Abstract
Benthic prokaryotes include Bacteria and Archaea and dominate densities of marine benthos. They play major roles in element cycles and heterotrophic, chemoautotrophic, and phototrophic carbon production. To understand how anthropogenic disturbances and climate change might affect these processes, better estimates of prokaryotic biomasses and densities are required. Hence, I developed the ProkaBioDen database, the largest open-access database of benthic prokaryotic biomasses and densities in marine surface sediments. In total, the database comprises 1,089 georeferenced benthic prokaryotic biomass and 1,875 density records extracted from 85 and 112 studies, respectively. I identified all references applying the procedures for systematic reviews and meta analyses and report prokaryotic biomasses as g C cm−3 sediment, g C g−1 sediment, and g C m−2. Density records are presented as cell cm−3 sediment, cell g−1 sediment/ sulfide/ vent precipitate, and cell m−2. This database should serve as reference to close sampling gaps in the future.
Measurement(s) | prokaryotic benthic biomass • prokaryotic benthic density |
Technology Type(s) | PLFA • microscopy • ATP |
Sample Characteristic - Organism | unclassified Bacteria |
Sample Characteristic - Environment | marine sediment • deep marine sediment • shallow marine sediment |
Sample Characteristic - Location | Pacific Ocean • Atlantic Ocean • Indian Ocean • Southern Ocean • Arctic Ocean |
Similar content being viewed by others
Background & Summary
Unicellular prokaryotes comprise the domains Bacteria and Archaea1. They have densities of 108 to 109 cells cm−3 2 in the upper 10 cm of sediment and are therefore the most abundant benthic organisms. On a regional scale, prokaryotic densities in surface sediments decrease with depth (e.g.,3). Globally, however, their densities and biomasses do not decline with increasing water depth4,5. Prokaryotes are major players in the global cycling of elements, such as carbon6,7,8,9,10,11,12, nitrogen13,14,15, phosphorus13,16,17 and sulfur18,19,20,21, and they dominate sediment community oxygen consumption (SCOC) in deep-sea ecosystems >3,500 m water depth22.
Depending on the environmental conditions, prokaryotes can be involved in the transfer of organic matter to higher trophic levels: In the oxygen minimum zone of the Arabian Sea (Indian Ocean), the transfer of labelled carbon, that was taken up by prokaryotes, to their metazoan meiobenthic and macrobenthic consumers is relatively inefficient6. In comparison, for an intertidal area of the Scheldt estuary (North Sea), a model combined with a pulse-chase tracer experiment estimated that 3% of the prokaryotic carbon production was grazed upon by meiobenthos and 24% of this carbon production was consumed by macrobenthos23. In the deep-sea sediments of the Fram Strait (N Atlantic) and of the Clarion-Clipperton Fracture Zone (equatorial Pacific), however, no direct transfer of labelled carbon from prokaryotes to metazoan meiobenthos or metazoan macrobenthos was detected8,24.
Bacteria in marine surface sediments, i.e., sediment layers ranging from the sediment surface to – depending on the study – approximately 5 cm to 20 cm depth, contribute between 30 and 70% to total prokaryotic densities25,26,27,28. This corresponds to about 1029 bacteria cells living on our planet (uncertainty: 10-fold)29, of which 3.5 × 1028 ± 0.9 × 1028 occur in deep-sea surface sediments4. Hence, the upper 50 cm of said sediment are estimated to contain 1.29 Pg C4 (1 Pg = 1 petagram = 1015g) bacterial carbon which is up to 99% of the total estimated marine bacterial biomass (1.30 Pg C, uncertainty: 10-fold29). Bacteria can alter their environment, such as benthic cyanobacteria that can form so-called “microbially induced sedimentary structures”30. Particularly long, filamentous bacteria, known as “cable bacteria”, are even able to conduct long-distance electron transport over several centimeters31,32,33,34,35.
