Abstract
Advances in culture-independent methods have meant that we can more readily detect and diagnose emerging infectious disease threats in humans and animals. Metagenomics is fast becoming a popular tool for detection and characterisation of novel bacterial pathogens in their environment, and is particularly useful for obligate intracellular bacteria such as Chlamydiae that require labour-intensive culturing. We have used this tool to investigate the microbial metagenomes of Chlamydia-positive cloaca and choana samples from snakes. The microbial complexity within these anatomical sites meant that despite previous detection of chlamydial 16S rRNA sequences by single-gene broad-range PCR, only a chlamydial plasmid could be detected in all samples, and a chlamydial chromosome in one sample. Comparative genomic analysis of the latter revealed it represented a novel taxon, Ca. Chlamydia corallus, with genetic differences in regards to purine and pyrimidine metabolism. Utilising statistical methods to relate plasmid phylogeny to the phylogeny of chromosomal sequences showed that the samples also contain additional novel strains of Ca. C. corallus and two putative novel species in the genus Chlamydia. This study highlights the value of metagenomics methods for rapid novel bacterial discovery and the insights it can provide into the biology of uncultivable intracellular bacteria such as Chlamydiae.
Similar content being viewed by others
Introduction
Recent advances in culture-independent molecular methods and diagnostics, coupled with an increased breadth and depth of sampling, have played a significant role in detecting emerging disease threats in humans and animals1,2,3,4. This method is advantageous over traditional single-gene pathogen detection methods in which nucleotide differences alone are, in some cases, not powerful enough to distinguish between closely related species5,6,7. Metagenomic sequencing also addresses challenges encountered in a clinical setting: a) if the putative pathogen is novel or unknown, b) if there are no established culture systems, or c) if culture is time-consuming and laborious (eg. for obligate intracellular pathogens such as Chlamydiae).
We recently used metagenomics to sequence and characterise the genome of a putative novel Candidatus species, Ca. Chlamydia sanzinia8, originating from a diverse group of chlamydial strains circulating among clinically healthy, captive snakes in Switzerland9. This species is closely related to Chlamydia pneumoniae and its genome encodes several chlamydial virulence markers such as a type three secretion system, translocated actin-recruiting phosphoprotein (Tarp) and chlamydial protease-like activity factor (CPAF)8. Elsewhere, chlamydiosis has been described in both wild and captive reptiles, including crocodiles, lizards and snakes, in broad geographical locations, with the impact of infection ranging from asymptomatic infections to severe disease9,10,11,12,13,14,15,16. Little or nothing is otherwise known about the the biological diversity of chlamydiae infecting these hosts.
Further, few studies have used metagenomics to describe the metagenome and microbiota of wild or non-model vertebrates, with most studies focussing on mammalian species and agriculturally important animals17, 18. Recently, groups have used culture-dependent and independent methods to characterise the microbiota of several anatomical sites in various reptile hosts, to uncover the diversity and function within these communities, and their potential impact on animal and human health19,20,21,22,23. We therefore aimed to assess the microbial diversity in Chlamydia-positive cloaca and choana samples from captive, asymptomatic snakes. In doing so, we also showed that metagenomics analysis is not only useful for novel chlamydial species discovery but that it also reveals key genomic differences between a novel chlamydial taxon and established species.
Results and Discussion
Snake choana and cloaca metagenome assembly and microbial composition
135,167,964 reads were obtained across two cloaca (G1/1679-8 and G2/2464-204) and three choana (G3/2742-324, G6/0661-435, and G7/2741-436) samples in which novel C. pneumoniae strains were detected by 16S rRNA sequencing. Reads were trimmed for quality and adapter sequence prior to de novo assembly and metagenome binning using SPAdes and MaxBin, respectively. 27,763–378,622 contigs were obtained for the samples (76–10,516 contigs over 1,000 bp) (Supplementary Information Table 1).
Metagenomic assessment revealed a high level of complexity in the samples, with such deep sequencing allowing us to simultaneously uncover the microflora of these sites, i.e. both putative novel bacteria and microbial eukaryotes residing in the choana and cloaca of these snakes. The cloaca samples harboured up to five bacterial species, and the choana samples up to two species based on recovery of full 16S rRNA sequences and partial or complete bacterial genomes (Table 1).
