Abstract
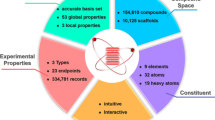
We here introduce the Aquamarine (AQM) dataset, an extensive quantum-mechanical (QM) dataset that contains the structural and electronic information of 59,783 low-and high-energy conformers of 1,653 molecules with a total number of atoms ranging from 2 to 92 (mean: 50.9), and containing up to 54 (mean: 28.2) non-hydrogen atoms. To gain insights into the solvent effects as well as collective dispersion interactions for drug-like molecules, we have performed QM calculations supplemented with a treatment of many-body dispersion (MBD) interactions of structures and properties in the gas phase and implicit water. Thus, AQM contains over 40 global and local physicochemical properties (including ground-state and response properties) per conformer computed at the tightly converged PBE0+MBD level of theory for gas-phase molecules, whereas PBE0+MBD with the modified Poisson-Boltzmann (MPB) model of water was used for solvated molecules. By addressing both molecule-solvent and dispersion interactions, AQM dataset can serve as a challenging benchmark for state-of-the-art machine learning methods for property modeling and de novo generation of large (solvated) molecules with pharmaceutical and biological relevance.
Similar content being viewed by others
Background & Summary
Introduction
In pharmaceutical research and development, computational chemistry can play an integral role in expediting candidate drugs into the clinic. Particularly, quantum-mechanical (QM) methods (e.g., density-functional theory (DFT), post-Hartree-Fock approaches, and quantum Monte Carlo) have been utilized to describe covalent and non-covalent interatomic interactions and to estimate diverse physicochemical properties of molecular systems1,2. QM methods can for instance be used to understand the reactivity of covalent binders3,4, evaluate conformational energy landscapes of ligands5, study the stability of potential active pharmaceutical ingredients, calculate theoretical acidity constants6, or calculating theoretical charges to more accurately capture electrostatic properties and surfaces. However, the computational cost and the challenge of conducting QM calculations at a large scale present a limitation to their widespread use in drug discovery pipelines. Scanning conformational landscapes through QM calculations performed at DFT levels of theory typically takes several hours for a single ligand of typical pharmaceutical size (e.g., 30-40 heavy atoms) on a single computer. As these calculations are readily parallelizable, supercomputers can be employed to enhance throughput enabling the screening of a few hundred compounds, but it remains challenging to perform these calculations at the scale of virtual libraries which can easily be composed of tens of thousands of compounds.
Accelerated QM methods have emerged as promising solutions in recent years, offering a balance between accuracy and computational efficiency. These can take the form of quantum fragmentation methods7,8, semi-empirical methods (e.g., parametric method series9, density functional tight-binding (DFTB)10,11 or its extended version (GFNn-xTB)12,13) as well as machine learning (ML) models14,15,16,17,18,19,20 capable of optimizing geometries or estimating physicochemical properties. The resulting acceleration enables researchers to include QM-based knowledge as a part of their workflow. Accordingly, relevant QM datasets of small organic molecules have widely assisted the development of ML-based approaches for a fast and accurate estimation of structural, vibrational, and electronic properties of complex organic molecules21,22,23,24. Among them, one can find QM725,26,27, QM928,29, QM7-X30, MD1715,31, MD2232, ANI-114,33, ANI-1x/ANI-1ccx34 and AIMNet-NSE35. While these QM datasets have significantly advanced the field of computational chemistry, they do exhibit certain limitations that stem from three important facts. First, they primarily consist of molecules that are considerably smaller than what is commonly encountered in modern medicinal chemistry. Second, their structures have been optimized using theoretical models that do not account for molecule-solvent and collective dispersion interactions. Lastly, they have not fully explored the vast conformational landscape inherent in these molecules. Especially, the interaction between the molecule and the chemical environment (i.e., solvent) is crucial when investigating molecules of pharmaceutically relevant size, as it is well-known that drug binding does not occur in vacuo but rather in solutio36. Indeed, there is extensive literature discussing the solvent effects on the properties of specific molecular systems37,38,39,40,41. For instance, Gorges et al.38 found that solvation can have a substantial effect of several cal/mol ⋅ K on the entropy of 25 commercially available drug molecules as a result of large conformational changes. The geometry, energetics, HOMO/LUMO energies, dipole moment, and polarizability of formaldehyde and thioformaldehyde have also been reported to change upon solvation in solvents of low polarity39. Similarly, the molecule-solvent interaction can affect the dynamic stability and antiviral inhibitory potential of Cissampeline40 as well as the chemical reaction type SN2 between Cl− and CH3Cl41. Omitting solvent effects can thus result in inappropriate treatment of conformations, tautomers, physicochemical properties, or molecular reactivity. In the computational modeling of molecular systems, solvents can either be considered implicitly or represented explicitly. However, owing to the computational cost and intricate nature of explicitly representing the solvent, most QM studies opt for the utilization of implicit solvent models such as conductor-like screening model for real solvents (COSMO-RS)42, modified Poisson-Boltzmann (MPB)43, and Generalized Born (GB)44 model augmented with the hydrophobic solvent accessible surface area term (GBSA)45.
Lately, to overcome the molecular size limitation within benchmark QM datasets, several efforts have been made to generate datasets that comprehensively explore the conformational space of large and flexible molecules along with their associated QM properties calculated in gas phase or solvent, see Table 1. For instance, the QMugs46 collection comprises 19 QM properties of circa 2 M gas-phase conformers of 665, 911 molecules with up to 100 non-hydrogen atoms computed using ωB97X-D47 density functional and the def2-SVP basis set. The OE6248 dataset covers 61, 489 molecules with up to 92 non-hydrogen atoms that were optimized in gas phase using PBE(tight) level of theory supplemented with Tkatchenko-Scheffler van der Waals (TS) interaction49. This dataset also contains 3 QM properties for 30, 876 structures evaluated using PBE0(tight) level of theory together with implicit water defined by the Multipole Expansion (MPE) model50. Regarding the vast GEOM collection (which stands for Geometric Ensemble Of Molecules)51, only 1.3 M conformers corresponding to 1, 511 BACE52 molecules were generated considering molecule-solvent interactions described by the analytical linearized Poisson-Boltzmann (ALPB)53 model of water. From here, 455, 000 conformers of 534 BACE molecules were selected and used for further geometry optimization calculations using r2scan-3c functional with C-PCM54 (which stands for conductor-like polarizable continuum model) implicit model of water and, posteriorly, 6 QM properties were collected. Moreover, Eastman et al.55 have recently introduced the SPICE dataset (which is short for Small-molecule/Protein Interaction Chemical Energies) that explicitly considers the interaction between the molecule and water molecules via Amber14 classical force field to get 1, 300 (equilibrium and non-equilibrium) structures of 26 amino acids. Energies, atomic forces, and other 6 QM properties were computed using the ωB97M-D3(BJ) functional and def2-TZVPPD basis set. Despite these efforts, challenges remain to enable a better understanding of solvent effects as well as collective dispersion interactions in the chemical space of large drug-like molecules, including: (i) assessing the accuracy and reliability of QM structures and properties with respect to the employed density-functional approximation, especially for larger and more flexible molecules in which van der Waals (vdW) and molecule-solvent interactions are stronger, (ii) offering a large set of molecular (global) and atom-in-a-molecule (local) physicochemical properties that would enable a comprehensive exploration of these interactions in structure-property and property-property relationships throughout chemical space, and (iii) providing accurate and reliable QM data that will enable the construction of models for describing covalent and non-covalent vdW interactions in large (solvated) molecules.