Archaea, whose most abundant phyla in the deep sea are Thaumarchaeota (53% of total Archaea density) and Euryarcheota (29% of Archaea density)36, account for <1 to 40% of prokaryotic densities in surface sediments25,27,37. Our planet is estimated to host 1028 marine benthic Archaea cells which is equivalent to 0.3 Pg C Archaea carbon (uncertainty: 13-fold)29.
Due to the prominent role of prokaryotes in the global carbon cycle, detailed knowledge about their biomasses and densities are necessary to understand how these microorganisms will be impacted by climate change and anthropogenic disturbances. Therefore, I prepared the open access “ProkaBioDen database”38 that, in comparison to preceding databases by Wei et al.39 and Rex et al.5, allows direct and free access to the data and transparently reports the selection process. It also covers the whole globe and not only the Atlantic Ocean and the Mediterranean Sea like in Danovaro et al.36 or is limited to specific water depths like in Danovaro et al.4.
The “ProkaBioDen database” lists 1,299 benthic prokaryotic biomass and 1,104 benthic prokaryotic density studies that were identified applying procedures for systematic reviews and meta analyses40. Based on this compilation, I extracted 1,089 georeferenced benthic prokaryotic biomass records and 1,875 georeferenced benthic prokaryotic density records from 85 and 112 chosen studies, respectively. I present benthic prokaryotic biomasses as g C cm−3 wet sediment, g C g−1 dry sediment, g C g−1 wet sediment, g C m−2 and benthic prokaryotic densities as cell cm−3 dry sediment, cell cm−3 wet sediment, cell g−1 dry sediment, cell g−1 dry sulfide, cell g−1 vent precipitate, cell g−1 wet sediment, cell m−2. All data further contain information about substrate type (e.g. Calyptogena sp. field, hydrothermal vent precipitate, iron oxidizing mat, mangrove, microbial mat, pogonophoran field, salt marsh, seagrass bed, sediment, sulfide chimney, sulfide oxidizing mat, sulfur band) and the methods applied to determine prokaryotic biomasses and densities and how researchers differentiated between Bacteria and Archaea densities. In this way, scientists can focus specifically on Archaea or Bacteria if they wish. The database is the first systematic open-access compilation of benthic Archaea and Bacteria densities and prokaryotic biomasses and densities and points towards undersampled geographic locations and water depth.
Methods
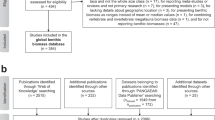
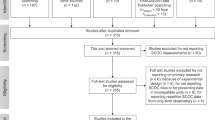
In March and June 2020, I compiled the “ProkaBio” part of the “ProkaBioDen database” applying the principles of “Preferred Reporting Items for Systematic Reviews and Meta-Analyses” (PRISMA)40. In the so-called “Identification” step, I identified 1,553 peer-reviewed articles in the Web of Science by using the key words “microb* biomass benth*”, “benthic prokaryotic biomass”, “benth* bacteria* biomass marin*”, and “Archaea biomass marin*”. Additionally, I found 138 publications in other sources, such as PANGAEA® Data Publisher (https://www.pangaea.de/) and peer-reviewed publications known to the author. After removing duplicate publications, I screened all titles and abstracts of 1,299 studies (Table 1; Fig. 1a; “Screening” step) and excluded 967 studies that did not report prokaryotic biomasses. In step 3, the so-called “Eligibility” step, I excluded in total 249 studies because they did not present prokaryotic biomasses in the marine sediment surface in standardizable units, i.e., in g C cm−3 wet sediment, g C g−1 wet sediment, g C g−1 dry sediment, or g C m−2. Furthermore, several studies lacked detailed geographical information about sampling stations or did not present primary research. Additional reasons for study exclusion were presenting prokaryotic biomasses for specific taxa instead of for all prokaryotes, being inaccessible, or introducing modelling, simulation, or experimental studies. In the final step, I included 85 studies from which I extracted 1,098 georeferenced benthic prokaryotic biomass records (Table 1, Fig. 1a).