Many of the BLAST hits of microflora species were known members of reptilian, piscine or mammalian microbiomes, for example Achromobacter sp. and Luteimonas sp. in the respiratory tract21, 22 and Serratia marcescens, Pseudomonas aeruginosa and Salmonella enterica in the cloaca23. Interestingly, Chitinophageaceae appeared to dominate the choana samples, which has not been described before. This discrepancy is most likely due to the different detection methods used19,20,21,22,23. It is unclear what role these bacteria are playing at these sites: S. enterica has been repeatedly described as a reptile pathogen and such a high level of abundance may provide evidence for this (Table 1). As the name suggests, the Chitinophagaceae members in the choana samples (which are 98.5% identical to each other) are rich in chitinases. The role for these bacteria and their enzymes in the choana/oral cavity are unknown, but they may contribute to the digestion of the exoskeleton of animals ingested as part of the snake’s diet. Interestingly, previous studies of reptile microbiota did not detect any Chlamydia species19,20,21,22,23. This highlights a strength of metagenomics and strongly suggests that it may be a pathogen rather than a commensal, however, additional in vitro and in vivo studies are obviously necessary to confirm this. We also detected rRNA sequences from a flagellate, Monocercomonas coluborum, in a choana sample (G1/1679-8), and a fungal species related to Sporothrix schenckii in a cloaca sample (G7/2741-436). Further metagenomic sequencing would clarify the presence, abundance and roles of these and other species in the choana and cloaca of snakes.
Despite methylated DNA depletion prior to MDA and sequencing, a host mitochondrial genome was recovered from each sample. It has been shown that mitochondrial DNA may not be methylated in all species24, and this is reflected in our data by the presence of these sequences at differing levels of coverage in each sample, combined with fragmented mitochondrial genomes for two species. The mitochondrial genomes were obtained on a single contig or over up to six contigs, with the read coverage ranging from 43x to 38,621x, accounting for ~0.07% to ~31% of the reads (Supplementary Information Table 2).
Chlamydial genome construction from a snake choana metagenome
Given the fact that more than one bacterial genome was present in these metagenomes, a full chlamydial genome could only be recovered from a single sample (G3/2472-324), the characteristics of which are summarised in Table 2 in comparison with other chlamydial genomes. The single metagenome containing chlamydial chromosomal contigs contained 4,445 contigs over 1,000 bp, seven of which were chlamydial and divided between six predicted chromosomal contigs and one predicted plasmid contig. The combined chromosomal contigs total 1,196,452 bp in length and were predicted to encode 1,076 genes. The GC content of 39.33% was comparable to other chlamydial genomes (Table 2). The plasmid contig was 7,522 bp, harboured the typical eight open reading frames and has a lower GC content than the chromosome (32.0%), as is expected for plasmids. The average read coverage across the chromosome and plasmid were ~110x and ~40,134x, respectively.
For the remaining four samples, despite lacking a chlamydial chromosome, a plasmid sequence could be recovered, presumably since chlamydial plasmids are found at copy numbers of two to ten times that of the chromosome25, 26 and because MDA may preferentially amplify the plasmid DNA8. The plasmid sequences ranged from 7,210 to 7,534 bp, with 5-14,471x average read coverage (Table 3).
Phylogenomic analysis of Ca. Chlamydia corallus within the Chlamydiaceae
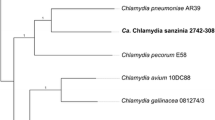
To assess the genetic relationship of Ca. Chlamydia corallus to other chlamydial species, we utilised the classification scheme published by Pillonel et al.5. Sequence homology within the 16S rRNA gene placed this novel taxon as a member of the order Chlamydiales, (99.2% identical to C. pneumoniae LPCoLN9). Sequence analysis of the additional genes show that this sample is closely related to C. pneumoniae but is sufficiently genetically different (ANI 90.16-90.31% with C. pneumoniae AR39 and LPCoLN) that, according to this scheme, G3/2742-324 should be classified as a novel species within the genus Chlamydia (Supplementary Information Table 3). To visualise the genetic relationships between this putative new chlamydial species with other members of the genus Chlamydia, a phylogenetic tree was constructed from a concatenation of the eleven gene alignments5. The resultant phylogenetic tree reiterates the distinct lineage formed by G3/2742-324, in a major cluster with Ca. Chlamydia sanzinia, C. pneumoniae and C. pecorum (Fig. 1).