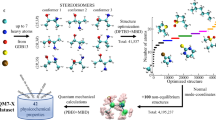
In this work, we introduce the Aquamarine (AQM) dataset with the aim of addressing these challenges. The current version of AQM contains an extensive conformational sampling of 1, 653 molecules with up to 54 (mean: 28.2) non-hydrogen atoms (including C, N, O, F, P, S, and Cl), producing a total of 59, 783 low-and high-energy conformers with a total number of atoms N ranging from 2 until 92 (mean: 50.9), see Fig. 1. In doing so, QM conformers were generated using the conformational search workflow implemented in CREST code56 (which is short for Conformer-Rotamer Ensemble Sampling Tool) that considers semi-empirical GFN2-xTB13 with GBSA implicit solvent model of water45. Since vdW interactions have a significant impact on the conformations of large molecules, we have optimized a set of representative conformers using third-order DFTB method10,57,58 (or DFTB3) supplemented with a treatment of many-body dispersion (MBD) interactions59,60,61,62 (see “Methods”). Moreover, to have a better understanding of solvent effects, we have performed these calculations in gas phase and in implicit water described by the GBSA model. For each of the (gas-phase and solvated) optimized conformers, AQM also includes an extensive number (over 40) of global (molecular) and local (atom-in-a-molecule) QM properties computed at a high level of theory that depends on the chemical environment used during the geometry optimization. The majority of QM properties for gas-phase structures were evaluated using non-empirical hybrid DFT with MBD interactions (i.e., PBE0+MBD) in conjunction with tightly-converged numeric atom-centered orbitals63. In addition, MPB implicit solvent model of water was considered to obtain the properties for solvated structures. Hence, we have two different AQM subsets, namely AQM-gas and AQM-sol, which contain the QM structural and property data of molecules in gas phase and implicit water, respectively. Based on its design, AQM holds the potential to enhance the comprehension of the influence of molecule-solvent and collective dispersion interactions in structure-property and property-property relationships of molecules of pharmaceutically relevant size and composition.
Scheme of the generation procedure of Aquamarine (AQM) dataset. We have here investigated molecules containing C, N, O, H, Cl, S, P, and F atoms. Then, we generated the conformers per molecule using the conformational search workflow implemented in CREST code which employs a metadynamics sampling with an additional genetic z-matrix crossing step. A set of representative conformers is selected using an energetic analysis considering the DFTB total energy Etot and many-body dispersion (MBD) energy EMBD, see “Methods”. Thus, the current version of AQM dataset contains 59, 783 low-and high-energy conformers of 1, 653 molecules up to 54 non-hydrogen atoms (mean: 28.2) with a total number of atoms ranging from 2 to 92 (mean: 50.9). All structures in the final set of conformers are optimized using DFTB3+MBD and DFTB3+MBD supplemented with the GBSA implicit solvent model of water. Afterward, over 40 molecular (global) and atom-in-a-molecule (local) QM properties of gas-phase and solvated structures are computed by performing single point calculations at the PBE0(tight)+MBD level of theory and further enhanced with the MPB implicit solvent model of water, respectively. Hence, we have two different AQM subsets, namely AQM-gas and AQM-sol.
Key advancements
Our main motivation for proposing AQM as a benchmark dataset is to advance in the development of the next generation of ML models that enable a fast and accurate property estimate and ideation of drug-like molecules synthesized in a chemical environment. In pursuit of this aim, we have ensured that the AQM dataset exhibits the following characteristics,
-
The idea of combining the CREST conformational search workflow with the subsequent DFTB3+MBD geometry optimization, both in gas phase and implicit water, has provided us with access to a more extensive and reliable exploration of low- and high-energy (compact/extended) conformers of large molecules. To the best of our knowledge, this procedure has not been considered in previous works. Also, notice that MBD interaction is a key factor in accurately describing and identifying diverse conformations in large molecular complexes, due to their anisotropic shapes.
-
The AQM dataset provides a more accurate set of over 40 global (molecular) and local (atom-in-a-molecule) QM properties of gas-phase and solvated structures when compared to already public datasets (see Table 1). These properties can assist in the estimation and comprehension of the impact of molecule-solvent interactions in the structure-property and property-property relationships of large molecules, e.g., via delta learning approach.
-
The property data stored in AQM-gas and AQM-sol can potentially be used to develop more robust QM descriptors for large molecules, enabling fast and accurate calculations of their physicochemical or biological properties. Moreover, such quantum property-based molecular descriptors are complementary to the widely used geometric descriptors and both can be used in synergy for estimating molecular observables measured in experiment.
-
The gas-phase and solvated conformations of AQM molecules, along with their highly accurate QM properties make the AQM dataset a valuable resource for in silico assisted methods.
Methods
Selection of representative chemistries
We sought to select a set of compounds from the public domain that approximate a typical corporate library including H, C, N, O, F, P, Cl, and S atoms. To this end, we sampled 5000 compounds from ChEMBL64 and compared them to the Johnson & Johnson Innovative Medicines corporate database. Compounds with molecular weights over 1200, more than 30 rotatable bonds, a quantitative estimate of drug-likeness (QED) score65 under 0.4, and heavy atom count over 200 were removed. Then, we got a reduced ChEMBL set of compounds with similar molecular weights, numbers of rotatable bonds, and fraction of sp3 to the corporate database (see Fig. S1 of the Supplementary Information (SI)). This subset was subjected to diversity selection, followed by manual inspection to remove molecules containing undesirable or unusual chemical substructures. As a result, we initially selected SMILES (which stands for Simplified Molecular Input Line Entry System) of molecular building blocks and typical lead-like compounds as well as a few protein degraders and macrocycles to produce a total of 2, 635 unique molecules with up to 60 non-hydrogen atoms (N ≤ 116). To have a more extensive sampling of the chemical space described by the selected SMILES, we generated all possible stereoisomers for each structure using the RDKit code—an open source toolkit developed for cheminformatics66,67. In our script, we generate unique stereoisomers with the option to perturb each stereocenter while keeping the same atomic connectivity (i.e., tautomers are not considered), yielding a larger number of isomers whose stability is later checked via quantum mechanical (QM) calculations. Accordingly, the new number of molecular structures considering the stereoisomers is circa 10 k. Initial 3D structures were subsequently generated with RDKit and optimized using the MMFF94 force field68,69,70,71,72.
Generation of molecular conformers
Conformational sampling plays a crucial role in the generation of AQM dataset. We have meticulously explored different conformational search workflows to identify the most effective approach for comprehensively sampling the potential energy surface (PES) and the molecular property space of large drug-like molecules (see “Technical Validation” and Sec. 2 of the SI). In doing so, we opted to use the approach implemented in CREST56 code which uses extensive sampling based on the much faster and yet reliable semi-empirical extended tight-binding method (GFN2-xTB)12,13 to generate 3D conformations. The semi-empirical energies and structures are thought to be more accurate than classical force fields, accounting for electronic effects, rare functional groups, and bond-breaking/formation of labile bonds13,46,51,73. Moreover, the CREST search algorithm is based on metadynamics (MTD), a well-established thermodynamic sampling approach that can efficiently explore the low-energy search space. The collective variables used for the MTD sampling are the atomic root-mean-squared deviation (RMSD) values between the previous structures on the PES of a given molecule56. The atomic RMSD values are introduced into the expression of the bias potential, which is used to compute the guiding forces. These forces are responsible for driving the structure further away from previous geometries, providing an extensive exploration of PES. Conformers are thus generated in an iterative manner of MTD and GFN2-xTB optimization, where those geometries are added to the conformer rotamer ensemble (CRE) that overcome certain energy ( > 12.0 kcal/mol) and root-mean-square deviation (ΔR > 0.1 Å) thresholds concerning the input structure. The procedure is restarted using the conformer as input if a new conformer has a lower energy than the input structure. The three conformers of lowest energy undergo two normal molecular dynamics (MD) simulations at 400K and 500K, which are used to sample low-energy barrier crossings, such as simple torsional motions. Finally, a genetic Z-matrix crossing algorithm is used and the results are added to the CRE. Then, a normal-type convergence optimization separates the geometries into conformers, rotamers, and duplicates, where duplicates are deleted and conformers and rotamers added to the CRE. Both geometry optimization and conformational search calculations were carried out considering implicit water described by the Generalized Born (GB) model augmented with the hydrophobic solvent accessible surface area term (GBSA)45. Finally, we obtained 2, 242, 490 conformers for the initial set of 2, 635 molecules.