In March and June 2020, I established the “ProkaDen” part of the database that consists of records of prokaryotic density as well as of density of Bacteria and of Archaea. Following the PRISMA approach40, I searched the Web of Science using the key words “marin* microb* abundance benth*”, “‘benthic bacteria’ abundance marin*”, “prokaryotic abundance marin*”, “prokaryotic density marin*”, “Archaea density abundance marin*”, “Archaea density marin*”, “Archaea abundance marin* benth*”, “Crenarchaea density abundance marin*”, “Crenarchaea density marin*”, “Crenarchaea abundance marin* benth*”, “Euryarchaea density abundance marin*”, and “Euryarchaea abundance marin* benth*” and found 1,204 peer-reviewed articles (Fig. 1b). I was aware of 171 additional studies that I included in the dataset which contained 1,104 studies after removing duplicates. In step 2 of the PRISMA approach (“Screening” step), I excluded 752 studies because they did not report benthic prokaryotic densities. In the “Eligibility” step, I furthermore omitted 239 studies because they did not present surface sediment prokaryotic densities or densities of a reduced number of prokaryotic taxa instead of reporting densities of all prokaryotes. I also removed studies that showed prokaryotic densities in poor-quality figures impeding data extraction and studies that listed densities which could not be converted to the common density units cell cm−3 dry sediment, cell cm−3 wet sediment, cell g−1 dry sediment, cell g−1 dry sulfide, cell g−1 vent precipitate, cell g−1 wet sediment, or cell m−2. I also excluded studies that reported experimental or culture studies and publications that I could not access. In the last step, I included 112 studies in the global benthic prokaryotic density database from which I extracted 1,875 georeferenced benthic prokaryotic density records (Table 1, Fig. 1a).
In 51% of the prokaryotic biomass studies and 34% of the prokaryotic density studies, the authors of the original publications did not report exact geographical coordinates (latitude, longitude) of the sampling stations. In these cases, I approximated the sampling locations using Google Maps based on maps from the original publications and indicated this with the label “approximated location”.
Prokaryotic biomasses were often not directly measured, but determined by extraction of bacterial adenosine triphosphate (ATP), extraction of bacterial phospholipid-derived fatty acid (PLFA), or by measuring prokaryotic densities. Subsequently, the authors of the original publications converted these data to prokaryotic biomasses using conversion factors (Table 2).
For cases where the prokaryotic biomasses and densities were not reported in the text or in tables, but were shown in figures, I extracted the data using ImageJ41.
Data Records
The “ProkaBioDen database” is an open access database in the Dryad Digital Repository and contains two txt.files, i.e., the List of studies for ProkaBio database and the List of studies for ProkaDen database, and two xlsx.files, i.e., the file ProkaBio database and the file ProkaDen database38. The List of studies files report all studies in alphabetical order (prokaryotic biomasses: 1,300 studies:, prokaryotic densities: 1,104 studies) that I identified in the “Identification” step of the systematic review after I eliminated duplicates. Each data entry in the “ProkaBioDen database” includes information about the region and the ocean where the samples were taken, the geographical location (latitude, longitude), the water depth (in m), and the depth range after Dunne et al.42. The authors of said study classified the ocean into near-shore areas from 0 to 50 m water depth, continental shelves from >50 to 200 m water depth, continental slopes from >200 to 2,000 m water depth, and continental rises and abyssal plains >2,000 m depth. The database includes biomass and density records for individual sediment layers and information about the thickness of said sediment layers and its specific upper and lower boundaries when a layer was sliced horizontally, but also biomass and density records for vertically integrated sediment profiles. Additionally, the database contains information about sediment type, median sediment grain size (µm), sediment density (g cm−3), and porosity, and whether prokaryotic densities were reported for total prokaryotes, Bacteria, or Archaea.
