was not clear in the ML and BI trees (Supplementary Fig. S2). The name of haplogroups were assigned as NE (northeast), ME (mid-east) and SE (southeast) in accordance with the geographic regions in which they were predominantly observed (Supplementary Tables S2–S5), which was also supported by the result of AMOVA (Tables S6). The ME and NE haplogroups formed a cluster, while SE was placed as a sister to that cluster (Supplementary Fig. S2). In the phylogenetic tree analysis using the other two loci (COI and 12S), the haplogroup SE was clearly isolated from others, but the distinction between ME and NE was somewhat difficult (Supplementary Figs S3–S4). The divergence between S. g. majimae and S. multimaculatus was estimated to be 1.83 (95% HPD 0.31–2.37) MYA (Fig. 2). Within S. multimaculatus, age estimates of 0.59 (95% HPD 0.38–0.81) MYA and 0.31 (95% HPD 0.19–0.45) MYA were allocated to the nodes leading to the separation of haplogroup SE and the divergence between NE and ME, respectively (Fig. 2). Our analysis of ancestral area reconstruction (BBM) showed that the common ancestor of both S. g. majimae and S. multimaculatus was inferred to be distributed in the southwestern Korean Peninsula and that the common ancestor of S. multimaculatus colonized in SE region for the first time before dispersion (Fig. 2).
Time-calibrated Bayesian tree reconstructed by BEAST using cyt b sequences. The 95% highest posterior confidence intervals of divergence times are indicated on every major node with blue bar. Ancestral area reconstruction was also inferred based on the Bayesian Binary Markov Chain Monte-Carlo (BBM) and incorporated in the tree. The color circle of nodes showed the most likely ancestral area; blue: western Korean Peninsula, pink: Taehwa and Hoeya River (region SE), red: Hyeongsan River (region ME).
Based on our NJ analysis results, each population was included in a single haplogroup. Six northern populations (MP, PN, BR, CS, YO and GG; NE region) only contained NE haplotypes in the three loci, except that a single SE haplotype of cyt b and a few haplotypes of S. g. majimae were found in population YO (Supplementary Tables S2–S5). Population HS (mid-east region) contained haplotypes allocated to haplogroup ME, whereas SE haplotypes were predominantly found in populations TH and HY (southeast region), though one ME haplotype in both COI and cyt b was found in population TH (Supplementary Tables S2–S5). Population DJ contained haplotypes that could be allocated to haplogroup NE, despite its geographical proximity to populations HS and TH (Supplementary Tables S2–S5). The haplotype composition in population JA was somewhat complicated because the haplotypes of ME and S. g. majimae coexisted (Supplementary Tables S2–S5). Analysis of RAG1 sequences (1,455 bp) based on the double peak calling of both species specific nucleotides revealed that twelve individuals in population JA and three individuals in population YO were hybrids of S. multimaculatus and S. g. majimae (Supplementary Tables S2–S5; Fig. S1).
The presence of three haplogroups was also evident upon network analysis with the combined datasets of the three mitochondrial loci (Fig. 3). In this analysis, haplogroups NE and ME were genetically close to each other, similar to the results of phylogenetic tree analyses, whereas haplogroup SE appeared to be closer to the haplotypes of S. g. majimae than the other two haplogroups (Fig. 3), indicating that the first founder of S. multimaculatus likely colonized the SE region. Network analysis with cyt b revealed a similar pattern of separation among the three haplogroups (Supplementary Fig. S5). However, the separation of haplogroup SE was only observed upon analysis with 12S. In addition, no noticeable separation was observed upon analysis with COI, despite a slight difference between NE and ME (Supplementary Fig. S5).
The unrooted network generated based on the haplotypes of three mitochondrial loci (COI, cyt b and 12S) from Squalidus multimaculatus and S. gracilis majimae. See Fig. 1 for the details about how to generate the map.
The intra-population genetic diversity was estimated and compared based on the combined dataset of three mitochondrial loci (Table 1; see also Supplementary Tables S7–S9). Populations YO (NE), HS (ME) and TH (SE) exhibited the highest level of genetic diversity among S. multimaculatus populations (Table 1). The remaining NE populations (MP, PN, BR, CS, GG and DJ) showed low intra-population genetic diversities (Table 1). The genetic diversity of population HY (SE) was also relatively low (Table 1). A moderate level of diversity was observed in population JA (Table 1). For S. g. majimae, population GH was higher than that of YA (Table 1).