Unlike other already public datasets of conformers of large molecules (see Table 1), we have here defined a method to select a set of representative conformers per molecule (i.e., per SMILE) instead of considering all conformers generated by CREST. The purpose of using this method is to filter out conformers that are similar in regions of the chemical space defined by the atomic structure, total energy Etot and many-body dispersion (MBD) energy EMBD. We have here considered EMBD due to its relevance in the definition of stability rankings in large molecules and molecular crystals59,60,61,62. Accordingly, our initial step involves determining clusters that consist of conformers exhibiting a root-mean-square deviation (ΔR) between their structures of less than 1.5 Å. ΔR is computed with the help of DockRMSD tool74. After clustering the conformers, we obtained Etot and EMBD of all conformers per cluster via single-point calculations using third-order self-consistent charge density functional tight binding (DFTB3)10,57,58 supplemented with a treatment of MBD interactions59,60,61,62, making use of 3ob parameters75,76. Then, we select the conformers with the most distinct values of both energies (i.e., Etot > 0.24 eV and EMBD > 0.048 eV) per cluster, see Fig. 1. This exemplifies a new approach for selecting conformers of large molecules within chemical space, utilizing an in-depth analysis of their electronic properties. To showcase its efficacy, we have considered only 1, 653 molecules ( ≈ 60% of total initial unique molecules) with up to 54 non-hydrogen atoms (N ≤ 92), reducing the number of conformers for these molecules from 280, 182 to 59, 783.
While this method does indeed yield a more diverse set of conformers, it remains essential to confirm the energetic and mechanical stability of these molecular structures, especially, taking into account the treatment of MBD interactions. To maintain consistency with our earlier publication of the QM7-X dataset for small organic molecules, we have conducted the geometry optimization calculations utilizing the DFTB3+MBD level of theory. Moreover, to construct a dataset that can be used to understand the influence of molecule-solvent interaction on the physicochemical properties of large drug-like molecules, our generation procedure considers the optimization of structures in gas phase and in implicit water described by the GBSA model, as implemented in the DFTB+ code77. We have stored the gas-phase and solvated optimized structures into two subsets named AQM-gas and AQM-sol, respectively. These DFTB calculations were performed by interfacing DFTB+ code with the Atomic Simulation Environment (ASE)78. Despite the majority of these molecular structures being identified as local minima at the DFTB3+MBD level, both in gas phase and in implicit water, it is worth mentioning that some of them are situated at saddle points on the respective PES.
Calculation of physicochemical properties
These ≈ 60 k DFTB optimized structures were now utilized for more accurate QM single-point calculations using dispersion-inclusive hybrid DFT. Energies, forces, and several other physicochemical properties (as detailed in Table 2) were calculated at a higher level of theory that varied depending on the chemical environment used in the structure optimization process. Property calculations for AQM-gas molecules were computed using PBE0+MBD59,79,80 level, while, for AQM-sol molecules, the modified Poisson-Boltzmann (MPB)43,81 model of water was also considered. The MPB model solves the size-modified Poisson-Boltzmann equation for the implicit inclusion of electrolytic solvation effects into DFT calculations. It also includes a model for the well-known Stern layer that separates the diffusing ions from the solvation cavity by introducing non-mean-field ion-solute interactions. For these calculations, we have used the FHI-aims code82,83 (version 221103) together with “tight” settings for basis functions and integration grids. Energies were converged to 10−6 eV and the accuracy of the forces was set to 10−4 eV/Å. The convergence criteria used during self-consistent field (SCF) optimizations were 10−3 eV for the sum of eigenvalues and 10−6 electrons/Å3 for the charge density.
The MBD energies and MBD atomic forces were here computed using the range-separated self-consistent screening (rsSCS) approach60, while the atomic C6 coefficients, isotropic atomic polarizabilities, molecular C6 coefficients and molecular polarizabilities (both isotropic and tensor) were obtained via the SCS approach59. Hirshfeld ratios correspond to the Hirshfeld volumes divided by the free atom volumes. The TS dispersion energy refers to the pairwise Tkatchenko-Scheffler (TS) dispersion energy in conjunction with the PBE0 functional49. The vdW radii were also obtained using the SCS approach via\({R}_{{\rm{vdW}}}={\left({\alpha }^{{\rm{SCS}}}/{\alpha }^{{\rm{TS}}}\right)}^{1/3}{R}_{{\rm{vdW}}}^{{\rm{TS}}}\), where αTS and \({R}_{{\rm{vdW}}}^{{\rm{TS}}}\) are the atomic polarizability and vdW radius computed according to the TS scheme, respectively. Atomization energies were obtained by subtracting the atomic PBE0 energies from the PBE0 total energy of each gas-phase and solvated molecular conformation (see Table S1 of the SI). The exact exchange energy is the amount of exact (or Hartree-Fock) exchange that has been admixed into the exchange-correlation energy.
Data Records
The AQM dataset is provided in two HDF5 files in a ZENODO.ORG data repository84. The QM structural and property data of the 59, 783 conformations corresponding to 1, 653 molecules in both gas phase and implicit water were stored in the AQM-gas.hdf5 and AQM-sol.hdf5 files, respectively. Additionally, we have uploaded the AQM-initial.hdf5 file which only contains the structural data of the 2, 242, 490 conformations corresponding to the initial set of 2, 635 molecules (obtained by using CREST code). One can also find there a README file with technical usage details and an example of how to access the information stored in AQM (see readAQM.py file).
HDF5 file format
Independent of the AQM subset, the information for each molecular structure is stored in a Python dictionary (dict) type containing all relevant properties and recorded in groups in HDF5 file format30. HDF5 keys to access the atomic numbers, atomic positions (coordinates), and physicochemical properties in each dictionary are provided in Table 2. The dimension of each array depends on the number of atoms N and the required property, e.g., for a methane (CH4) molecule, ’atNUM’ is a 1D array of N = 5 elements ([6, 1, 1, 1, 1]) while ’atXYZ’ is a 2D array comprised of N = 5 rows and three columns (x, y, z coordinates). All structures are labeled as Geom-mr-ct, where r enumerates the SMILE strings and t the considered conformer. Note that the indices t used in the AQM dataset reflect the order in which a given structure was generated and do not correspond to sorted xTB/DFTB (or DFT) total energies.