Technical Validation
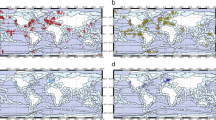
In the database, 40% of the benthic prokaryotic biomass samples originated in the Mediterranean Sea, 34% in the Atlantic, and 11% in the Arctic Ocean (Fig. 2). Most benthic prokaryotic density samples were taken in the Mediterranean Sea (42%), the Atlantic (27%), and the Arctic Ocean (15%), and also benthic Bacteria and Archaea densities were mainly sampled in the Mediterranean Sea (Bacteria: 62%, Archaea: 65%) and the Atlantic Ocean (Bacteria: 15%, Archaea: 17%) (Fig. 3). Both, benthic prokaryotic biomasses and densities were predominantly sampled in the northern hemisphere north of 1°N (biomass: 87%, density: 90%), whereas the southern hemisphere was seriously undersampled (Fig. 4 left panel and Fig. 5). Almost no samples were collected in the Indian Ocean (biomass: 7%, density: 1%) and the Southern Ocean (biomass: 2%, density: 1%). Hence, benthic prokaryote samples are biased towards the northern hemisphere and particularly towards the Mediterranean Sea and the North Atlantic.
Benthic prokaryotic biomasses were mostly quantified in the near-shore areas at <50 m water depth (54% of all samples, Fig. 4 right panel) that encompass 2% of the global ocean floor42. In comparison, only 15% of all benthic prokaryotic biomass samples were taken at the continental rise/in abyssal plains that contribute 89% to the global ocean floor area42. Benthic prokaryotic densities were sampled to 39% in near-shore areas, to 8% at continental shelves, to 31% at continental slopes, and to 22% at the continental rise/in abyssal plains (Fig. 6). Benthic Bacteria and Archaea density samples were mostly taken at >200 m water depth (i.e., continental slope: 34 and 31%, respectively; continental rise and abyssal plains: 45 and 50%, respectively).
About half of the benthic prokaryotic biomass (52%) and two-third of the density (67%) records were surface sediment records. 32% (benthic prokaryotic biomass) to 48% (benthic prokaryotic density) of these surface sediment layers stretched from 0 to 1 cm. The thinnest sediment layers had thicknesses of 0.3 cm and the thickest sediment layers reached to 20 cm below the sediment surface.
References
Madigan, M., Martinko, J., Stahl, D. & Clark, D. Brock - Biology of Microorganisms. (Pearson Education, Inc., 2012).
Jørgensen, B. B. & Boetius, A. Feast and famine — microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5, 770–781 (2007).
Bak, R. P. M. & Nieuwland, G. Seasonal variation in bacterial and flagellate communities of deep-sea sediments in a monsoonal upwelling system. Deep. Res. II 44, 1281–1292 (1997).
Danovaro, R., Corinaldesi, C., Rastelli, E. & Dell’Anno, A. Towards a better quantitative assessment of the relevance of deep-sea viruses, Bacteria and Archaea in the functioning of the ocean seafloor. Aquat. Microb. Ecol. 75, 81–90 (2015).
Rex, M. A. et al. Global bathymetric patterns of standing stock and body size in the deep-sea benthos. Mar. Ecol. Prog. Ser. 317, 1–8 (2006).
Pozzato, L., van Oevelen, D., Moodley, L., Soetaert, K. & Middelburg, J. J. Sink or link? The bacterial role in benthic carbon cycling in the Arabian Sea’s oxygen minimum zone. Biogeosciences 10, 6879–6891 (2013).
Glud, R. N. & Mathias, M. Virus and bacteria dynamics of a coastal sediment: Implication for benthic carbon cycling. Limnol. Oceanogr. 49, 2073–2081 (2004).
Sweetman, A. K. et al. Key role of bacteria in the short‐term cycling of carbon at the abyssal seafloor in a low particulate organic carbon flux region of the eastern Pacific Ocean. Limnol. Oceanogr. 64, 694–713 (2019).
van Oevelen, D., Moodley, L., Soetaert, K. & Middelburg, J. J. The trophic significance of bacterial carbon in a marine intertidal sediment: Results of an in situ stable isotope labeling study. Limnol. Oceanogr. 51, 2349–2359 (2006).
Valentine, D. L. Emerging topics in marine methane biogeochemistry. Ann. Rev. Mar. Sci. 3, 147–171 (2011).
Molari, M., Manini, E. & Dell’Anno, A. Dark inorganic carbon fixation sustains the functioning of benthic deep-sea ecosystems. Global Biogeochem. Cycles 27, 212–221 (2013).