Neutrality tests revealed that haplogroups NE and SE had significant negative Fu’s Fs and Tajima’s D values (Table 2; see also Supplementary Tables S10–S12), indicating the signature of demographic expansion in the sizes of those haplogroups following diversification and colonization. The Tajima’s D and Fu’s Fs values of haplogroup ME were negative but not significant; thus, the neutrality model could not be rejected (Table 2; see also Supplementary Tables S10–S12). Upon mismatch analyses, haplogroups NE and SE showed a clearly unimodal pattern, confirming the results based on the estimation using the Fu’s Fs and Tajima’s D values (Fig. 4). A bimodal shape was exhibited in haplogroup ME, indicating that its size has been relatively stationary since colonization (Fig. 4). Our Bayesian skyline plot analyses showed that the effective population sizes of haplogroups NE and ME increased by about 100 KYA (Fig. 4). Haplogroup SE underwent gradual decline in population size and subsequent rapid increase at about 20 KYA.
Discussion
In our phylogentic analyses, Squalidus gracilis majimae was resolved to be the closest sister to S. multimaculatus. Although our analysis does not take into account extinct taxa, when solely comparing all of the existing species in Squalidus, it is conceivable that S. multimaculatus on the east coast originated from colonization by individuals of S. g. majimae or its derived populations. The results of our haplotype network and phylogenetic analyses showed that S. multimaculatus populations could be divided into three major haplogroups, NE, ME and SE. The NE and ME haplogroups closely clustered with each other, while SE was rather separated, but showed relatively higher affinity to S. g. majimae. These results, combined with past geological changes, could provide the basis for verifying which of the three hypothetical dispersal paths described in view of existing literatures and geographical environment is most plausible.
The first hypothetical route was established based on the possibility of migration from the major freshwater systems on the Korean Peninsula (south and west) to the Samcheok-Osip River on the east coast by headwater capture16,29. Since the Samcheok-Osip River is located somewhat northward of the known natural distribution range of S. multimaculatus (from PN to JA), it is unreasonable to regard this river as a starting point for colonization. Although S. multimaculatus individuals were observed in the Samcheok-Osip River as well as several other northern rivers (including population MP) in our field investigations and previous studies30,31,32, those individuals probably existed because of artificial transplanting given that the presence of this species has never been known until a few decades ago, despite intensive field investigations29,30,33. Furthermore, considering the results of our phylogenetic tree analyses that showed NE and ME were clustered together and SE was placed as a sister group, it is difficult to explain how this species could spread towards southern east coastal rivers.
The second hypothetical route was established based on the fact that the Yangsan Fault Zone between the Nakdong River and rivers along the southern east coast has undergone active crustal fluctuations34,35,36, and two freshwater systems (the Nakdong River and southern east coastal rivers) were likely to have fused together during the geological changes (Fig. 5). In this fault zone, the strongest activities occurred about 1 million years ago34,35,36, which is the same time at which S. g. majimae and S. multimaculatus were estimated to have diverged in our analyses. The haplotypes of populations TH and HY (SE) exhibited the highest affinity to the haplotypes observed in S. g. majimae in our haplotype network analyses, indicating that ancestral S. multimaculatus founders colonized the SE region for the first time in the east coast. As shown in the phylogenetic clustering pattern, it is conceivable that S. multimaculatus individuals naturally migrated north from this point. This second hypothesis can also be supported by the fact that species inhabiting the Nakdong River, such as Cobitis hankugensis (Cobitidae), Iksookimia longicorpa (Cobitidae) and Liobagrus sp. (Liobagridae), are also present in SE regional rivers37,38,39. Although it is known that a historical migration occurred between the Hyeongsan River (population HS) and the Nakdong River through the Yangsan Fault40, our data did not verify this possibility.
The colonization route of Squalidus multimaculatus inferred based on our genetic results and previous geologic studies. Dashed lines on the ocean indicate the range of the continental shelf developed along the east coast on the Korean Peninsula. Blue lines indicate the virtual paleo-drainage system existed along the coast when the sea levels were lower than at present designed to illustrate the confluences inferred. Gray arrows indicate the historical dispersal route of S. multimaculatus or its ancestors through Yangsan (H2; YFZ) and Ulsan (UFZ) Fault Zones. See Fig. 1 for the details about how to generate the map.