Technical Validation
A significant challenge in simulating the physicochemical properties of large drug-like molecules lies in the fact that, in experiments, their conformations and electronic structures are influenced by interactions with the surrounding solvent. However, the standard approach in contemporary QM simulations involves running them in gas phase, without accounting for molecule-solvent interactions. Unlike another recently published dataset of large molecules (see Table 1), AQM dataset considers the molecule-solvent interactions as well as a treatment of van der Waals (vdW) interactions in its generation procedure—two important physical and chemical effects in determining structural conformations and stability rankings of molecules of pharmaceutically relevant size. As mentioned above, the AQM comprises the structural and electronic data of 59, 783 gas-phase and solvated (low-and high-energy) conformers of 1, 653 molecules with up to 54 non-hydrogen atoms (N ≤ 92), including C, N, O, F, P, S and Cl. The structures of solvated conformers were obtained using DFTB3+MBD method supplemented with the GBSA implicit model of water. This model has been successfully used in the study of free solvation energies of neutral/ionic molecules45 and the folding of short peptides85. Whereas, the level of theory selected to compute the QM properties per conformer was PBE0+MBD supplemented with the MPB implicit model of water. The MPB model has been shown to provide a more accurate description in the study of diverse electrochemical reactions86,87,88,89. In all calculations, many-body dispersion (MBD) interactions have been included to deal with long-range interactions that are not adequately represented by the baseline level of theory. These advanced theoretical models have thus generated a more accurate collection of molecular and atom-in-a-molecule (as well as ground state and response) QM properties of conformers in implicit water, which are stored in AQM-sol. Moreover, when integrated with the property information in AQM-gas, these data can assist in fine-tuning ML models for the precise estimation of electronic properties of solvated molecules, e.g., via a delta learning approach.
An essential step in the generation of AQM dataset involved the thoughtful selection of the conformational search workflow. Here, we exhaustively analyzed the sampling method implemented in four different codes: CREST56, Maestro, Omega90 and RDKit66,91. The last three codes are of standard use in cheminformatics, primarily relying on stochastic algorithms for their application. They explore the conformational space of molecules very sparsely through a combination of pre-defined distances and stochastic samples92 and can miss many low-energy conformations. Moreover, in most standalone applications, conformer energies are typically computed using classical force fields without the incorporation of solvent models, which made these values rather inaccurate93 (for more details of these methods see Sec. 2 of the SI). On the contrary, CREST code utilizes a robust sampling strategy, leveraging the semi-empirical extended tight-binding method (GFN2-xTB)13, supplemented by the GBSA implicit solvent model of water, in order to generate more reliable 3D conformations compared to those obtained using classical force fields (see “Methods”). The conformational search workflow implemented in CREST provides access to a more extensive exploration of low- and high-energy conformers, generating molecular structures inaccessible by distance geometry methods (see Fig. 2)46,51,73.
Comparison of the performance of conformational search workflows. (a) Variation of the averaged number of clusters ⟨M⟩ as a function of the root-mean-square error ΔR between conformers generated by the four conformational search workflows examined in this work (see legend on top). The average was taken over 18 randomly selected compositions with approximately N = 50 atoms. (b) Two-dimensional property space defined by EAT and EMBD of the set of representative conformers obtained after applying the criteria defined in “Methods”. The energy calculations were performed using DFTB3+MBD method. Examples of structures only generated by CREST code are inserted in the graph.
To gain a better understanding of the influence of these sampling methods on the conformational search of large drug-like molecules, we analyzed the structural and energetic data of conformers corresponding to 18 randomly selected compositions, each containing approximately N = 50 atoms. Fig. 2(a) displays the output for the variation of the averaged number of clusters, denoted as ⟨M⟩, as a function of the root-mean-square deviation, ΔR, among conformers that constitute a cluster. This calculation was first done per molecule and then averaged over the 18 cases. This showed that more diverse conformers become part of the same cluster when ΔR increases, resulting in a reduction of the number of distinct clusters. The decrease in ⟨M⟩ for Maestro and Omega is faster compared to CREST, which may indicate that conformational search workflows based on distance geometry methods are insufficient for probing the PES of more flexible molecules. However, thanks to the random ensemble option for conformer generation, RDKit produces a larger ⟨M⟩ than CREST for ΔR > 1.0 Å. To examine this result further, we compute EAT and EMBD for a set of representative conformers per molecule (see selection criterion in “Methods”) using DFTB3+MBD level of theory. Notice that the resulting total number of conformers depends on the code employed for their generation, i.e., CREST → 3747 conformers, Maestro → 100 conformers, Omega → 204 conformers and RDKit → 1872 conformers. Thus, we have found that, compared to other methods, the conformers generated by CREST code show a more organized coverage of the energetic space defined by DFTB-EAT and DFTB-EMBD (see the well-defined cluster in Fig. 2(b)). Moreover, the larger coverage of DFTB-EMBD values is a clear indicator that CREST sampling method can generate conformations of increased complexity, characterized by a more folded structure and a stronger dispersion interaction, as illustrated with the inserted structures in Fig. 2(b). This underscores the relevance of taking vdW interactions into account when generating and identifying conformers of large drug-like molecules. Similarly, this provides compelling evidence of the efficient conformational search workflow implemented in CREST code, which improves coverage of both conformational and property molecular space. It is noteworthy that the calculations executed by the CREST code incurred higher computational expenses compared to those carried out by chemoinformatics codes.
After generating all conformers and selecting the representative conformers for each molecule, we proceed to optimize the molecular structures. This optimization is carried out using the DFTB3+MBD level of theory in the gas phase as well as in implicit water described by the GBSA model. Once this optimization step is complete, we obtain the final set of molecular structures for AQM-gas (gas phase) and AQM-sol (implicit water) subsets. To quantitatively analyze the impact of molecule-solvent interaction on the structure of AQM molecules, we have computed ΔR between the molecular structures stored in AQM-gas and AQM-sol. Indeed, Fig. 3(a) displays the size dependence of the averaged ΔR (blue dots) together with the total range of ΔR values spanned at different N (blue shadow). The results show that the structures of small molecules (N ≤ 20 atoms) present ⟨ΔR⟩ < 0.1 Å, i.e., they are minimally affected by the interaction with the solvent. In contrast, when dealing with molecules with N > 40 atoms, solvent effects become significantly more pronounced, and, as a result, we observe greater deviations in the ΔR values (> 2.0 Å). A similar effect can be observed when comparing the gyration radius Rg for gas-phase and solvated molecular structures, see Fig. 3(b). Notice that there also are large compounds (N ≈ 90) characterized by extensively constrained structures, which remain unaffected by the interaction with implicit water. These findings hold significant relevance in the context of advancing QM-based pipelines for the creation of datasets of molecules of pharmaceutical relevant size. Particularly in cases where the research objectives encompass the generation of non-equilibrium structures for training ML force fields since the PES of these molecules will be largely modified by solvent effects—a phenomenon that can be inferred from the outcomes of the geometry optimization process.
Effect of solvation on the atomic structures. (a) Size dependence of averaged root-mean-square deviation ⟨ΔR⟩ between geometries optimized using DFTB3+MBD method in gas phase and in implicit water described by the GBSA model. We also show the range of ΔR values spanned at different N (blue shadow). (b) Correlation plot between the gyration radius Rg of gas-phase and solvated conformations. Datapoints are colored with respect to the number of non-hydrogen atoms. To perform these analyses, we have considered the 59, 783 low-and high-energy conformers contained in AQM-gas and AQM-sol. In each panel, select conformations were inserted to highlight the effect of solvent on the molecular structure. Solvated structures are represented with pink balls.