Middelburg, J. J. et al. The fate of intertidal microphytobenthos carbon: An in situ 13C-labeling study. Limnol. Oceanogr. 45, 1224–1234 (2000).
Hensen, C., Zabel, M. & Schulz, H. N. Benthic cycling of oxygen, nitrogen and phosphorus. in Marine Geochemistry (eds. Schulz, H. D. & Zabel, M.) 207–240 (Springer-Verlag, 2006).
Dalsgaard, T., Thamdrup, B. & Canfield, D. E. Anaerobic ammonium oxidation (anammox) in the marine environment. Res. Microbiol. 156, 457–464 (2005).
Canfield, D. E., Kristensen, E. & Thamdrup B. The Nitrogen Cycle. in Aquatic Geomicrobiology vol. 48, 205–267 (Elsevier Inc., 2005).
Schink, B. & Friedrich, M. Phosphite oxidation by sulphate reduction. Nature 406, 37 (2000).
Canfield, D. E., Kristensen, E. & Thamdrup, B. The phosphorus cycle. in Aquatic Geomicrobiology vol. 48, 419–440 (Elsevier Inc., 2005).
Jørgensen, B. B. A Thiosulfate Shunt in the Sulfur Cycle of Marine Sediments. Science 249, 152–154 (1990).
Jørgensen, B. B. & Bak, F. Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark). Appl. Environ. Microbiol. 57, 847–856 (1991).
Thamdrup, B., Finster, K., Hansen, J. W. & Bak, F. Bacterial disproportionation of elemental sulfur coupled to chemical reduction of iron or manganese. Appl. Environ. Microbiol. 59, 101–108 (1993).
Jørgensen, B. B., Findlay, A. J. & Pellerin, A. The biogeochemical sulfur cycle of marine sediments. Front. Microbiol. 10, 849 (2019).
Glud, R. N. Oxygen dynamics of marine sediments. Mar. Biol. Res. 4, 243–289 (2008).
van Oevelen, D., Middelburg, J. J., Soetaert, K. & Moodley, L. The fate of bacterial carbon in an intertidal sediment: Modeling an in situ isotope tracer experiment. Limnol. Oceanogr. 51, 1302–1314 (2006).
Guilini, K. et al. Nutritional importance of benthic bacteria for deep-sea nematodes from the Arctic ice margin: Results of an isotope tracer experiment. Limnol. Oceanogr. 55, 1977–1989 (2010).
Baldrighi, E. et al. From microbes to macrofauna: An integrated study of deep benthic communities and their response to environmental variables along the Malta Escarpment (Ionian Sea). Sci. Mar. 77, 625–639 (2013).
Llobet-Brossa, E., Rosselló-Mora, R. & Amann, R. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl. Environ. Microbiol. 64, 2691–2696 (1998).
Lösekann, T. et al. Diversity and abundance of aerobic and anaerobic methane oxidizers at the Haakon Mosby Mud Volcano, Barents Sea. Appl. Environ. Microbiol. 73, 3348–3362 (2007).
Hollingsworth, A. L., Jones, D. O. B. & Young, C. R. Spatial variability of abyssal nitrifying microbes in the North-Eastern Clarion-Clipperton Zone. Front. Mar. Sci. 8, 663420 (2021).
Bar-On, Y. M., Phillips, R. & Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. 115, 6506–6511 (2018).
Noffke, N., Gerdes, G. & Klenke, T. Benthic cyanobacteria and their influence on the sedimentary dynamics of peritidal depositional systems (siliciclastic, evaporitic salty, and evaporitic carbonatic). Earth-Science Rev. 62, 163–176 (2003).
Nielsen, L. P., Risgaard-Petersen, N., Fossing, H., Christensen, P. B. & Sayama, M. Electric currents couple spatially separated biogeochemical processes in marine sediment. Nature 463, 1071–1074 (2010).
Burdorf, L. D. W. et al. Long-distance electron transport occurs globally in marine sediments. Biogeosciences 14, 683–701 (2017).