Phylogenetic position of the novel taxon, Ca. Chlamydia corallus G3/2742-324 within the Chlamydiaceae. Phylogenetically informative marker genes were retrieved from each genome, concatenated and aligned using MAFFT, prior to tree construction using FastTree. Numbers on the branches indicate support values.
Based on the nucleotide identities of the analysed genes highlighted in the classification scheme5, G3/2742-324 should be classified as a novel Candidatus species within the genus Chlamydia. We propose for it the name Candidatus Chlamydia corallus (strain G3/2742-324), so named for the genus of the Amazon Basin emerald tree boa (Corallus batesii) in which it was detected. Ca. Chlamydia corallus was detected in the choana of a clinically healthy, captive snake in Switzerland.
Plasmid-based diversity within chlamydial species infecting snakes
Although culture-independent genome sequencing failed to resolve whole genome sequences for the all samples in this study, an extra-chromosomal plasmid was detected in all five metagenomes. An approximately 7.5 Kbp contig from G1/1679-8, G2/2464-204, G3/2742-324, G7/2741-436, and an approximately 7.2 Kbp contig from G6/0661-435, showed a BLASTn hit against the C. pneumoniae LPCoLN plasmid. The sequence homology between these sequences and C. pneumoniae LPCoLN plasmid was 78.76–86.09% and among each other was 77.45–96.8%.
Almost all chlamydial species, but not all strains, are known to carry a plasmid, and the nucleotide and amino acid sequences are highly conserved between species26. The presence of a plasmid has been suggested to contribute to the pathogenicity or tissue tropism of the chlamydial species27, 28, and plasmid proteins are used for diagnostic targets and vaccine candidates. The chlamydial plasmid is normally organised with eight open reading frames (ORFs), encoding for genes involved in plasmid maintenance and glycogen synthesis29. Alignment of the plasmid sequences revealed conservation of these ORFs, with the exception of a gap in the coding region for helicase in the sequence for G6/0661-435, which resulted in a partial plasmid sequence (data not shown).
Previous research has also shown that there is a co-evolution between the chromosome and plasmid sequences for the chlamydial species26, 30, so in the absence of chlamydial chromosomal genetic data for these additional samples, we performed phylogenetic analysis on the nucleotide sequences across the plasmid in order to assess the genetic relationship of all strains obtained from the metagenomes in this study. In agreement with the tree constructed from its chromosomal loci, phylogenetic analysis revealed that Ca. Chlamydia corallus clusters with but is genetically distinct from C. pneumoniae. (Fig. 2). Plasmid sequences from G6/0661-435 and G7/2741-436 can also be found in this clade. Notably, G1/1679-8 and G2/2464-204 form two additional branches distinct from Ca. C. corallus, C. pneumoniae and Ca. C. sanzinia, sharing 84.09% of their nucleotides with each other and 72.27% to 83.51% with these three species.
As no typing scheme exists to distinguish species based on plasmid sequence analysis or phylogeny, we used linear regression analysis to assess the relationships between chromosome and plasmid pair-wise patristic distances (sum of branch lengths) within the Chlamydiaceae (Supplementary Information Table 4 and Supplementary Information Fig. 1). Based on the phylogenetic markers used in this study, at the strain level (eg. C. pneumoniae LPCoLN and N16; C. pecorum MC/MarsBar and L1), patristic distances for the chromosome are 0, while for the plasmid they are 0.01–0.02. For closely related species pairs such as C. caviae and C. felis or C. suis and C. trachomatis, chromosomal and plasmid patristic distances are 0.12–0.16 and 0.20–0.22, respectively. For more distantly related species pairs such as C. trachomatis and C. psittaci, or C. pecorum and C. muridarum, chromosomal and plasmid patristic distances are higher: 0.28–0.33 and 0.52–0.57 (Supplementary Information Table 4). For the sequences obtained in this study, branch lengths between G6/0661–435 and G7/2741–436, and between these two samples and G3/2742–324, are equivalent to those at the strain level (0.0), as are their extrapolated chromosomal patristic distances based on the curve (Supplementary Information Table 4, Supplementary Information Fig. 1). On the other hand, the plasmid and extrapolated chromosomal patristic distances between G1/1679-8, G2/2464-204 and C. pneumoniae of 0.18–0.21 and 0.11–0.13, respectively, are slightly lower than those of C. caviae and C. felis, but not as close as that of C. psittaci and C. abortus, highlighting their relatedness to each other (Fig. 2). Meanwhile, their branch lengths with most other members of the genus is 0.36–0.41, which is comparable to most other pair-wise distances (Supplementary Information Table 4), thus may represent two distinct novel species.