The AQM dataset considers an extensive array of more than 40 distinct molecular (global) and atom-in-a-molecule (local) QM properties, which were computed to gain insights into the effect of molecule-solvent interactions on structure-property and property-property relationships of large molecules. In Table 2, we list the properties of gas-phase and solvated molecules, derived from QM calculations performed at the PBE0+MBD level (AQM-gas) and further enhanced with the MPB implicit solvent model of water (AQM-sol), respectively. PBE0+MBD has been chosen as our baseline level of theory for property calculations due to its well-established accuracy and reliability, demonstrated in the description of intramolecular degrees of freedom as well as intermolecular interactions in organic molecular dimers, supramolecular complexes, and molecular crystals59,79,80,94,95,96,97. The use of these DFT methods also provides interesting insights into the effect of molecule-solvent interactions on the potential energy surface of molecules, highlighting the importance of the dataset generation procedure. In Fig. S2 of the SI, one can see that the energy range and the energetic ranking of molecules in AQM-gas and AQM-sol are different, but further analysis is required. We therefore consider that our QM calculations are suitable to validate the quality of future research utilizing the AQM dataset.
To understand the relevance of accessing QM data for solvated molecules, we first discuss the influence of implicit water on extensive and intensive molecular QM properties. In doing so, as an illustrative example, we have analyzed the 2D property space defined by two contrasting properties98,99 such as the isotropic molecular polarizability α and the HOMO-LUMO gap Egap (i.e., \(\left(\alpha ,{E}_{{\rm{gap}}}\right)\)-space) for the 59, 783 conformations in AQM-gas and AQM-sol as well as the set of most stable conformer per molecule in AQM-sol (only 1, 653 conformations), see Fig. 4(a). For comparison, the values corresponding to QM7-X equilibrium molecules are also plotted (green circles). Our findings reveal that AQM molecules exhibit a significantly broader coverage of the α range, surpassing QM7-X molecules by a factor of 6. This expanded coverage is attributed to the inherently extensive character of α. Whereas, Egap range now covers molecules with circa 2.5 eV of energy gap, and the mean value for the entire dataset reduced from 7.0 eV to 4.5 eV, see distribution plots on the top panel of Fig. 4(a). The slight differences between the \(\left(\alpha ,{E}_{{\rm{gap}}}\right)\)-space covered by AQM-gas and AQM-sol may be attributed to compensation between the pronounced fluctuations in α, which are predominantly observed in molecules with \(\alpha > 300\,{a}_{0}^{3}\), and the more sensitive behavior of Egap to the presence of implicit water, as displayed in the correlation plots in Fig. 4(b). Accordingly, these findings could carry crucial implications in the “freedom of design” when searching for large drug-like molecules with targeted \(\left(\alpha ,{E}_{{\rm{gap}}}\right)\) values98,99,100. Notice that the conformational sampling per molecule largely improved the coverage of both properties, connecting isolated regions associated with a single molecular structure with specific size and chemical composition. Fig. 4(b) also shows the correlation plots between HOMO energy EHOMO and total dipole moment Ds of the 59, 783 conformers contained in AQM-gas and AQM-sol. Thus, it becomes evident that intensive properties are more sensitive to the incorporation of molecule-solvent interactions in the QM calculations when contrasted with extensive properties.
Influence of solvent in select global (molecular) QM properties considered in the AQM dataset. (a) 2D projections of the high-dimensional AQM property space defined by isotropic molecular polarizability αs and HOMO-LUMO gap Egap, i.e., \(\left({\alpha }_{{\rm{s}}},{E}_{{\rm{gap}}}\right)\)-space. We show the property values of 59, 783 low-and high-energy conformers contained in AQM-gas (blue circles) and in AQM-sol (red circles) as well as the corresponding values for the most stable conformer of the 1, 653 unique molecules in AQM-sol (cyan circles). For comparison, we also present the values obtained for QM7-X equilibrium molecules (green circles). The frequency plot (distribution) associated with each property and subset of molecules is plotted on the top and right-side panels of the graph. (b) Correlation plots between the property values corresponding to 59, 783 gas-phase and solvated structures. Here, we have considered extensive and intensive QM properties such as HOMO-LUMO gap Egap, HOMO energy EHOMO, isotropic molecular polarizability αs, and scalar dipole moment Ds.
On the other hand, atom-in-a-molecule QM properties can provide important insight into the distinct chemical environments within large drug-like molecules. To illustrate this, Fig. 5(a) shows the 2D property space defined by Hirshfeld charges qH and atomic polarizabilities \({\widetilde{\alpha }}_{{\rm{s}}}\) (i.e., \(\left({q}_{{\rm{H}}},{\widetilde{\alpha }}_{{\rm{s}}}\right)\)-space) for the 59, 783 conformations in AQM-gas and AQM-sol. For comparison, the values corresponding to QM7-X equilibrium molecules are also plotted (green circles). Fig. 5(a) shows the existence of well-defined clusters that are mostly related to a specific atom type. The slight overlap between these clusters is a clear example of the need to develop more robust geometric and electronic descriptors capable of effectively representing intricate chemical environments in large drug-like molecules for ML applications. Furthermore, our calculations have revealed that implicit solvation has a pronounced influence on qH values, particularly for heavier atoms such as P, S, and Cl—relevant atoms in the design of pharmaceutical compounds as well as in the determination of their physicochemical and biological properties. Certainly, the molecule-solvent interaction has a stronger effect on the local properties compared to global ones, as illustrated by the correlation plots in Fig. 5(b). This becomes more evident by observing the atomic forces Ftot, where the significant variations in values can strongly affect the accuracy of ML force fields when applied to run the dynamics of large molecules.
Influence of solvent in select local (atom-in-a-molecule) QM properties considered in the AQM dataset. (a) 2D projections of the high-dimensional AQM property space defined by Hirshfeld charges qH and atomic polarizabilities \({\widetilde{\alpha }}_{{\rm{s}}}\), i.e., \(\left({q}_{{\rm{H}}},{\widetilde{\alpha }}_{{\rm{s}}}\right)\)-space. We show the property values of 59, 783 low-and high-energy conformers contained in AQM-gas (blue circles) and AQM-sol (red circles). For comparison, we also present the values obtained for QM7-X equilibrium molecules (green circles). The arrows in the graph highlight the region covered per local chemical environment corresponding to each atom type (X). The frequency plot (distribution) associated with each property and subset of molecules is plotted on the top and right-side panels of the graph. (b) Correlation plots between the property values corresponding to 59, 783 gas-phase and solvated structures. Here, we have considered local QM properties such as total atomic forces Ftot, Hirshfeld charges qH, atomic polarizabilities \({\widetilde{\alpha }}_{{\rm{s}}}\), and atomic Hirshfeld dipole moment DH,s.
Up to now, we have been focused on the impact of considering molecule-solvent interaction when computing molecular/atom-in-a-molecule QM properties of AQM molecules. However, the data of the non-electrostatic part of solvation energy due to molecule-solvent interaction Enelec in conformers contained in AQM-sol can also be crucial for having a better understanding of the solvation effect on structure-property and property-property relationships of large molecules. As an example, Fig. 6(a) shows the correlation plot between Enelec and dispersion interaction energy Edisp calculated using two well-established methods: many-body dispersion (MBD) and Tkatchenko-Scheffler (TS). The datapoints are colored according to the gyration radius Rg of each solvated structure. The high degree of correlation between these properties underscores the importance of considering both molecule-solvent and dispersion interactions when investigating large molecules, as we did to generate AQM dataset. Moreover, the growing difference in energies obtained by MBD and TS methods, particularly as the system size increases, highlights the significant influence of many-body interactions in the energetic description of these compounds. To further elucidate the role of both interactions in the generation of AQM, we have selected the molecule C29H39N5O3S2 (N = 78 atoms, ID in dataset: 2070) with 720 conformers and then examined their respective energy values. These conformers show a difference in dispersion energies ΔEdisp of up to ≈1.0 eV, where the smallest ΔEdisp values correspond to more compact molecular structures while the largest ΔEdisp values are observed for more extended ones (see Fig. 6(b)). Besides presenting a size dependence, the data plotted in Fig. 6(c) demonstrate that, similar to EMBD, Enelec also depends on the structural conformation of molecules. These findings show the significance of both interactions in the generation of a robust QM dataset comprising large and more flexible molecules that bear pharmaceutical relevance.