In SE, the genetic diversity of population TH was much higher than that of HY, presumably indicating that this species (or its common ancestor) first colonized the Taehwa River (TH; Fig. 5) and then moved to the Hoeya River (HY), though the contemporary diversity estimates may not represent the past demographics. The mouths of the two rivers are geographically adjacent, and they were likely occasionally linked during the sea level fluctuation in the last glacial maxima (Fig. 5)41,42,43,44. This inference is also supported by the ichthyofaunal similarity between the two rivers38,45. Our EBSP results indicated that the dispersal within the SE region was most active around the late Pleistocene (20 KYA), because sudden demographic growth was noticeable during this period, following the gradual decline since 200 KYA. How could S. multimaculatus that colonized the SE region disperse to the river in the north? Headwater capture has also likely contributed to the migration of S. multimaculatus founders to the Hyeongsan River (population HS). The Ulsan Fault Zone is located between the Hyeongsan River and the Taehwa River (population TH), and the watersheds of both rivers are in geographical proximity (Fig. 5). The divergence time of haplogroup ME and NE from SE was estimated to be 430 KYA in our analyses, which completely coincides with the active period of this fault35,46. Taken together, these findings indicate that the headwater capture between those two rivers was a starting point of northward dispersal of S. multimaculatus.
Population HS exhibited an exceptionally high level of diversity in all three loci examined. In addition, haplogroup ME did not show any demographic decline or sudden fluctuations in our EBSP results. This was probably related to the physical habitat in the area inhabited by this population. Although most east coastal rivers are small and subject to weather-sensitive physical changes, even during a short-term period, the Hyeongsan River is large, conceivably providing stable and wide habitats that maintain a large-sized population. Since there is no fault zone from the Hyeongsan River towards the north, headwater capture might not have contributed to colonization in the NE region. Instead, there is a well-developed continental shelf, that is likely to have served as a place for the coalescence among river mouths. The historical northward migration from ME to NE is supported by the finding that natural populations of Iksookimia yongdokensis (Cobitidae) are observed in both the ME and NE region, but not the SE region25.
The third hypothetical route was made by the assumption that frequent estuary coalescence between south coastal rivers (including the Nakdong River) and southern east coastal rivers probably contributed to the migration of freshwater species. If this assumption is correct, the Jangan River (population JA; ME), which is located where the south and east coast meet, might have served as a bridge for the historical colonization of S. multimaculatus to the east coast. However, our results did not provide any evidence supporting such an assumption. The population JA did not exhibit the highest genetic affinity to population HY (in the Hoeya River; SE), located in the immediate north, but was closer to population HS (Hyeongsan River; ME). As discussed below, haplogroup ME did not show the highest genetic affinity to S. g. majimae. In addition to our results, we made an effort to find evidence of water connection around this area. No geological evidence has been known about the connection, for example between the Jangan River and any from SE region. Since the headwaters are not geographically adjacent around this area, the possible water connection might be the estuary coalescence responding to sea level fluctuation. However, the continental shelf on the coast is too narrow to imagine the effect of sea level change. Except for S. multimaculatus, there was no ichthyofaunal commonality between the Janan River and SE rivers.
In our analyses, several populations showed quite unexpected genetic characteristics. First, population DJ was part of haplogroup NE in our analyses, despite its geographical proximity to SE or ME populations, which cannot be explained solely based on the biogeographical inference. This may be the results of artificial introduction, though this prediction should be examined by empirical studies in the future. Second, as already mentioned, MP was also considered to be created by artificial introduction. In the Myeongpa River (MP), there are several cyprinid species that were not observed in other adjacent rivers, including Zacco platypus, Z. koreanus and Aphyocypris chinensis, indicating that some massive introduction might have occured31,32. However, the results of this study do not clearly indicate NE rivers from which this population originated. In particular, two cyt b gene haplotypes, NE04 and NE06, observed in population MP were not present in any of the NE populations analyzed in the present study, indicating the possibility of transplantation from the river(s) that was (were) not analyzed in this study.
Population JA contained the haplotypes of both S. multimaculatus and S. g. majimae. Where did the individuals of this population come from? S. multimaculatus haplotypes in this population most likely originated from population HS. Only a single ME haplotype was observed, indicating that a small number of individuals were introduced to this population probably from the population HS. A single SE haplotype was also observed in this population, which may be the evidence of additional introductions from SE regional rivers, though the possibility of natural existence could also be regarded. However, the haplotype diversity of S. g. majimae was somewhat higher than that expected to be formed by artificial introduction of a small number of individuals. Therefore, it is conceivable that S. g. majimae individuals might have existed naturally prior to the introduction of S. multimaculatus in this river. In addition, some S. g. majimae haplotypes in this population were not observed in two populations from the Nakdong River, indicating the possibility of introduction from other river(s). However, caution is needed when drawing conclusions regarding the historical origins of this population, because sampling has not been performed throughout the Nakdong River. A low frequency of S. g. majimae haplotypes was also observed in the population JA, indicating that a small number of S. g. majimae individuals were introduced in this population. Examination with the nuclear marker (RAG1) showed clear evidence of active hybridization between S. g. majimae and S. multimaculatus in both populations.