These data also fit with initial testing results, in which G1/1679-8 and G2/2464-204 could not be definitively assigned to a species based on a Chlamydiaceae ArrayTube assay designed to detect established species8. G1/1679-8 was identified as a Chlamydia species, and G2/2464-204 did not yield a conclusive result (Table 3)9. G3/2742-324, G6/0661-435 and G7/2741-436 were designated as C. pneumoniae, which is in line with their close plasmid nucleotide identity. This suggests the assay is less specific when taxa are so closely related, but is robust enough to detect novel species.
Given the above, the phylogenetic position of G1/1679-8 and G2/2464-204 among other species and their evolutionary distances from other species and strains provide strong evidence of additional species-level diversity within the Chlamydiaceae. These data provide (a) evidence that, for some taxa, 16S rRNA sequencing is not sufficient to speciate, (b) validation of the use of genome sequencing to further investigate genetic diversity within and/or between populations, and (c) evidence for the use of plasmid sequence to assess diversity and phylogeny of novel chlamydial species for which plasmids are ubiquitous.
Genetic differences within the plasticity zone of Ca. Chlamydia corallus
In order to further characterise the genome of the novel species, Ca. Chlamydia corallus, in comparison to other chlamydial species, the plasticity zone (PZ) region was analysed. The plasticity zone is a unique region within the Chlamydia genome that has been associated with host adaptation for some chlamydial species6, 31. The well-known variability between the chlamydial species within this region makes it an appropriate target for understanding the factors that might have influenced the tissue tropism of Ca. Chlamydia corallus.
The plasticity zone of Ca. Chlamydia corallus is approximately 13,700 bp in size and composed of genes required for several biochemical pathways such as Acetyl-CoA-carboxylase (accBC), purine and pyrimidine synthesis genes (guaABadd) and the MAC/perforin gene, as seen in Fig. 3. When compared with other chlamydial species, the plasticity zone harboured by Ca. Chlamydia corallus is structurally most similar to the human-isolated strain of C. pneumoniae AR39 (Fig. 3). Both species have a slightly smaller plasticity zone than other species, due to the absence of any cytotoxin, which is present in C. psittaci and duplicated in C. pecorum 32, 33. The main difference between the PZs of Ca. C. corallus and C. pneumoniae appears to be fragmented or truncated hypothetical proteins in AR39, and the absence of a putative lipoprotein in Ca. Chlamydia corallus, which is present in both strains of C. pneumoniae (Fig. 3)34.
Comparison of plasticity zone regions encoded by Ca. Chlamydia corallus and related chlamydial species. Figure constructed using Easy Fig54. Grey shading represents tBLASTx matches (see BLAST identity scale). Coloured arrows represent coding regions (see legend).
Compared to the other known chlamydial species infecting snakes, the plasticity zone of Ca. Chlamydia corallus was genetically variable from Ca. Chlamydia sanzinia (Fig. 3). For instance, the MAC/perforin complex gene was not detected in the plasticity zone of Ca. Chlamydia sanzinia8. The function of the MAC/perforin gene in the chlamydial species is unknown, but has been suggested to contribute to the pathogenesis of these species35. Additional differences in the PZs of Ca. Chlamydia sanzinia and Ca. Chlamydia corallus lie in the purine ribonucleotide biosynthesis pathways, as highlighted in Fig. 3. The purine ribonucleotide biosynthesis (guaABadd) cluster, detected in Ca. Chlamydia corallus, plays a critical role in both de novo and salvage pathways for purine synthesis in prokaryotes36. This cluster is present in some chlamydial species33, 34, 37, but was never detected in the other recently described snake chlamydia, Ca. Chlamydia sanzinia8. Chlamydial species that do not encode for this gene cluster are most likely able to synthesise purines through alternative pathways38. Its absence in other bacterial species, such as Helicobacter pylori, has been found to have an effect on their rate of growth36. The absence of the guaABadd genes in several of the chlamydial species, however, indicates that these genes are not needed by the chlamydial species; its absence in Ca. Chlamydia sanzinia also suggests that these genes are not necessary for the chlamydial species to establish infection in snakes8, 37.
