Abstract
The seeds of Arabidopsis thaliana become encapsulated by a layer of mucilage when imbibed. This polysaccharide-rich hydrogel is constituted of two layers, an outer layer that can be easily extracted with water and an inner layer that must be examined in situ in order to study its properties and structure in a non-destructive manner or disintegrated through hydrolysis or physical means in order to analyze its constituents. Mucilage production is an adaptive trait and we have exploited 19 natural accessions previously found to have atypical and varied outer mucilage characteristics. A detailed study using biochemical, histological and Time-Domain NMR analyses has been used to generate three related datasets covering 33 traits measured in four biological replicates. This data will be a rich resource for genetic, biochemical, structural and functional analyses investigating mucilage constituent polysaccharides or their role as adaptive traits.
Measurement(s) | intrinsic viscosity • galacturonic acid • seed and inner mucilage layer width • Transverse Spin Relaxation Time • Transverse Spin Relaxation intensity • mucilage • neutral sugar content • mass per seed • sample mass |
Technology Type(s) | size-exclusion chromatography • colorimetry • immunolabeling method • confocal microscopy • nuclear magnetic resonance spectroscopy • Time-domain NMR • biochemical, histological and Time-Domain NMR analyses • biochemical method using orcinol • microbalance |
Sample Characteristic - Organism | Arabidopsis thaliana |
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.13320593
Similar content being viewed by others
Background & Summary
Seeds of a number of plant species, including Arabidopsis (Arabidopsis thaliana), become surrounded by sticky mucilage when imbibed. A range of roles has been suggested for this polysaccharide-rich hydrogel, such as aiding germination, dispersion, seedling growth or interaction with soil microorganisms reviewed by Yang et al.1. In the reference accession for the model plant Arabidopsis, Columbia (Col-0), the major component of mucilage is the pectin rhamnogalacturonan I (RG-I), which is organized in two distinct layers that differ in their polysaccharide composition and structure2. This suggests that the outer water-soluble and inner adherent layer could perform different ecophysiological functions2.
Although Arabidopsis is used widely as a model for geneticists, it is a widespread weed whose native range covers most of Europe to central Asia. As mucilage is an adaptive trait its functional advantages are likely to influence the dynamics and evolution of natural Arabidopsis populations. Natural variation in the outer water-soluble layer of Arabidopsis mucilage was recently reported for 306 natural Arabidopsis accessions3,4. Large variations were observed in the amount and properties of the constituent polysaccharides. Nonetheless, the composition of the outer mucilage layer was stable between genotypes with RG-I always being the major constituent of outer mucilage. Analysis of the inner mucilage layer is more complex as the polysaccharides are tightly adhered to the seed surface and hydrolysis of the biopolymers into fragments is required. To date, the detailed composition of the inner mucilage layer has only been determined for a limited number of accessions used to generate induced mutant collections (Col-0, Col-2)2,5. As the inner mucilage layer can be observed using the cytochemical stain ruthenium red the visual aspect of inner mucilage was previously examined for 280 accessions6. This method identified fifty variants that differed in the size of the inner mucilage layer. Observation with ruthenium red can, however, only give an indication of major differences in the width of the inner mucilage layer and this is not necessarily an indication of more or less polysaccharides as the hydrophilic properties, molar mass and conformation of the pectin polymers can alter the volume they occupy7,8. Moreover, seed size can vary between natural variants and the volume of the mucilage layer may appear bigger or smaller due to these differences. Furthermore, loss of adhesion of inner layer pectin in the muci70 mutant, was recently proposed to be linked to modified macromolecular characteristics as outer mucilage RG-I polymers were shorter in this mutant4, which suggests that polymer length contributes to adhesion through intermolecular entanglement. This highlights that much is still unknown concerning the physicochemical requirements for the formation of the inner layer.
To study the variability in mucilage traits in more detail, we have carried out a detailed characterization of both inner and outer mucilage traits for 19 natural variants identified previously as exhibiting atypical outer mucilage macromolecular properties4, these included the reference accession Col-0 (Table 1).