Conclusion
In our genetic analyses, we inferred the route of colonization and distribution origin of a freshwater fish species at the southeast tip of East Asia, though further comparative studies are required to confirm this result. The first colonization of S. multimaculatus ancestral population likely occurred through confluence caused by headwater capture during geological changes in landscape structures. In the rivers around mid- and northern east coast regions where the continental shelf is wider than the southern part of the eastern coast, estuary coalescence could be presumed to be the main route of migration and colonization of this species. However, our study also showed the potential loss of such historical imprints that have been created over a million years because of anthropogenic interference.
Materials and Methods
Sampling and sequencing
All samples used were collected according to the Inland Water Fisheries Act and Wildlife Protection and Management Act of the Republic of Korea. The entire procedure of this study was approved by the Yeungnam University Institutional Animal Care and Use Committee (Protocol # 2015013). A total of 209 individuals of Squalidus multimaculatus were collected from 11 east coastal rivers on the Korean Peninsula (Fig. 1; Table 1). A total of 40 individuals of S. gracilis majimae were also collected from two spots located on the Nakdong River (Fig. 1; Table 1). Number of scales above the lateral-line was rapidly counted for all individuals collected, because this characteristic was known to be different between S. multimaculatus and S. g. majimae25,27,28. All individuals collected were released back to their original habitats after removing a small fragment of tissue from each caudal fin. All tissue samples were preserved in 99% ethanol until they were used for DNA extraction and further genetic analyses. Whole genomic DNA was extracted using a Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA).
Three mitochondrial loci, cytochrome b (cyt b), cytochrome oxidase subunit 1 (COI) and 12S rRNA (12S), were analyzed in this study (Supplementary Table S1). The nuclear RAG1 gene was also analyzed only for detection of hybrid individuals (by checking the existence of double peaks of S. multimaculatus and S. g. majimae specific nucleotides) known to occur in several S. multimaculatus populations28 (Supplementary Fig. S1). PCR was performed in a 25 µl mixture containing 1 µl DNA template, 1× Taq Buffer, 0.2 mM dNTPs, 2.5 µM of each primer and 2.5 units of Taq DNA polymerase (Genetbio, Daejeon, South Korea). Thermal cycling was performed under a program consisting of initial denaturation at 94 °C for 2 m, 35 cycles of 94 °C for 30 s, 54 °C (or 56 °C for COI) for 30 s and 72 °C for 30 s and then final extension at 72 °C for 10 m. Amplified products were purified with the PrimePrep PCR Purification Kit (Genetbio, Daejeon, South Korea) and sent to Genotech (Daejeon, South Korea) for sequencing, which was conducted in an ABI3730xl DNA analyzer (Applied Biosystems, Foster City, CA) with the same primer sets used in our PCR reactions. The residues of DNA samples were deposited at the Department of Life Sciences, Yeungnam University.
Diversity estimation
All of the haplotype sequences were deposited in GenBank (Accession number: KY486515–486635). DNA sequences analyzed for all four loci (including RAG1) were initially edited using Geneious 9.147, after which they were manually checked for erroneous base calls. Multiple sequence alignments were performed using ClustalW48,49 implemented in MEGA650. Genetic diversity indices of three mitochondrial loci, including nucleotide diversity (π)51, number of haplotypes (h)51, haplotype diversity (hd)51 and number of segregating sites (S)52, were estimated using DnaSP 5.153.
Phylogenetic analyses
Phylogenetic trees were generated under three different algorithms, Bayesian inference (BI), maximum likelihood (ML) and neighbor-joining (NJ). All three mitochondrial loci were used in the reconstruction of phylogenetic tree. Since the cyt b sequence is the most widely reported in fish species, however, the phylogenetic tree using key species as outgroup could only be reconstructed using this locus. The outgroup species were selected from Cyprinidae as follows: five Hemibarbus species including H. labeo (DQ267432), H. maculatus (JN003322), H. barbus (AB070241), H. mylodon (DQ225265), H. longirostris (DQ267422), eight Squalidus species including S. japonicas (JN003320; Japan), S. chankaensis (JN003321; China, Mongol and Russia), S. chankaensis tsuchigae (KY486514; Korea), S. japonicus coreanus (KY486513; Korea), S. atromaculatus (AY952333; China and South Asia), S. nitens (AY952986; China), S. argentatus (JQ421222; China and South Asia) and S. gracilis gracilis (AF375866; Japan), Rhodeus pseudosericeus (KF410783), Tinca tinca (NC008648) and Zacco seiboldii (NC008653). Jmodeltest254,55 was implemented in CIPRES portal version 3.156 to identify the most appropriate nucleotide substitution models for BI and ML based on the Akaike information criterion (AIC)57. The best fitting model was found to be TIM3 + I + G.