Relevance of dispersion and molecule-solvent interactions in drug-like molecules. (a) Variation of the non-electrostatic free energy Enelec as a function of the dispersion energy Edisp for all solvated structures in AQM-sol. We have considered two different dispersion models in this analysis: Many-body dispersion (MBD) and Tkatchenko-Scheffler (TS). (b) Frequency plot of the difference between MBD and TS energies (i.e., ΔEdisp = EMBD − ETS) for the 720 low-and high-energy conformers of the molecule C29H39N5O3S2 (ID in dataset: 2070). (c) Variation of Enelec as a function of EMBD for all conformers of the molecule C29H39N5O3S2. The color code in panels (a) and (c) is with respect to the gyration radius Rg of each solvated structure. We have inserted select conformations in panels (b) and (c) to highlight the effect on the molecular structure of reducing or increasing the magnitude of both properties.
In summary, we demonstrated that the extensive structural and property data contained in AQM dataset hold the potential to enhance the understanding of how molecular-solvent interactions influence both structure-property and property-property relationships of large drug-like molecules. As such, AQM may in some cases be employed as a benchmark dataset for direct/delta learning and generative methods, estimating the properties of pharmaceutical compounds in solution from their gas-phase counterparts.
Code availability
The initial structure generation was carried out using RDKit 2020.09.566,67. Further structure optimization and the creation of conformers were performed by utilizing CREST13,56 and DFTB+10,77 codes together with ASE78. Note that all necessary features regarding the utilized DFTB3+MBD (with and without GBSA implicit solvent) approach are available in the current DFTB+ version11. All DFT calculations were carried out using FHI-aims82 (version 221103).
Additional conformer generation experiments were performed with RDKit 2020.09.5, OpenEye’s Omega 4.0.0.490 and Schrodinger’s Maestro suite v2020-4 (see SI for detailed procedures).
References
Friesner, R. A. ab initio quantum chemistry: Methodology and applications. Proc. Natl. Acad. Sci. 102, 6648–6653 (2005).
Marzari, N., Ferretti, A. & Wolverton, C. Electronic-structure methods for materials design. Nat. Mater. 20, 736–749 (2021).
Palazzesi, F., Grundl, M. A., Pautsch, A., Weber, A. & Tautermann, C. S. A fast ab initio predictor tool for covalent reactivity estimation of acrylamides. J. Chem. Inf. Model 59, 3565–3571 (2019).
Mihalovits, L. M., Ferenczy, G. G. & Keserũ, G. M. Affinity and selectivity assessment of covalent inhibitors by free energy calculations. J. Chem. Inf. Model 60, 6579–6594 (2020).
Hofmans, S. et al. Tozasertib analogues as inhibitors of necroptotic cell death. J. Medicinal Chem 61, 1895–1920 (2018).
Prasad, S., Huang, J., Zeng, Q. & Brooks, B. R. An explicit-solvent hybrid QM and MM approach for predicting pKa of small molecules in SAMPL6 challenge. J. Comput. Mol. Des. 32, 1191–1201 (2018).
Raghavachari, K. & Saha, A. Accurate composite and fragment-based quantum chemical models for large molecules. Chem. Rev. 115, 5643–5677 (2015).
Pruitt, S. R., Bertoni, C., Brorsen, K. R. & Gordon, M. S. Efficient and accurate fragmentation methods. Acc. Chem. Res. 47, 2786–2794 (2014).
Stewart, J. J. P. Optimization of parameters for semiempirical methods II. applications. J. Comput. Chem. 10, 221–264 (1989).
Seifert, G., Porezag, D. & Frauenheim, T. Calculations of molecules, clusters, and solids with a simplified LCAO-DFT-LDA scheme. Int. J. Quantum Chem. 58, 185–192 (1996).
Hourahine, B. et al. DFTB+, a software package for efficient approximate density functional theory based atomistic simulations. J. Chem. Phys 152, 124101 (2020).
Bannwarth, C. et al. Extended tight-binding quantum chemistry methods. WIREs Comput. Mol. Sci. 11, e1493 (2021).
Bannwarth, C., Ehlert, S. & Grimme, S. GFN2-xTB—an accurate and broadly parametrized self-consistent tight-binding quantum chemical method with multipole electrostatics and density-dependent dispersion contributions. J. Chem. Theory Comput. 15, 1652–1671 (2019).
Smith, J. S., Isayev, O. & Roitberg, A. E. ANI-1: An extensible neural network potential with DFT accuracy at force field computational cost. Chem. Sci. 8, 3192–3203 (2017).
Chmiela, S., Sauceda, H. E., Müller, K.-R. & Tkatchenko, A. Towards exact molecular dynamics simulations with machine-learned force fields. Nat. Commun. 9, 3887 (2018).
Schütt, K. T., Arbabzadah, F., Chmiela, S., Müller, K. R. & Tkatchenko, A. Quantum-chemical insights from deep tensor neural networks. Nat. Commun. 8, 13890 (2017).
Unke, O. T. et al. Spookynet: Learning force fields with electronic degrees of freedom and nonlocal effects. Nat. Commun. 12, 7273 (2021).
Batzner, S. et al. E (3)-equivariant graph neural networks for data-efficient and accurate interatomic potentials. Nat. Commun. 13, 2453 (2022).
Musaelian, A. et al. Learning local equivariant representations for large-scale atomistic dynamics. Nat. Commun. 14, 579 (2023).
Batatia, I. et al. (eds.) Advances in Neural Information Processing Systems, vol. 35, 11423–11436 (Curran Associates, Inc., 2022).
Huang, B., von Rudorff, G. F. & von Lilienfeld, O. A. The central role of density functional theory in the AI age. Science 381, 170–175 (2023).
Kulik, H. J. et al. Roadmap on machine learning in electronic structure. Electron. Struct 4, 023004 (2022).
Stöhr, M., Medrano Sandonas, L. & Tkatchenko, A. Accurate many-body repulsive potentials for density-functional tight binding from deep tensor neural networks. J. Phys. Chem. Lett 11, 6835–6843 (2020).
Qiao, Z., Welborn, M., Anandkumar, A., Manby, F. R. & Miller, T. F. OrbNet: Deep learning for quantum chemistry using symmetry-adapted atomic-orbital features. J. Chem. Phys 153, 124111 (2020).
Blum, L. C. & Reymond, J.-L. 970 million druglike small molecules for virtual screening in the chemical universe database GDB-13. J. Am. Chem. Soc. 131, 8732–8733 (2009).
Montavon, G. et al. Machine learning of molecular electronic properties in chemical compound space. New J. Phys. 15, 095003 (2013).
Yang, Y. et al. Quantum mechanical static dipole polarizabilities in the QM7b and AlphaML showcase databases. Sci. Data 6, 152 (2019).
Ruddigkeit, L., van Deursen, R., Blum, L. C. & Reymond, J.-L. Enumeration of 166 billion organic small molecules in the chemical universe database GDB-17. J. Chem. Inf. Model. 52, 2864–2875 (2012).
Ramakrishnan, R., Dral, P. O., Rupp, M. & von Lilienfeld, O. A. Quantum chemistry structures and properties of 134 kilo molecules. Sci. Data 1, 140022 (2014).