Histological, biochemical and Time-Domain NMR (TD-NMR) analyses were used to generate three datasets: dataset 1 contains 182 899 variables for 33 mucilage and seed traits, dataset 2 comprises raw NMR data files and dataset 3 is 4560 values measured following microscope acquisition. In addition to confirming previous values for outer mucilage composition and intrinsic viscosity (IV), the size of the hydrated inner mucilage layer, seed and mucilage width were each measured on images acquired following labeling of the seed surface with a cellulose specific stain (DR23) and the periphery of the inner mucilage layer with an antibody recognizing RG-I epitopes (INRA RU19). The amount of the major RG-I sugars in the inner layer was determined following hydrolysis of pectin polymers with rhamnogalacturonan hydrolase. Finally, to obtain information about the mobility of water in interaction with macromolecules in different compartments of seed and mucilage, TD-NMR was carried out on dry seeds and over a period of 23 h of imbibition using either intact seeds or seeds pre-treated to remove outer mucilage. The different steps in data production are summarized in Fig. 1.
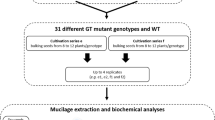
Schematic representation of the production of the datasets 1, 2 and 3 for mucilage and seed traits for Arabidopsis accessions with atypical outer mucilage. Data was generated using four seed lots generated from bulks of independent plants that had been produced at two different times corresponding to series c or d. Analyses of the sugar composition, macromolecular properties, water mobility during imbibition, mucilage and seed width, for the 19 accessions generated raw and treated data available in three datasets.
Methods
Plant material and growth conditions
The 19 accessions used in this study (Table 1) were obtained from the Versailles Arabidopsis stock center (http://publiclines.inra.fr/naturalAccession/index) and are listed by their Versailles identification number in a four-digit format (i.e. 0001 for accession AV1). These were chosen from 306 outlier accessions analysed previously4 and included accessions with extreme phenotypes for each of the four macromolecular traits examined, with certain also exhibiting atypical mucilage amounts or composition. Plants were grown in a chamber with 65% relative humidity and 170 µmol m−2 s−1 of light and for the first three weeks with a 16 h photoperiod at 21 °C and 8 h dark at 18 °C, followed by 6 weeks at 6 °C with an 8 h photoperiod to synchronise flowering when subsequently returned to a 16 h photoperiod. Plants were grown in compost (Tref substrates) following a randomized sowing plan in two independent series of plants grown together, with twenty-four plants of each genotype per series. To differentiate these from the seed stocks produced previously to study outer mucilage traits3 these were termed series c, grown from November 2014 to March 2015 and series d from April to August 2015. Four independent biological replicates were produced for analyses by bulking seed harvested from different plants. These were assigned sample codes c1, c2, d1 and d2 corresponding to two independent lots derived from bulks of 10 to 12 plants from series c or d, respectively (Fig. 2). Ten seeds from each lot were weighed using a Sartorius M2P microbalance.
Biochemistry
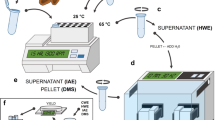
Outer mucilage (sample type 0) was water-extracted (4 mL) from seeds (200 mg) and analysed as described previously3 (Fig. 3a). Briefly, after 3 h of head-over-tail mixing at 20 °C and centrifugation (8000 g, 5 min), water extracts were filtered through a disposable glass microfilter (13 mm diameter, 2.7 μm pore size) and analysed colorimetrically for galacturonic acid (GalA) and total neutral sugar (NS) contents10,11. Both quantification methods used are based on the ability of sugars to be converted into furfuric derivates in the presence of hot sulfuric acid. Furfuric derivates can then condense with various phenolic compounds to produce a colored complex that can be quantified colorimetrically. Acidic sugars can be quantified specifically using meta-hydroxy biphenyl (mphenyl-phenol or 3 phenyl-phenol)12 while neutral sugars can be quantified using orcinol (3,5 dihydroxytoluene)13. These methods have been automated10,11. Extracts were also analysed for their intrinsic viscosity (IV) by high-performance size exclusion chromatography (HPSEC) coupled to a differential refractometer and a differential pressure viscometer. Seeds remaining after water-extraction were rinsed three times with 8 mL of water and then used to extract inner mucilage (sample type 1) (Fig. 3a). Water volume was adjusted to 2 mL and 2 mL of 100 mM sodium acetate buffer pH 4.5 were added. Twenty µL of rhamnogalacturonan hydrolase at 10 mg mL−1 in cold 50 mM sodium acetate buffer pH 4.5 were added. The rhamnogalacturonan hydrolase (EC 3.2.1.171, glycoside hydrolase family 28) used was purified from a technical preparation of Aspergillus aculeatus as described in Schols et al.14. The reaction was incubated at 40 °C for 16 h. After centrifugation (8000 g, 5 min), supernatants were carefully removed for GalA analysis by colorimetry.