BI analysis was conducted using MrBayes 3.258 implemented in the CIPRES portal. Each BI analysis consisted of two parallel runs of 80 million Markov Chain Monte Carlo (MCMC) generations with sampling every 1,000 steps. The consensus tree for each data set was generated after omitting the first 25% of sampled trees as burn-in. The node confidence in the BI tree was presented with the Bayesian posterior probabilities. The convergence and stationarity of chains were determined based on whether the effective sampling size (ESS) reached more than 200 using TRACER 1.659. The ML tree was created using RAxML GUI 1.560 under the GTRGAMMA model with 1000 bootstrap replicates. NJ analysis was performed using MEGA6 under the Kimura 2-parameter model (K2P)61. Node confidence in the NJ tree was assessed based on 1,000 bootstrap replicates.
Hierarchical genetic structuring was analyzed by assessing the relative contributions of among-haplogroup, among-population and within-population components using an analysis of molecular variance (AMOVA), which was performed using Arlequin 3.562. The unrooted haplotype network was initially generated for four different datasets (cyt b, COI, 12S and combined) based on the connection limit above 0.95 in probability using TCS 1.263. When the link was disconnected between haplogroups, PopART64 was applied to generate the connection.
Divergence time estimation
Divergence time estimation among phylogenetic groups was conducted in BEAST 2.3.065 using cyt b sequences in the CIPRES portal. The BEAST analysis was run using a strict and an uncorrelated lognormal relaxed molecular clock to examine the variation of rate across branches. Posterior distributions of parameters were estimated using 40 million MCMC generations, with samples drawn every 1,000th steps under the model chosen as the most-fit, TIM3 + I + G. The initial 10% of samples were discarded as burn-in. The analysis was conducted using a Yule speciation and a birth-death model for the tree prior to examine the sensitivity of the results to the choice of tree prior. The genus Hemibarbus was known to be a sister to Squalidus66 and its fossil record (5 MYA)67 was available as the calibration point to constrain the ages. This record was assigned in the form of uniform prior to provide a minimum age for the separation of Hemibarbus and Squalidus. TRACER 1.659 was used to confirm whether ESS exceeded 200 and the MCMC parameters of runs converged on the same stationary point with unimodal distribution. A consensus tree was generated after discarding the first 20% of trees as burn-in in TreeAnnotator 2.3.065, and was visualized in FigTree 1.4.268. The node credibilities of the consensus tree were evaluated by posterior probabilities. To examine dispersal scenario for S. multimaculatus, the distributional areas of common ancestor was inferred using the Bayesian binary MCMC method (BBM)69 implemented in RASP70. BEAST tree was used as an input for this analysis. Six areas are assigned based on the distributional area of the species used in the BEAST tree: (A) China; (B) Korean Peninsula excluding east coastal rivers; (C) the Taehwa and Hoeya River; (D) the Hyeongsan River; (E) the Myeongpa, Pyeonghae-Namdae, Baegrok, Chuksan, Youngdeok-Osip and Gokgang River; (F) Japan.
Demographic history
Historical demographic changes of phylogenetic groups were examined based on Tajima’s D71,72, Fu’s Fs73, mismatch distribution74 and an extended Bayesian skyline plot (EBSP)75. Tajima’s D and Fu’s Fs were quantified using Arlequin 3.562 by coalescent simulations under the infinite-sites model. The mismatch distribution for each phylogenetic group was analyzed to examine the possibility of sudden population expansion76 in Arlequin. Finally, EBSP were generated independently for three mitochondrial loci using BEAST 1.8.277 to provide evidence of the historical expansion of geographic groups. BEAST was run for 60 million MCMC generations, with sampling every 5000th tree, based on TIM3 + I + G and a strict clock model (1% pairwise divergence per million years)78. TRACER 1.659 was used to confirm whether ESS reached above 200 and the MCMC parameters of runs converged on the same stationary point with unimodal distribution.
References
Ward, R. D., Woodwark, M. & Skibinski, D. O. F. A comparison of genetic diversity levels in marine, freshwater, and anadromous fishes. J. Fish Biol. 44, 213–232 (1994).