Hoja, J. et al. QM7-X, a comprehensive dataset of quantum-mechanical properties spanning the chemical space of small organic molecules. Sci. Data 8, 43 (2021).
Schütt, K. T., Sauceda, H. E., Kindermans, P.-J., Tkatchenko, A. & Müller, K.-R. SchNet – a deep learning architecture for molecules and materials. J. Chem. Phys 148, 241722 (2018).
Chmiela, S. et al. Accurate global machine learning force fields for molecules with hundreds of atoms. Sci. Adv. 9, eadf0873 (2023).
Smith, J. S., Isayev, O. & Roitberg, A. E. ANI-1, A data set of 20 million calculated off-equilibrium conformations for organic molecules. Sci. Data 4, 170193 (2017).
Smith, J. S. et al. The ANI-1ccx and ANI-1x data sets, coupled-cluster and density functional theory properties for molecules. Sci. Data 7, 134 (2020).
Zubatyuk, R., Smith, J. S., Nebgen, B. T., Tretiak, S. & Isayev, O. Teaching a neural network to attach and detach electrons from molecules. Nat. Commun. 12, 4870 (2021).
Decherchi, S. & Cavalli, A. Thermodynamics and kinetics of drug-target binding by molecular simulation. Chem. Rev. 120, 12788–12833 (2020).
Hirata, F. Molecular theory of solvation, vol. 24 (Springer Science & Business Media, 2003).
Gorges, J., Grimme, S., Hansen, A. & Pracht, P. Towards understanding solvation effects on the conformational entropy of non-rigid molecules. Phys. Chem. Chem. Phys. 24, 12249–12259 (2022).
Matczak, P. & Domagała, M. Heteroatom and solvent effects on molecular properties of formaldehyde and thioformaldehyde symmetrically disubstituted with heterocyclic groups C4H3Y (where Y= O–Po). J. Mol. Model. 23, 1–11 (2017).
Odey, M. O. et al. Unraveling the impact of polar solvation on the molecular geometry, spectroscopy (ft-ir, uv, nmr), reactivity (elf, nbo, homo-lumo) and antiviral inhibitory potential of cissampeline by molecular docking approach. Chem. Phys. Impact 7, 100346 (2023).
Ensing, B., Meijer, E. J., Blöchl, P. & Baerends, E. J. Solvation effects on the sn 2 reaction between ch3cl and cl-in water. J. Phys. Chem. A 105, 3300–3310 (2001).
Klamt, A. Conductor-like screening model for real solvents: A new approach to the quantitative calculation of solvation phenomena. J. Phys. Chem 99, 2224–2235 (1995).
Ringe, S., Oberhofer, H., Hille, C., Matera, S. & Reuter, K. Function-space-based solution scheme for the size-modified poisson–boltzmann equation in full-potential DFT. J. Chem. Theory Comput. 12, 4052–4066 (2016).
Onufriev, A. V. & Case, D. A. Generalized born implicit solvent models for biomolecules. Annu. Rev. Biophys. 48, 275–296 (2019).
Xie, L. & Liu, H. The treatment of solvation by a generalized born model and a self-consistent charge-density functional theory-based tight-binding method. J. Comput. Chem 23, 1404–1415 (2002).
Isert, C., Atz, K., Jiménez-Luna, J. & Schneider, G. QMugs, quantum mechanical properties of drug-like molecules. Sci. Data 9, 273 (2022).
Chai, J.-D. & Head-Gordon, M. Systematic optimization of long-range corrected hybrid density functionals. J. Chem. Phys 128, 084106 (2008).
Stuke, A. et al. Atomic structures and orbital energies of 61,489 crystal-forming organic molecules. Sci. Data 7, 58 (2020).
Tkatchenko, A. & Scheffler, M. Accurate molecular van der Waals interactions from ground-state electron density and free-atom reference data. Phy. Rev. Lett. 102, 073005 (2009).
Sinstein, M. et al. Efficient implicit solvation method for full potential DFT. J. Chem. Theory Comput. 13, 5582–5603 (2017).
Axelrod, S. & Gómez-Bombarelli, R. GEOM, energy-annotated molecular conformations for property prediction and molecular generation. Sci. Data 9, 185 (2022).
Subramanian, G., Ramsundar, B., Pande, V. & Denny, R. A. Computational modeling of β-secretase 1 (BACE-1) inhibitors using ligand based approaches. J. Chem. Inf. Model. 56, 1936–1949 (2016).
Ehlert, S., Stahn, M., Spicher, S. & Grimme, S. Robust and efficient implicit solvation model for fast semiempirical methods. J. Chem. Theory Comput. 17, 4250–4261 (2021).
Barone, V. & Cossi, M. Quantum calculation of molecular energies and energy gradients in solution by a conductor solvent model. J. Phys. Chem. A 102, 1995–2001 (1998).
Eastman, P. et al. SPICE, A Dataset of Drug-like Molecules and Peptides for Training Machine Learning Potentials. Sci. Data 10, 11 (2023).
Pracht, P., Bohle, F. & Grimme, S. Automated exploration of the low-energy chemical space with fast quantum chemical methods. Phys. Chem. Chem. Phys. 22, 7169–7192 (2020).
Elstner, M. et al. Self-consistent-charge density-functional tight-binding method for simulations of complex materials properties. Phys. Rev. B 58, 7260–7268 (1998).
Gaus, M., Cui, Q. & Elstner, M. DFTB3: Extension of the self-consistent-charge density-functional tight-binding method (SCC-DFTB). J. Chem. Theory Comput. 7, 931–948 (2011).
Tkatchenko, A., DiStasio, R. A. Jr, Car, R. & Scheffler, M. Accurate and efficient method for many-body van der Waals interactions. Phys. Rev. Lett. 108, 236402 (2012).
Ambrosetti, A., Reilly, A. M., DiStasio, R. A. Jr & Tkatchenko, A. Long-range correlation energy calculated from coupled atomic response functions. J. Chem. Phys 140, 18A508 (2014).
Stöhr, M., Michelitsch, G. S., Tully, J. C., Reuter, K. & Maurer, R. J. Communication: Charge-population based dispersion interactions for molecules and materials. J. Chem. Phys 144, 151101 (2016).
Mortazavi, M., Brandenburg, J. G., Maurer, R. J. & Tkatchenko, A. Structure and stability of molecular crystals with many-body dispersion-inclusive density functional tight binding. J. Phys. Chem. Lett 9, 399–405 (2018).
Havu, V., Blum, V., Havu, P. & Scheffler, M. Efficient O(N) integration for all-electron electronic structure calculation using numeric basis functions. J. Comput. Phys 228, 8367–8379 (2009).
Gaulton, A. et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40, D1100–D1107 (2012).
Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S. & Hopkins, A. L. Quantifying the chemical beauty of drugs. Nat. Chem. 4, 90–98 (2012).
Landrum, G. et al. RDKit: Open-source cheminformatics. https://www.rdkit.org (2020).
Landrum, G. et al. rdkit/rdkit: 2020_03_1 (q1 2020) release https://doi.org/10.5281/zenodo.3732262 (2020).
Halgren, T. A. Merck molecular force field. I. basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 17, 490–519 (1996).
Halgren, T. A. Merck molecular force field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. J. Comput. Chem 17, 520–552 (1996).
Halgren, T. A. Merck molecular force field. III. molecular geometries and vibrational frequencies for MMFF94. J. Comput. Chem. 17, 553–586 (1996).