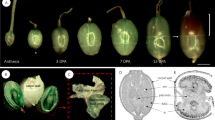
Summary of methodology and experimental workflow used to generate dataset. Extraction procedure used to generate samples for biochemical and NMR analyses (a), picture of NMR tubes filled with dried or imbibed seeds (b), confocal optical section through a seed immunolabelled with the antibody (INRA RU1) that binds to rhamnogalacturonan 1 with cellulose co-stained with Direct Red 23 showing how mucilage and seed width (MSW) or seed width (SW) were measured (c).
Histochemical staining and immunolabeling of inner mucilage
Seed and inner mucilage layer size (Fig. 3c) were determined following immunolabelling and staining of seeds with an anti-RG-I antibody (INRA-RU19) and the cellulose specific fluorescent dye Direct Red 23 (DR23) essentially as previously described15, except that 1% (w/v) powdered milk was used for the blocking solution and seeds were mounted for observation directly in the DR23 counterstain. As outer mucilage is lost during the immunolabelling procedure seeds analysed correspond to sample type 5. The INRA-RU1 antibody labels the periphery of the inner mucilage while DR23 labels the cellulose within the inner mucilage and the cell walls on the seed surface. Observations was performed with a Zeiss LSM710 confocal microscope using 488 nm or 561 nm lasers to excite Alexa Fluor 488® or DR23, respectively. Fluorescence emission was detected between 500 and 550 nm for Alexa fluor 488® and 565 and 640 nm for DR23. For each seed lot, measurements were obtained from 30 seeds using Zen software (dataset 316) and the mean value calculated (dataset 117).
Time-domain NMR
Seeds were either analysed directly (sample type 3 or 6) or after removal of water-soluble mucilage (sample type 2 or 5). The latter were prepared by mixing 350 mg of seeds in 10 mL of water for 3 h at 20 °C. Extracts were then centrifuged at 8000 g for 3 min and supernatants carefully removed. Seeds were rinsed four times with 10 mL of water and freeze-dried. Dehydrated seeds with (sample type 3) or without soluble mucilage (sample type 2) were stored at room temperature before being analysed by TD-NMR in dry state or imbibed in water (Fig. 3b).
A Time-Domain spectrometer (Minispec BRUKER, Germany) operating at 0.47 T (resonance frequency of 20 MHz) was used to measure T2 relaxation times. The temperature of samples was regulated at 20 °C with a temperature control device (±0.1 °C) connected to a calibrated optical fiber (Optoprim; France). The NMR tubes were filled with dry seeds or dry seeds and water (Fig. 3b) as previously described18. Tubes were then weighed and hermetically sealed. Acquisitions of T2 were carried out first on dry seeds and then from 3 min (t0) to 23 h (H23) of imbibition. The FID-CPMG sequence used the following parameters: a 90° pulse close to 2.8 µs, a dwell time of 0.4 µs for a FID duration of 150 µs, 16 scans, a recycle delay of 5 s, an echo time of 0.2 ms with 5000 or 16000 data points, depending on the genotype and/or the seed state (dry, with or without soluble mucilage).