Avise, J. C. Phylogeography: the history and formation of species. (Harvard university press, 2000).
Gyllensten, U. The genetic structure of fish: differences in the intraspecific distribution of biochemical genetic variation between marine, anadromous, and freshwater species. J. Fish Biol. 26, 691–699 (1985).
Avise, J. C. et al. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 18, 489–522 (1987).
Hurwood, D. A. & Hughes, J. M. Phylogeography of the freshwater fish, Mogurnda adspersa, in streams of northeastern Queensland, Australia: evidence for altered drainage patterns. Mol. Ecol. 7, 1507–1517 (1998).
Jeon, H. & Suk, H. Y. Pseudo but actually genuine: Rhodeus pseudosericeus provides insight into the phylogeographic history of the Amur bitterling. Animal Cells Syst. 18, 275–281 (2014).
Wong, B. B. M., Keogh, J. S. & McGlashan, D. J. Current and historical patterns of drainage connectivity in eastern Australia inferred from population genetic structuring in a widespread freshwater fish Pseudomugil signifer (Pseudomugilidae). Mol. Ecol. 13, 391–401 (2004).
Unmack, P. J., Hammer, M. P., Adams, M., Johnson, J. B. & Dowling, T. E. The role of continental shelf width in determining freshwater phylogeographic patterns in south‐eastern Australian pygmy perches (Teleostei: Percichthyidae). Mol. Ecol. 22, 1683–1699 (2013).
Kitamura, A., Takano, O., Takata, H. & Omote, H. Late Pliocene–early Pleistocene paleoceanographic evolution of the Sea of Japan. Palaeogeogr. Palaeoclimatol. Palaeoecol. 172, 81–98 (2001).
Unmack, P. J. Biogeography of Australian freshwater fishes. J. Biogeogr. 28, 1053–1089 (2001).
Thacker, C. E., Unmack, P. J., Matsui, L. & Rifenbark, N. Comparative phylogeography of five sympatric Hypseleotris species (Teleostei: Eleotridae) in south‐eastern Australia reveals a complex pattern of drainage basin exchanges with little congruence across species. J. Biogeogr. 34, 1518–1533 (2007).
Lima, S. M. et al. Headwater capture evidenced by paleo-rivers reconstruction and population genetic structure of the armored catfish (Pareiorhaphis garbei) in the Serra do Mar mountains of southeastern Brazil. Front. Genet. 8, 199 (2017).
Thomaz, A. T., Malabarba, L. R., Bonatto, S. L. & Knowles, L. L. Testing the effect of palaeodrainages versus habitat stability on genetic divergence in riverine systems: study of a Neotropical fish of the Brazilian coastal Atlantic Forest. J. Biogeogr. 42, 2389–2401 (2015).
Zúñiga-Vega, J. J., Ingley, S. J., Unmack, P. J. & Johnson, J. B. Do freshwater ecoregions and continental shelf width predict patterns of historical gene flow in the freshwater fish Poecilia butleri? Biol. J. Linn. Soc. 112, 399–416 (2014).
Xu, W. et al. Phylogeographical Analysis of a cold-temperate freshwater fish, the amur sleeper (Perccottus glenii) in the amur and Liaohe River basins of northeast asia. Zoolog. Sci. 31, 671–679 (2014).
Kim, D., Hirt, M. V., Won, Y. & Simons, A. M. Small fishes crossed a large mountain range: Quaternary stream capture events and freshwater fishes on both sides of the Taebaek Mountains. Integr. Zool. 12, 292–302 (2017).
Bishop, P. Drainage rearrangement by river capture, beheading and diversion. Prog. Phys. Geogr. 19, 449–473 (1995).
Burridge, C. P., Craw, D. & Waters, J. M. An empirical test of freshwater vicariance via river capture. Mol. Ecol. 16, 1883–1895 (2007).
Carrea, C., Anderson, L. V., Craw, D., Waters, J. M. & Burridge, C. P. The significance of past interdrainage connectivity for studies of diversity, distribution and movement of freshwater-limited taxa within a catchment. J. Biogeogr. 41, 536–547 (2014).
Xu, D., Liu, X., Zhang, X., Li, T. & Chen, B. China offshore geology. (Geological publishing house, 1997).
Yoo, D., Park, S., Sunwoo, D. & Oh, J. Evolution and chronology of late Pleistocene shelf-perched lowstand wedges in the Korea Strait. J. Asian Earth Sci. 22, 29–39 (2003).
Ren, M. & Shi, Y. Sediment discharge of the Yellow River (China) and its effect on the sedimentation of the Bohai and the Yellow Sea. Cont. Shelf Res. 6, 785–810 (1986).