No tryptophan operon (trpAB) was detected in the plasticity zone or other genomic regions of Ca. C. corallus. Tryptophan is a necessary amino acid for chlamydial growth39, however, host cell defence mechanisms against chlamydial infections exist in which interferon gamma (IFN-γ) production depletes intracellular tryptophan stores40. Certain strains of C. trachomatis encode for an intact tryptophan operon (trpAB), which is absent or incomplete in other chlamydial species37, suggesting that not all chlamydial species are able to synthesise tryptophan. For urogenital strains of C. trachomatis, the vaginal microflora is believed to provide indole, allowing for synthesis of tryptophan39. The absence of a tryptophan operon would suggest that Ca. C. corallus either has alternative pathways for synthesising tryptophan or is possibly completely auxotrophic for tryptophan. Notably, neither Ca. C. corallus nor Ca. C. sanzinia encode for an aromatic amino acid hydroxylase, which has been suggested to contribute to tryptophan metabolism in the absence of trpAB. As has been suggested for C. trachomatis 39, the diverse microflora in these snakes may provide nutrients or substrates for chlamydial synthesis of amino acids. Metagenomic mining revealed several tryptophan synthesis pathway or rescue genes encoded by the bacteria present in the cloaca and choana samples, for example, tryptophan synthase, tryptophanase and indole-3-glycerol phosphatase were detected among the samples, encoded by Achromobacter sp., Serratia marcescens. Clostridium sp., Salmonella enterica and Chitinophagaceae. Previous studies also describe the presence of indole-producing bacteria at these sites21,22,23.
In the current study, we have used culture-independent metagenome analysis to investigate the microbial metagenome of snake choana and cloaca samples. In doing so, we have shown that this method provides a wealth of biological information for novel species discovery through microbial community profiling, and have described the presence of highly abundant bacterial species at these sites, some of which have not previously been described. The animal and public health implications of these findings are unknown, but the repeated observations of human pathogens in the microflora of snakes21, 22 and other reptiles warrants further investigation.
The metagenomic method used is particularly useful for characterising novel species or strains with no reference genome such as novel uncultivable bacteria (eg. members of the phylum Chlamydiae). The complexity within these anatomical sites meant that despite previous detection of chlamydial 16S rRNA sequences by PCR, only a chlamydial plasmid could be detected in all samples, and a single chlamydial chromosome. Nonetheless, comparative analysis of the novel chlamydial species with other Chlamydia sp. revealed genetic differences in regards to purine and pyrimidine metabolism. The detection of chlamydial plasmids in all samples, which was only possible using this method, highlights additional diversity within the Chlamydiae, which appears to be a growing trend with increased breadth and depth of sampling and advances in molecular techniques. Further studies, such as metatranscriptomic analysis would better elucidate the complex role of the microbiota on chlamydial pathogenesis and vice versa.
Materials and Methods
Sample preparation
Suspected novel genotypes (n = 5) of C. pneumoniae were recently detected in collections of captive snakes in Switzerland8. Clinical swabs were taken from either the cloana or choana of clinically healthy snakes, and DNA was extracted as previously described8. All samples were subjected to host methylated DNA depletion prior to multiple displacement amplification, as previously described9.
Ethics approval and consent to participate
The collection and molecular analysis of the snake samples was approved and performed in accordance with the relevant guidelines and regulations of the Veterinary Office of Canton Zurich (authorization no. ZH010/15).
Metagenome assembly and analysis
Deep sequencing was carried out on an Illumina NextSeq at the Australian Genome Research Facility using 150 bp paired-end reads. Read quality was assessed through FastQC v.011.2 and reads were trimmed for adaptors and quality using Trimmomatic v.30541. Reads were assembled into contigs using SPAdes v.3.1.1 in metagenome mode with default kmer values (21, 33, 55)42. Each assembly was assessed through QUAST43. To obtain chlamydial contigs from the assembled metagenome, contigs were subject to BLAST analysis against an in-house chlamydial genome database, and subsequent analysis against the NCBI nucleotide database. Contigs with hits against chlamydial sequences were automatically annotated using RAST44 and manually curated in Artemis45.