Transverse relaxation data were analyzed using the following model:
where T2i and T2j are the proton relaxation times of the solid phase of seeds and those of the more mobile populations (water and oil protons), respectively. The corresponding NMR signal intensities were Ii and Ij. Dry seeds samples were characterized by four T2 components (T2_1, _2, _3, _4). Compared to previous studies performed on the reference accession Col-0 and two Arabidopsis insertion mutants18,19, the present analyses acquired with a longer FID signal, made it possible to identify an additional T2 component at around 100 µs for imbibed seeds termed T2_2a while the previously identified component was termed T2_2b so that in total six T2 (T2_1, _2a, _2b, _3, _4, _5) were identified for imbibed seeds. Seeds where outer mucilage had been previously removed by extraction (see above) resulted in the loss of the longer T2 relaxation time T2_5 and the splitting of T2_3 into T2_3a and T2_3b (Table 2). Each of these T2 components could be assigned to populations of protons in water or oil having different mobilities and proportions18.
Data Records
All three data records use the same sample nomenclature for input and this is explained in Fig. 2 and Table 2.
Data record 1
The dataset described here contains values obtained from biochemical, microscopy and TD-NMR analyses and has been published on the Data INRAE site17. The raw data used to generate mean values for MSW and SW or values for time T2 and intensity I of components 1, 2a, 2b, 3, 4 and 5 at t0 and H23 and for the sample type 6, are available in datasets described in data records 3 and 2, respectively. An overview of the data set is in shown in Table 3 with the following nine columns:
-
1.
sample_code: the sample code (see Fig. 2)
-
2.
accession: the accession number
-
3.
cultivation_series: c or d
-
4.
seed_lot: Biological replicate 1 or 2
-
5.
sample type: the sample type analyzed (0, 1, 2, 3, 5, 6) (see Fig. 2)
-
6.
variable: the code of the variable (see Table 2 for the description)
-
7.
value: the measured value
-
8.
date: the date of acquisition in format (year-month-day) or NA if no time points.
-
9.
time: the time of acquisition in format (hour:minute:second) or NA if no time points
Data record 2
Dataset 2 consists of NMR raw data files whose name is the combination of the dataset number, the seed lot (2), the cultivation series (c or d) followed by “imb-“, the imbibition time (t0 or H23) and the accession code. Data correspond to the TD-NMR raw data of intact seeds imbibed in water (code 6) at initial (imb-t0) or final (imb-24H) imbibition time. The format is supplier imposed (.dps) but can be read by any application using tabular formats. The files comprise three columns: the first indicates the total number of recorded data points, the second acquisition time (in ms), while the third column gives the NMR signal intensity (in arbitrary units). Data were recorded using a Time-Domain spectrometer (Minispec BRUKER, Germany) operating at 0.47 T (resonance frequency of 20 MHz) at 20 °C. The Free Induction Decay-Car Purcell Meiboom Gill (FID-CPMG) pulse train acquisition sequence used the following parameters: a 90° pulse close to 2.8 µs, a dwell time of 0.4 µs for a FID duration of 150 µs, 16 scans, a recycle delay of 5 s, an echo time of 0.2 ms with 8000 (imb-t0) or 5000 (imb-24H) data points. The dataset is accessible on the Data INRAE site19.
Data record 3
The values for individual measurements of the two traits seed width and mucilage and seed width used to generate the mean values in dataset 1 are listed as shown in the overview Table 4. In addition to the nine columns found in dataset 1 an additional column indicates the number of the seed measured. The dataset comprises 4560 values corresponding to measurements of 30 seeds from each seed lot. The dataset is accessible on the Data INRAE site16.