Bjerg, J. T. et al. Long-distance electron transport in individual, living cable bacteria. Proc. Natl. Acad. Sci. 115, 5786–5791 (2018).
Meysman, F. J. R. Cable bacteria take a new breath using long-distance electricity. Trends Microbiol. 26, 411–422 (2018).
Pfeffer, C. et al. Filamentous bacteria transport electrons over centimetre distances. Nature 491, 218–221 (2012).
Danovaro, R., Molari, M., Corinaldesi, C. & Dell’Anno, A. Macroecological drivers of archaea and bacteria in benthic deep-sea ecosystems. Sci. Adv. 2, e1500961 (2016).
Molari, M. & Manini, E. Reliability of CARD-FISH procedure for enumeration of Archaea in deep-sea surficial sediments. Curr. Microbiol. 64, 242–250 (2012).
Stratmann, T. The ProkaBioDen database, a global database of benthic prokaryotic biomasses and densities in the marine realm. Dryad Digital Repository https://doi.org/10.5061/dryad.wm37pvmnv (2022).
Wei, C.-L. et al. Global patterns and predictions of seafloor biomass using random forests. PLoS One 5, e15323 (2010).
Moher, D., Liberati, A., Tetzlaff, J. & Altman, D. G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 6, e1000097 (2009).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Dunne, J. P., Sarmiento, J. L. & Gnanadesikan, A. A synthesis of global particle export from the surface ocean and cycling through the ocean interior and on the seafloor. Global Biogeochem. Cycles 21, GB4006 (2007).
Stratmann, T. The ProkaBioDen database, a global database of benthic prokaryotic biomass and densities in the marine realm - R code. Zenodo https://doi.org/10.5281/zenodo.6264038 (2022).
Bjørnsen, P. Bacterioplankton growth yield in continuous seawater cultures. Mar. Ecol. Prog. Ser. 30, 191–196 (1986).
Børsheim, K. Y. & Bratbak, G. Cell volume to cell carbon conversion factors for a bacterivorous Monas sp. enriched from seawater. Mar. Ecol. Prog. Ser. 36, 171–175 (1987).
Fuhrman, J. A. & Azam, F. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: Evaluation and field results. Mar. Biol. 66, 109–120 (1982).
Fukuda, R., Ogawa, H., Nagata, T. & Koike, I. Direct determination of carbon and nitrogen contents of natural bacterial assemblages in marine environments. Appl. Environ. Microbiol. 64, 3352–3358 (1998).
Grossmann, S. & Reichardt, W. Impact of Arenicola marina on bacteria in intertidal sediments. Mar. Ecol. Prog. Ser. 77, 85–93 (1991).
Simon, M. & Azam, F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51, 201–213 (1989).
Kröncke, I., Tan, T. & Stein, R. High benthic bacteria standing stock in deep Arctic basins. Polar Biol. 14, 423–428 (1994).
Lee, S. & Fuhrman, J. A. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl. Environ. Microbiol. 53, 1298–1303 (1987).
Meyer-Reil, L.-A., Faubel, A., Graf, F. & Thiel, H. Aspects of benthic community structure and metabolism. in Sea water-interactions in coastal waters (eds. Rumohr, J., Walger, E. & Zeitschel, B.) 69–110 (Springer Verlag, 1987).
Moriarty, D.J. W. Techniques for estimating bacterial growth rates and production of biomass in aquatic environments. in Techniques in Microbial Ecology (eds. Grigorova, R. & Norris, J. R.) vol. 22, 211–234 (Elsevier, 1990).
Norland, S. The relationship between biomass and volume of bacteria. in Handbook of methods in aquatic microbial ecology 303–307 (CRC Press, 1993).
Torréton, J.-P. Importance des bactéries hétérotrophes aérobies dans une lagune eutrophe tropicale (Lagune Ebrié, Côte d’Ivoire). PhD-Thesis (Université d’Aix-Marseille II, 1991).
Børsheim, K. Y., Bratbak, G. & Heldal, M. Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl. Environ. Microbiol. 56, 352–356 (1990).