Metaxa was employed initially to assess the species richness within the resulting metagenomes, detecting ribosomal RNA subunits of various origins46. MaxBin was used to construct partial or complete draft genomes for the microbial species detected in the samples and determine genome completeness for each assembly47.
Burrows-Wheeler aligner, SAMtools and BEDtools were used to map reads and assess read coverage across the various metagenomic components48,49,50.
Phylogenetic analysis
The genetic relationships of the novel species described in this study to other chlamydial species was assessed using the classification system published by Pillonel et al.5. Individual genes were extracted from the assembled genome and established chlamydial species, including Simkania negevensis as an out group. Extracted genes were concatenated and aligned using MAFFT51 and a phylogenetic tree based on the resulting alignment was constructed using FastTree52; both were performed in Geneious v7.153.
Plasmid phylogeny was performed based on the alignment of nucleotide sequences using MAFFT51 and tree construction using FastTree52. In order to include plasmid sequences from all possible species, nucleotide sequences were re-ordered and large gaps were removed so that each resulting plasmid sequence was 5,522–6,170 bp, thus comparable to C. suis plasmid which lacks the parA and pgp-6 genes.
Data availability
The metagenomic sequence data obtained for the Ca. Chlamydia corallus chromosome and plasmid was deposited in Genbank under accession numbers as part of bioproject PRJNA312988.
References
Miller, R. R., Montoya, V., Gardy, J. L., Patrick, D. M. & Tang, P. Metagenomics for pathogen detection in public health. Genome Med 5(81), gm485, doi:10.1186/gm485 (2013).
Lu, X. et al. Bacterial pathogens and community composition in advanced sewage treatment systems revealed by metagenomics analysis based on high-throughput sequencing. PLoS One 10(0125549), 0125549, doi:10.1371/journal.pone.0125549 (2015).
Nieuwenhuijse, D. F. & Koopmans, M. P. Metagenomic Sequencing for Surveillance of Food- and Waterborne Viral Diseases. Front Microbiol 8, 230, doi:10.3389/fmicb.2017.00230 (2017).
Taylor-Brown, A. & Polkinghorne, A. New and emerging chlamydial infections of creatures great and small, in press. New Microbes New Infect, doi:10.1016/j.nmni.2017.04.004 (2017).
Pillonel, T., Bertelli, C., Salamin, N. & Greub, G. Taxogenomics of the order Chlamydiales. Int J Syst Evol Microbiol 65, 1381–1393 (2015).
Bachmann, N. L., Polkinghorne, A. & Timms, P. Chlamydia genomics: providing novel insights into chlamydial biology. Trends Microbiol 22, 464–472 (2014).
Rajendhran, J. & Gunasekaran, P. Microbial phylogeny and diversity: small subunit ribosomal RNA sequence analysis and beyond. Microbiol Res 166, 99–110 (2011).
Taylor-Brown, A., Bachmann, N. L., Borel, N. & Polkinghorne, A. Culture-independent genomic characterisation of Candidatus Chlamydia sanzinia, a novel uncultivated bacterium infecting snakes. BMC Genomics 17, 710, doi:10.1186/s12864-016-3055-x (2016).
Taylor-Brown, A., Rüegg, S., Polkinghorne, A. & Borel, N. Characterisation of Chlamydia pneumoniae and other novel chlamydial infections in captive snakes. Vet Microbiol 178, 88–93 (2015).
Bodetti, T. J. et al. Molecular Evidence to Support the Expansion of the Hostrange of Chlamydophila pneumoniae to Include Reptiles as Well as Humans, Horses, Koalas and Amphibians. Syst Appl Microbiol 25, 146–152 (2002).
Jacobson, E. R., Heard, D. & Andersen, A. Identification of Chlamydophila pneumoniae in an Emerald Tree Boa, Corallus caninus. J Vet Diagn Invest 16, 153–154 (2004).