Technical Validation
The technical quality of the dataset was validated through the use of four biological replicates of seed lots in the different analyses; replicates were produced from plants cultivated in a randomised format to ensure that any environmental effects from their position within the growth chamber, or from plant neighbours, were minimised. The reproducibility of results was examined for biochemical and histological analyses based on the variation between the four replicates with the highest variation observed being under 7% (Table 5). Furthermore, certain analyses carried out on outer mucilage are equivalent to those previously carried out by Poulain et al.3, notably for GalA, NS and IV, and these were compared to validate the reliability of measurements and seed lots (Fig. 4). An excellent proportionality between values was observed for all 3 variables with a R2 of 0.82, 0.80 and 0.96, respectively. For quantification of sugar concentrations, a standard curve was established using standard solutions of Rha or GalA at 20, 40, 60, 80, and 100 µg/ml, which were measured both before and after a series of samples to confirm technical rigour. HP-SEC columns were calibrated for IV using both a calibrant and a standard sample passed at the beginning, middle and end of a series of samples to check that no drift occurred over time.
Comparison of outer mucilage variables obtained in this study to previously obtained values to validate the reliability of measurements obtained. Mean values for GalA (mg/g seed), NS (mg/g seed) and IV (mL/g) obtained for samples analysed here (y axis) are plotted versus those previously obtained from seed lots generated from plants cultivated independently for the same accessions3 (x axis). Error bars, ±SE (n = 4).
The NMR spectrometer underwent a daily control procedure in accordance with the manufacturer’s recommendations. In addition, the T2 relaxation time and intensity of a reference sample (mineral oil) were controlled each day at spectrometer temperature (around 40 °C). The optical fiber used to regulate sample temperature to 20 °C was calibrated before a series of measurements. In order to validate NMR results, data processing were performed using two different methods that were expected to converge: discrete20 and continuous maximum entropy (MEM21,22). Each of these methods was performed using two different codes listed below (Table 6). Moreover, the T2 times and amplitudes obtained with Col-0 samples used here were compared for reproducibility with those obtained previously with different Col-0 samples18. The reliable acquisition of images by the confocal microscope is certified through annual recalibration of the system, parfocality and light, head scan lens focalisation and collimator by Zeiss, France.
Code availability
The different available software and the versions used to acquire and process data presented in the datasets are summarized in Table 6.
References
Yang, X., Baskin, J., Baskin, C. & Huang, Z. More than just a coating: ecological importance, taxonomic occurrence and phylogenetic relationships of seed coat mucilage. Perspectives in Plant Ecology, Evolution and Systematics 14, 434–442, https://doi.org/10.1016/j.ppees.2012.09.002 (2012).
Macquet, A., Ralet, M. C., Kronenberger, J., Marion-Poll, A. & North, H. M. In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiology 48, 984–999 (2007).
Poulain, D., Botran, L., North, H. M. & Ralet, M. C. Composition and physicochemical properties of outer mucilage from seeds of Arabidopsis natural accessions. AoB Plants 11, plz031, https://doi.org/10.1093/aobpla/plz031 (2019).
Fabrissin, I. et al. Natural Variation Reveals a Key Role for Rhamnogalacturonan I in Seed Outer Mucilage and Underlying Genes. Plant Physiol 181, 1498–1518, https://doi.org/10.1104/pp.19.00763 (2019).
Huang, J. et al. The Arabidopsis transcription factor LUH/MUM1 is required for extrusion of seed coat mucilage. Plant Physiology 156, 491–502, https://doi.org/10.1104/pp.111.172023 (2011).
Voiniciuc, C. et al. Extensive Natural Variation in Arabidopsis Seed Mucilage Structure. Front Plant Sci 7, 803, https://doi.org/10.3389/fpls.2016.00803 (2016).
Macquet, A. et al. A naturally occurring mutation in an Arabidopsis accession affects a beta-D-galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. The Plant Cell 19, 3990–4006, https://doi.org/10.1105/tpc.107.050179 (2007).
Ralet, M. C. et al. Reduced number of homogalacturonan domains in pectins of an Arabidopsis mutant enhances the flexibility of the polymer. Biomacromolecules 9, 1454–1460, https://doi.org/10.1021/bm701321g (2008).