Yoo, D. et al. Seismic stratigraphy and depositional history of late Quaternary deposits in a tide-dominated setting: An example from the eastern Yellow Sea. Mar. Pet. Geol. 73, 212–227 (2016).
Nishimura, S. Formation of the Sea of Japan. (Tsukiji-Shokan, 1974)
Kim, I. S. Illustrated encyclopedia of fauna & flora of Korea, vol. 37, freshwater fishes, (Ministry of education, 1997).
Kwan, Y., Kim, D., Ko, M., Lee, W. & Won, Y. Multi-locus phylogenetic analyses support the monophyly and the Miocene diversification of Iksookimia (Teleostei: Cypriniformes: Cobitidae). Syst. Biodivers. 1–8 (2017).
Hosoya, K. & Jeon, S. R. A new cyprinid fish, Squalidus multimaculatus, from small rivers on the eastern slope of the Taebaik Mountain Chain. Korean J. Limnol. 1051, 41–49 (1984).
Lee, Y. J., Bae, H., Jeon, H., Kim, D. & Suk, H. Y. Human-mediated processes affecting distribution and genetic structure of Squalidus multimaculatus, a freshwater cyprinid with small spatial range. Animal Cells Syst. 1–9; https://doi.org/10.1080/19768354.2017.1371074 (2017).
Choi, K. C. On the geographical distribution of fresh-water fishes south of DMZ in Korea. Korean J. Limnol. 6, 29–36 (1973).
Choi, J., Byeun, H. & Cho, K. Studies on stream conditions and fish community in Osip stream (Samchuk county). Korean J. Limnol. 28, 263–270 (1995).
Park, S. et al. TheFish Fauna and Disturbance of Geographical Distributions in the Eastern Civilian Control Line of Korea. J. Environ. Impact Assess. 22, 27–37 (2013).
Ko, M., Moon, S. & Bang, I. Fish community structure and inhabiting status of endangered species in Baebong stream. Korean J. Ecol. Environ 46, 192–204 (2013).
Choi, K. C. The nature of Gangwon. (The education committee of Gangwon-do, 1986).
Lee, B. J., Choi, S. J., Chwae, U. & Ryoo, C. R. Characteristics of the Quaternary faulting of the Wolpyeong, Yangsan, SE Korea. J. Geol. Soc. Korea 35, 179–188 (1999).
Kyung, J. Paleoseismological study and evaluation of maximum earthquake magnitude along the Yangsan and Ulsan Fault Zones in the Southeastern Part of Korea. Geophys. Geophys. Explor. 13, 187–197 (2010).
Kim, H. et al. The offshore Yangsan fault activity in the Quaternary, SE Korea: Analysis of high-resolution seismic profiles. Tectonophysics 693, 85–95 (2016).
Kim, J., Yang, H., Cho, Y., Kim, Y. & Cho, M. Monitoring of pathogens and characteristics of fish community in the Taewha River. Korean J. Environ. Biol. 28, (2010).
Chae, B. & Yoon, H. A new distributional record of Cobitis hankugensis (Cobitidae, Pisces) in the Hoeya river, Korea. J. Nat. Park Res. 1, 101–103 (2010).
Kim, S., Kim, H. & Park, J. A new species of torrent catfish, Liobagrus hyeongsanensis (Teleostei: Siluriformes: Amblycipitidae), from Korea. Zootaxa 4007, 267–275 (2015).
Bae, H. & Suk, H. Y. Population genetic structure and colonization history of short ninespine sticklebacks (Pungitius kaibarae). Ecol. Evol. 5, 3075–3089 (2015).
Fairbanks, R. G. A 17,000-year glacio-eustatic sea level record: influence of glacial melting rates on the Younger Dryas event and deep-ocean circulation. Nature 342, 637–642 (1989).
Lambeck, K., Yokoyama, Y. & Purcell, T. Into and out of the Last Glacial Maximum: sea-level change during Oxygen Isotope Stages 3 and 2. Quat. Sci. Rev. 21, 343–360 (2002).
Rohling, E. J., Fenton, M., Jorissen, F. J. & Bertrand, P. Magnitudes of sea-level lowstands of the past 500,000 years. Nature 394, 162–165 (1998).
Lambeck, K., Esat, T. M. & Potter, E.-K. Links between climate and sea levels for the past three million years. Nature 419, 199–206 (2002).
Seo, J., Lim, I., Kim, H. & Lee, H. K. Status of fish inhabitation and distribution of eight abundant species in relation with water quality in streams and rivers, Ulsan city. Korean J. Limnol. 41, 283–293 (2008).