Duyl FCvan Ko, F. & van Kopp, A. Seasonal patterns of bacterial production and biomass in intertidal sediments of the western Dutch Wadden Sea. Mar. Ecol. Prog. Ser. 59, 249–261 (1990).
Williams, P. M. & Carlucci, A. F. Bacterial utilization of organic matter in the deep sea. Nature 262, 810–811 (1976).
Bratbak, G. & Dundas, I. Bacterial dry matter content and biomass estimations. Appl. Environ. Microbiol. 48, 755–757 (1984).
Christaki, U. Régulations trophiques et interactions microbiennes en milieu pélagique, relation avec les métazoires. (Université d’Aix-Marseille II, 1995).
Danovaro, R. Prokaryotic biomass in marine sediments. in Methods for the Study of Deep-Sea Sediments, Their Functioning and Biodiversity 331–336 (CRC Press - Taylor & Francis Group, 2010).
Deming, J. W. & Yager, P. L. Natural Bacterial Assemblages in Deep-Sea Sediments: Towards a Global View. in Deep-Sea Food Chains and the Global Carbon Cycle 11–27, https://doi.org/10.1007/978-94-011-2452-2_2 (Springer Netherlands, 1992).
Edler, L. Recommendations on the method for marine biological studies in the Baltic Sea. Phytoplankton and chlorophyll. in The Baltic Marine Biologist Working Group (ed. Edler, L.) 1–38 (1979).
Fichez, R. Composition and fate of organic matter in submarine cave sediments; implications for the biogeochemical cycle of organic carbon. Oceanol. Acta 14, 369–377 (1991).
Fry, J. C. Determination of biomass. in Methods in Aquatic Bacteriology 27–72 (John Wiley & Sons, 1990).
Boschker, H. T. S. & Middelburg, J. J. Stable isotopes and biomarkers in microbial ecology. FEMS Microbiol. Ecol. 40, 85–95 (2002).
Brinch-Iversen, J. & King, G. M. Effects of substrate concentration, growth state, and oxygen availability on relationships among bacterial carbon, nitrogen and phospholipid phosphorus content. FEMS Microbiol. Lett. 74, 345–355 (1990).
Findlay, R. H. & Dobbs, F. C. Quantitative description of microbial communities using lipid analysis. in Handbook of Methods in Aquatic Microbial Ecology (eds. Kemp, P. F., Sherr, B. F., Sherr, E. B. & Cole, J. J.) 271–284 (Lewis Publishers, 1993).
Oakes, J. M., Eyre, B. D., Middelburg, J. J. & Boschker, H. T. S. Composition, production, and loss of carbohydrates in subtropical shallow subtidal sandy sediments: Rapid processing and long-term retention revealed by 13 C-labeling. Limnol. Oceanogr. 55, 2126–2138 (2010).
Rajendran, N., Suwa, Y. & Urushigawa, Y. Distribution of phospholipid ester-linked fatty acid biomarkers for bacteria in the sediment of Ise Bay, Japan. Mar. Chem. 42, 39–56 (1993).
Karl, D. M. Cellular nucleotide measurements and applications in microbial ecology. Microbiol. Rev. 44, 739–796 (1980).
Raghukumar, S., Chandramohan, D. & Ramaiah, N. Contribution of the thraustochytrid Corallochytrium limacisporum Raghu-kumar to microbial biomass in coral reef lagoons. Indian. J. Mar. Sci. 16, 122–125 (1987).
Acknowledgements
This study received funding by JPI Oceans – Impacts of deep-sea nodule mining project “Mining Impact 2” from the Dutch Research council (NWO-ALW grant 856.18.003) and from the research programs NWO-Rubicon (grant 019.182EN.012) and NWO Talent Program Veni (grant VI.Veni.212.211).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author declares no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stratmann, T. The ProkaBioDen database, a global database of benthic prokaryotic biomasses and densities in the marine realm. Sci Data 9, 179 (2022). https://doi.org/10.1038/s41597-022-01281-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-022-01281-x
- Springer Nature Limited