Ralet, M. C., Tranquet, O., Poulain, D., Moise, A. & Guillon, F. Monoclonal antibodies to rhamnogalacturonan I backbone. Planta 231, 1373–1383, https://doi.org/10.1007/s00425-010-1116-y (2010).
Thibault, J.-F. Automatisation du dosage des substances pectiques par la méthode au métahydroxydiphényle. Lebensm Wiss Technol 12, 247–251 (1979).
Tollier, M. T. & Robin, J. P. Adaptation de la méthode à l’orcinol sulfurique au dosage automatique des glucides neutres totaux: conditions d’application aux extraits d’origine végétale. Ann Technol Agric 28, 1–15 (1979).
Blumenkrantz, N. & Asboe-Hansen, G. New methods for quantitative determination of uronic acids. Analytical Biochem 54, 484–489 (1973).
Brückner, J. Estimation of monosaccharides by the orcinol-sulphuric acid reaction. Biochem J 60, 200–205 (1955).
Schols, H. A., Geraerds, C. C. J. M., Searle-van Leeuwen, M. J. F., Kormelink, F. J. M. & Voragen, A. G. J. Rhamnogalacturonase: A novel enzyme that degrades the hairy regions of pectins. Carbohydrate Res 206, 105–115 (1990).
Voiniciuc, C. et al. MUCILAGE-RELATED10 Produces Galactoglucomannan That Maintains Pectin and Cellulose Architecture in Arabidopsis Seed Mucilage. Plant Physiol 169, 403–420, https://doi.org/10.1104/pp.15.00851 (2015).
Berger, A., Sallé, C. & North, H. M. Measurements of inner mucilage and seed width for Arabidopsis natural accessions - dataset 3. Portail Data INRAE https://doi.org/10.15454/LBUN4X (2021).
Cambert, M. et al. Seed mucilage traits for Arabidopsis thaliana natural accessions with atypical outer mucilage - dataset 1. Portail Data INRAE https://doi.org/10.15454/1MZ1ZC (2021).
Saez-Aguayo, S. et al. Local evolution of seed flotation in Arabidopsis. PLoS Genet 10, e1004221, https://doi.org/10.1371/journal.pgen.1004221 (2014).
Rondeau-Mouro, C. et al. T2 relaxation time raw data of water-imbibed Arabidopsis seeds - dataset 2. Portail Data INRAE https://doi.org/10.15454/EYABB2 (2021).
Marquardt, D. W. An Algorithm for Least-Squares Estimation of Nonlinear Parameters. Journal of the Society for Industrial and Applied Mathematics 11, 431–441 (1963).
Gull, S. F. In Maximum Entropy and Bayesian Methods: Cambridge, England, 1988 (ed J. Skilling) 53–71 (Springer Netherlands, 1989).
Skilling, J. Maximum Entropy and Bayesian Methods. J. Skilling edn, (Kluwer Academic Publishers, 1989).
Acknowledgements
This work was supported by the Agence Nationale de Recherche program (grant number: ANR-14-CE19-0001). The IJPB benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007). The NMR experiments were performed using the NMR facilities of the AgroScans Platform (Rennes, France).
Author information
Authors and Affiliations
Contributions
H.M.N. and C.S. produced seeds. M.C.R. performed biochemical analyses and A.B. histological analyses and measurements from acquired images. T.C. prepared seeds for NMR analyses of seeds without soluble mucilage (sample type 2). NMR analyses have been performed by M.C., S.E. and C.R.M., H.M.N., D.C., C.R.M. and M.C.R. formatted metadata and published data records in the datainrae repository. H.M.N., C.R.M. and M.C.R. wrote the manuscript. All authors read and improved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
About this article
Cite this article
Cambert, M., Berger, A., Sallé, C. et al. Datasets of seed mucilage traits for Arabidopsis thaliana natural accessions with atypical outer mucilage. Sci Data 8, 79 (2021). https://doi.org/10.1038/s41597-021-00857-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-021-00857-3
- Springer Nature Limited