Halgren, T. A. & Nachbar, R. B. Merck molecular force field. IV. conformational energies and geometries for MMFF94. J. Comput. Chem. 17, 587–615 (1996).
Halgren, T. A. Merck molecular force field. V. extension of MMFF94 using experimental data, additional computational data, and empirical rules. J. Comput. Chem 17, 616–641 (1996).
Cremer, J., Medrano Sandonas, L., Tkatchenko, A., Clevert, D.-A. & De Fabritiis, G. Equivariant graph neural networks for toxicity prediction. Chem. Res. Toxicol. 36, 1561–1573 (2023).
Bell, E. W. & Zhang, Y. DockRMSD: an open-source tool for atom mapping and RMSD calculation of symmetric molecules through graph isomorphism. J. Cheminformatics 11, 40 (2019).
Gaus, M., Goez, A. & Elstner, M. Parametrization and benchmark of DFTB3 for organic molecules. J. Chem. Theory Comput. 9, 338–354 (2013).
Gaus, M., Lu, X., Elstner, M. & Cui, Q. Parameterization of DFTB3/3OB for sulfur and phosphorus for chemical and biological applications. J. Chem. Theory Comput. 10, 1518–1537 (2014).
Aradi, B., Hourahine, B. & Frauenheim, T. DFTB+, a sparse matrix-based implementation of the DFTB method. J. Phys. Chem. A 111, 5678–5684 (2007).
Larsen, A. H. et al. The atomic simulation environment—a python library for working with atoms. J. Phys. Condens. Matter 29, 273002 (2017).
Perdew, J. P., Ernzerhof, M. & Burke, K. Rationale for mixing exact exchange with density functional approximations. J. Chem. Phys 105, 9982–9985 (1996).
Adamo, C. & Barone, V. Toward reliable density functional methods without adjustable parameters: The PBE0 model. J. Chem. Phys. 110, 6158–6170 (1999).
Ringe, S., Oberhofer, H. & Reuter, K. Transferable ionic parameters for first-principles Poisson-Boltzmann solvation calculations: Neutral solutes in aqueous monovalent salt solutions. J. Chem. Phys 146, 134103 (2017).
Blum, V. et al. Ab initio molecular simulations with numeric atom-centered orbitals. Comp. Phys. Commun. 180, 2175–2196 (2009).
Ren, X. et al. Resolution-of-identity approach to Hartree–Fock, hybrid density functionals, RPA, MP2 and GW with numeric atom-centered orbital basis functions. New J. Phys. 14, 053020 (2012).
Medrano Sandonas, L. et al. Aquamarine: Quantum-mechanical exploration of conformers and solvent effects in large drug-like molecules. ZENODO https://doi.org/10.5281/zenodo.10208010 (2024).
Ho, B. K. & Dill, K. A. Folding very short peptides using molecular dynamics. PLOS Comput. Biol. 2, 1–10 (2006).
Ringe, S. et al. Understanding cation effects in electrochemical CO2 reduction. Energy Environ. Sci. 12, 3001–3014 (2019).
Abidi, N., Lim, K. R. G., Seh, Z. W. & Steinmann, S. N. Atomistic modeling of electrocatalysis: Are we there yet? WIREs Comput. Mol. Sci. 11, e1499 (2021).
Gauthier, J. A. et al. Unified approach to implicit and explicit solvent simulations of electrochemical reaction energetics. J. Chem. Theory Comput. 15, 6895–6906 (2019).
Ringe, S., Hörmann, N. G., Oberhofer, H. & Reuter, K. Implicit solvation methods for catalysis at electrified interfaces. Chem. Rev. 122, 10777–10820 (2022).
Hawkins, P. C., Skillman, A. G., Warren, G. L., Ellingson, B. A. & Stahl, M. T. Conformer generation with omega: algorithm and validation using high quality structures from the protein databank and cambridge structural database. J. Chem. Inf. Model 50, 572–584 (2010).
Wang, S., Witek, J., Landrum, G. A. & Riniker, S. Improving conformer generation for small rings and macrocycles based on distance geometry and experimental torsional-angle preferences. J. Chem. Inf. Model 60, 2044–2058 (2020).
Spellmeyer, D. C., Wong, A. K., Bower, M. J. & Blaney, J. M. Conformational analysis using distance geometry methods. J. Mol. Graph. Model. 15, 18–36 (1997).
Kanal, I. Y., Keith, J. A. & Hutchison, G. R. A sobering assessment of small-molecule force field methods for low energy conformer predictions. Int. J. Quantum Chem. 118, e25512 (2018).
Ernzerhof, M. & Scuseria, G. E. Assessment of the Perdew–Burke–Ernzerhof exchange-correlation functional. J. Chem. Phys. 110, 5029–5036 (1999).
Lynch, B. J. & Truhlar, D. G. Robust and affordable multicoefficient methods for thermochemistry and thermochemical kinetics: the MCCM/3 suite and SAC/3. J. Phys. Chem. A 107, 3898–3906 (2003).
Reilly, A. M. & Tkatchenko, A. Understanding the role of vibrations, exact exchange, and many-body van der Waals interactions in the cohesive properties of molecular crystals. J. Chem. Phys 139, 024705 (2013).
Hoja, J. et al. Reliable and practical computational description of molecular crystal polymorphs. Sci. Adv. 5, eaau3338 (2019).
Góger, S., Medrano Sandonas, L., Müller, C. & Tkatchenko, A. Data-driven tailoring of molecular dipole polarizability and frontier orbital energies in chemical compound space. Phys. Chem. Chem. Phys. 25, 22211–22222 (2023).
Medrano Sandonas, L. et al. “Freedom of design” in chemical compound space: towards rational in silico design of molecules with targeted quantum-mechanical properties. Chem. Sci. 14, 10702–10717 (2023).
Fallani, A., Medrano Sandonas, L. & Tkatchenko, A. Enabling inverse design in chemical compound space: Mapping quantum properties to structures for small organic molecules. ArXiv https://doi.org/10.48550/arXiv.2309.00506 (2023).
Acknowledgements
LMS and AT acknowledge financial support from Janssen Pharmaceuticals (Aquamarine project). AF and MH are grateful for financial support from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Innovative Training Network - European Industrial Doctorate grant agreement No 956832, “Advanced Machine Learning for Innovative Drug Discovery” (AIDD). The results presented in this publication have been partially obtained using the HPC facilities of the University of Luxembourg and Meluxina supercomputer (PoC project). This research also used computational resources provided by the Center for Information Services and High-Performance Computing (ZIH) at TU Dresden.
Author information
Authors and Affiliations
Contributions
D.V.R. and J.V. selected relevant compounds from public datasets to include in the dataset, with input from G.T. L.M.S. generated the 3D molecular structures with CREST/xTB and DFTB3+MBD. D.V.R., L.P.B., and D.H. generated the additional molecular structures with RDKit, Maestro, and Omega. LMS performed the PBE0+MBD calculations in gas phase and implicit water for all structures. L.M.S. and D.V.R. designed and wrote the manuscript. A.F. and M.H. contributed to the curation and technical validation of the dataset. A.T., J.K.W., and H.C. supervised and revised all stages of the work. All authors discussed the results and contributed to the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Medrano Sandonas, L., Van Rompaey, D., Fallani, A. et al. Dataset for quantum-mechanical exploration of conformers and solvent effects in large drug-like molecules. Sci Data 11, 742 (2024). https://doi.org/10.1038/s41597-024-03521-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03521-8
- Springer Nature Limited
This article is cited by
-
Inverse mapping of quantum properties to structures for chemical space of small organic molecules
Nature Communications (2024)