Han, S., Park, J. & Kim, Y. Evolution modelling of the Yangsan-Ulsan fault system with stress changes. J. Geol. Soc. Korea 45, 361–377 (2009).
Kearse, M. et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. bioinformatics 23, 2947–2948 (2007).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Nei, M. Molecular evolutionary genetics. (Columbia university press, 1987).
Watterson, G. A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7, 256–276 (1975).
Librado, P. & Rozas, J. DnaSPv5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772 (2012).
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003).
Miller, M. A., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. in Gateway Computing Environments Workshop (GCE), 2010 1–8 (Ieee, 2010).
Akaike, H. A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19, 716–723 (1974).
Ronquist, F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Rambaut, A. Tracer v1. 4. http//beast. bio. ed. ac. uk/Tracer (2007).
Silvestro, D. & Michalak, I. raxmlGUI: a graphical front-end for RAxML. Org. Divers. Evol. 12, 335–337 (2012).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16, 111–120 (1980).
Excoffier, L. & Lischer, H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
Clement, M., Posada, D. & Crandall, K. A. TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1659 (2000).
Leigh, J. W. & Bryant, D. popart: full‐feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116 (2015).
Bouckaert, R. et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 (2014).
Tang, K. L. et al. Phylogeny of the gudgeons (Teleostei: Cyprinidae: Gobioninae). Mol. Phylogenet. Evol. 61, 103–24 (2011).
Zhou, J. J. The cyprinidae fossils from middle miocene of shanwang basin. Vertebr. Palasiat. 28, 95–127 (1990).
Rambaut, A. FigTreev1. 4. Mol. Evol. phylogenetics Epidemiol. Edinburgh, UK Univ. Edinburgh, Inst. Evol. Biol. (2012).
Nylander, J. A. A., Olsson, U., Alström, P. & Sanmartín, I. Accounting for phylogenetic uncertainty in biogeography: a Bayesian approach to dispersal-vicariance analysis of the thrushes (Aves: Turdus). Syst. Biol. 57, 257–268 (2008).
Yu, Y., Harris, A. J., Blair, C. & He, X. RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Mol. Phylogenet. Evol. 87, 46–49 (2015).
Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989).
Tajima, F. The effect of change in population size on DNA polymorphism. Genetics 123, 597–601 (1989).
Fu, Y. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925 (1997).
Rogers, A. R. & Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569 (1992).
Heled, J. & Drummond, A. J. Bayesian inference of population size history from multiple loci. BMC Evol. Biol. 8, 289 (2008).
Rogers, A. R. Genetic evidence for a Pleistocene population explosion. Evolution (N. Y). 49, 608–615 (1995).
Drummond, A. J., Suchard, M. A., Xie, D. & Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (2012).
Yu, D., Chen, M., Tang, Q., Li, X. & Liu, H. Geological events and Pliocene climate fluctuations explain the phylogeographical pattern of the cold water fish Rhynchocypris oxycephalus (Cypriniformes: Cyprinidae) in China. BMC Evol. Biol. 14, 225 (2014).
Acknowledgements
We thank Hari Won and Seul-Ki Park for providing experimental assistance. This research was supported by the grant “The Genetic Evaluation of Important Biological Resources (NIBR No. 2013–02–003)” funded by the National Institute of Biological Resources, Ministry of Environment and the grant from the National Research Foundation of Korea (Grant No. 2015R1D1A2A01058987).
Author information
Authors and Affiliations
Contributions
H.J. and H.Y.S. wrote the main manuscript text and prepared figures. J.A., D.K., Y.J.L. and H.B. collected the data. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jeon, HB., Kim, DY., Lee, Y.J. et al. The genetic structure of Squalidus multimaculatus revealing the historical pattern of serial colonization on the tip of East Asian continent. Sci Rep 8, 10629 (2018). https://doi.org/10.1038/s41598-018-28340-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-28340-x
- Springer Nature Limited
This article is cited by
-
Phylogeography of the Korean endemic Coreoleuciscus (Cypriniformes: Gobionidae): the genetic evidence of colonization through Eurasian continent to the Korean Peninsula during Late Plio-Pleistocene
Genes & Genomics (2022)
-
Evidence of an ancient connectivity and biogeodispersal of a bitterling species, Rhodeus notatus, across the Korean Peninsula
Scientific Reports (2020)
-
Molecular phylogenetic, population genetic and demographic studies of Nodularia douglasiae and Nodularia breviconcha based on CO1 and 16S rRNA
Scientific Reports (2020)