Soldati, G. et al. Detection of mycobacteria and chlamydiae in granulomatous inflammation of reptiles: a retrospective study. Vet Pathol 41, 388–397 (2004).
Huchzermeyer, F. W., Langelet, E. & Putterill, J. F. An outbreak of chlamydiosis in farmed Indopacific crocodiles (Crocodylus porosus). J S Afr Vet Assoc 79, 99–100 (2008).
Frutos, M. C., Monetti, M. S., Re, V. E. & Cuffini, C. G. Molecular evidence of Chlamydophila pneumoniae infection in reptiles in Argentina. Rev Argent Microbiol 46, 45–48 (2014).
Rüegg, S. R., Regenscheit, N., Origgi, F. C., Kaiser, C. & Borel, N. Detection of Chlamydia pneumoniae in a collection of captive snakes and response to treatment with marbofloxacin. Vet J 205, 424–426 (2015).
Shilton, C. M. et al. Diagnostic investigation of new disease syndromes in farmed Australian saltwater crocodiles (Crocodylus porosus) reveals associations with herpesviral infection. J Vet Diagn Invest 28, 279–290 (2016).
Hanning, I. & Diaz-Sanchez, S. The functionality of the gastrointestinal microbiome in non-human animals. Microbiome 3, 51, doi:10.1186/s40168-015-0113-6 (2015).
Yoon, S. S., Kim, E. K. & Lee, W. J. Functional genomic and metagenomic approaches to understanding gut microbiota-animal mutualism. Curr Opin Microbiol 24, 38–46 (2015).
Costello, E. K., Gordon, J. I., Secor, S. M. & Knight, R. Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J 4, 1375–1385 (2010).
Keenan, S. W., Engel, A. S. & Elsey, R. M. The alligator gut microbiome and implications for archosaur symbioses. Sci Rep 3, 2877, doi:10.1038/srep02877 (2013).
Schmidt, V. et al. Detection of pathogens in Boidae and Pythonidae with and without respiratory disease. Vet Rec 172, 236, doi:10.1136/vr.100972 (2013).
Plenz, B., Schmidt, V., Grosse-Herrenthey, A., Kruger, M. & Pees, M. Characterisation of the aerobic bacterial flora of boid snakes: application of MALDI-TOF mass spectrometry. Vet Rec 176, 285, doi:10.1136/vr.102580 (2015).
McLaughlin, R. W., Cochran, P. A. & Dowd, S. E. Metagenomic analysis of the gut microbiota of the Timber Rattlesnake. Crotalus horridus. Mol Biol Rep 42, 1187–1195 (2015).
Hong, E. E., Okitsu, C. Y., Smith, A. D. & Hsieh, C. L. Regionally specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Mol Cell Biol 33, 2683–2690 (2013).
Pickett, M. A., Everson, J. S., Pead, P. J. & Clarke, I. N. The plasmids of Chlamydia trachomatis and Chlamydophila pneumoniae (N16): accurate determination of copy number and the paradoxical effect of plasmid-curing agents. Microbiology 151, 893–903 (2005).
Jelocnik, M. et al. Molecular characterisation of the Chlamydia pecorum plasmid from porcine, ovine, bovine, and koala strains indicates plasmid-strain co-evolution. PeerJ 4, e1661, doi:10.7717/peerj.1661 (2016).
Zhong, G. Chlamydial Plasmid-Dependent Pathogenicity. Trends Microbiol 25, 141–152 (2016).
Chen, J. et al. Intrauterine infection with plasmid-free Chlamydia muridarum reveals a critical role of the plasmid in chlamydial ascension and establishes a model for evaluating plasmid-independent pathogenicity. Infect Immun 83, 2583–2592 (2015).
Song, L. et al. Chlamydia trachomatis Plasmid-Encoded Pgp4 Is a Transcriptional Regulator of Virulence-Associated Genes. Infect Immun 81, 636–644 (2013).
Seth-Smith, H. M. et al. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 10, 239, doi:10.1186/1471-2164-10-239 (2009).
Roulis, E. et al. Comparative genomic analysis of human Chlamydia pneumoniae isolates from respiratory, brain and cardiac tissues. Genomics 106, 373–383 (2015).
Jelocnik, M. et al. Genetic diversity in the plasticity zone and the presence of the chlamydial plasmid differentiates Chlamydia pecorum strains from pigs, sheep, cattle, and koalas. BMC Genomics 16, 893, doi:10.1186/s12864-015-2053-8 (2015).
Voigt, A., Schöfl, G. & Saluz, H. P. The Chlamydia psittaci Genome: A Comparative Analysis of Intracellular Pathogens. PLoS ONE 7, e35097, doi:10.1371/journal.pone.0035097 (2012).
Read, T. D. et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res 28, 397–1406 (2000).
Taylor, L. D., Nelson, D. E., Dorward, D. W., Whitmire, W. M. & Caldwell, H. D. Biological Characterization of Chlamydia trachomatis Plasticity Zone MACPF Domain Family Protein CT153. Infect Immun 78, 2691–2699 (2010).
Liechti, G. & Goldberg, J. B. Helicobacter pylori Relies Primarily on the Purine Salvage Pathway for Purine Nucleotide Biosynthesis. J Bacteriol 194, 839–854 (2012).
Mitchell, C. M. et al. Comparison of koala LPCoLN and human strains of Chlamydia pneumoniae highlights extended genetic diversity in the species. BMC Genomics 11, 1–10 (2010).
Gil, R., Silva, F. J., Peretó, J. & Moya, A. Determination of the Core of a Minimal Bacterial Gene Set. Microbiol Mol Biol Rev 68, 518–537 (2004).
Ziklo, N., Huston, W. M., Hocking, J. S. & Timms, P. Chlamydia trachomatis Genital Tract Infections: When Host Immune Response and the Microbiome Collide. Trends Microbiol 24, 750–765 (2016).
Akers, J. C. & Tan, M. Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis. J Bacteriol 188, 4236–4243 (2006).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19, 455–477 (2012).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Aziz, R. K. et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75, doi:10.1186/1471-2164-9-75 (2008).
Rutherford, K. et al. Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945 (2000).
Bengtsson J. et al. Metaxa: a software tool for automated detection and discrimination among ribosomal small subunit (12S/16S/18S) sequences of archaea, bacteria, eukaryotes, mitochondria, and chloroplasts in metagenomes and environmental sequencing datasets. Antonie van Leeuwenhoek 100 (2011).
Wu, Y.-W., Tang, Y.-H., Tringe, S. G., Simmons, B. A. & Singer, S. W. MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2, 1–18 (2014).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Katoh, K., Misawa, K., Kuma, K. Ä. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30, 3059–3066 (2002).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Genome Biol Evol 26, 1641–1650 (2009).
Kearse, M. et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012).
Sullivan, M. J., Petty, N. K. & Beatson, S. A. Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010 (2011).
Acknowledgements
We thank Simon Rüegg for sample collection and Carmen Kaiser for DNA extraction and initial PCR screening. We thank Helena Seth-Smith for her advice on the preparation of our samples for culture-independent genome sequencing as well as downstream analysis. This work was funded by a University of the Sunshine Coast Faculty Research Grant.
Author information
Authors and Affiliations
Contributions
A.T.B. managed the project, conducted the laboratory experiments and bioinformatics analysis, and prepared the manuscript; L.S. conducted bioinformatics analysis, prepared the figures and manuscript; N.B. provided samples and reviewed the manuscript; A.P. managed the project and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Taylor-Brown, A., Spang, L., Borel, N. et al. Culture-independent metagenomics supports discovery of uncultivable bacteria within the genus Chlamydia . Sci Rep 7, 10661 (2017). https://doi.org/10.1038/s41598-017-10757-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-10757-5
- Springer Nature Limited
This article is cited by
-
A current perspective on polycyclic aromatic hydrocarbons contamination and their bioremediation aspects
Environmental Earth Sciences (2024)
-
Gene gain facilitated endosymbiotic evolution of Chlamydiae
Nature Microbiology (2023)
-
An in silico analysis of rpoB mutations to affect Chlamydia trachomatis sensitivity to rifamycin
Journal of Genetic Engineering and Biotechnology (2022)
-
Genomic diversity and biosynthetic capabilities of sponge-associated chlamydiae
The ISME Journal (2022)
-
Whole genome de novo sequencing and comparative genomic analyses suggests that Chlamydia psittaci strain 84/2334 should be reclassified as Chlamydia abortus species
BMC Genomics (2021)