Abstract
The emergence of new structures can often be linked to the evolution of novel cell types that follows the rewiring of developmental gene regulatory subnetworks. Vertebrates are characterized by a complex body plan compared to the other chordate clades and the question remains of whether and how the emergence of vertebrate morphological innovations can be related to the appearance of new embryonic cell populations. We previously proposed, by studying mesoderm development in the cephalochordate amphioxus, a scenario for the evolution of the vertebrate head mesoderm. To further test this scenario at the cell population level, we used scRNA-seq to construct a cell atlas of the amphioxus neurula, stage at which the main mesodermal compartments are specified. Our data allowed us to validate the presence of a prechordal-plate like territory in amphioxus. Additionally, the transcriptomic profile of somite cell populations supports the homology between specific territories of amphioxus somites and vertebrate cranial/pharyngeal and lateral plate mesoderm. Finally, our work provides evidence that the appearance of the specific mesodermal structures of the vertebrate head was associated to both segregation of pre-existing cell populations, and co-option of new genes for the control of myogenesis.
Similar content being viewed by others
Introduction
Chordates are an animal clade characterized by the presence of a notochord (in at least one stage of their life cycle)1 and that include vertebrates, tunicates (or urochordates), and cephalochordates (i.e. amphioxus). Even if tunicates are phylogenetically more closely related to vertebrates2 and share with them some morphological features absent in amphioxus3, they show developmental modalities and a genomic content and organization that have diverged considerably from the chordate ancestral state4. On the other hand, amphioxus exhibit relatively conserved morphological, developmental, and genomic characteristics, and represent a model of choice for studying chordate evolution and the emergence of vertebrate novelties5,6.
The gastrula of cephalochordates has two germ layers: the ectoderm, which forms the epidermis and the central nervous system, and the internal mesendoderm, which develops into mesodermal structures in the dorsal part, and into endodermal structures in the ventral region7. Unlike vertebrates, the mesoderm is first simply divided during neurulation into the axial territory forming the notochord, and the paraxial domain that becomes completely segmented into somites from the most anterior to the posterior part of the embryo. In vertebrates, in addition to the notochord and somites, the mesoderm is subdivided into other territories: the lateral plate mesoderm in the trunk that forms several structures among which part of the heart and circulatory system, blood cells, fin buds or excretory organs8; and the prechordal plate (axial) and cranial/pharyngeal (non-axial, unsegmented) mesoderm in the anterior region that form head muscles and part of the heart9,10. If we consider that the amphioxus mesoderm organization could resemble that of the chordate ancestor, these mesodermal territories represent vertebrate-specific traits that contributed to the acquisition of particular structures, including the complex vertebrate head.
Although the entire paraxial mesoderm of amphioxus is segmented into somites, which first form epithelial spheres during neurulation, these segments elongate during development towards the ventral region where the cells intermingle between the endoderm and the epidermis11. The orthologs of several genes expressed in the vertebrate lateral plate mesoderm (or derivatives) are expressed in this ventral region of the amphioxus somites, suggesting homology between these two embryonic territories11,12,13,14. This hypothesis was further supported by transgenesis experiments showing that the regulatory region of draculin, a gene specifically expressed in the entire lateral plate mesoderm of zebrafish, is able to drive expression of a reporter gene in cells of the ventral region of amphioxus somites15. On the other hand, two studies, based on electron microscopy and in situ hybridization, suggested the compartmentalization of amphioxus somites not into a myotome part and non-myotome part, but into four regions that would be homologous to the myotome, the sclerotome, the dermomyotome and the lateral plate mesoderm of vertebrates16,17. Finally, gene expression and functional data also showed that the first amphioxus somite pair differs from the other somites and that it contributes to the formation of the excretory organ of the larva and of putative hematopoietic cells14,18,19.
Concerning the axial mesoderm, although the notochord extends along the entire anteroposterior axis of the amphioxus, we and others have shown that the anterior region exhibits cellular behaviors and gene expression that differ from those of the central and posterior notochord20,21,22. Based on these observations and our previous work on the control of somitogenesis in amphioxus, we have proposed a multi-step scenario for the evolution of the vertebrate anterior mesoderm22,23. The first step consists of the segregation of the ventral mesoderm from the paraxial mesoderm and the loss of its segmentation. This implies that the ventral part of amphioxus somites is homologous to the vertebrate lateral plate mesoderm as previously suggested11,12,13,14,15,16,17. The second step corresponds to the loss of the paraxial mesoderm in the anterior part of the embryo. This would have enabled the relaxation of the developmental constraints imposed by the anterior somites and the remodeling of the axial and lateral plate mesoderm, resulting in the appearance of the prechordal plate and cranial/pharyngeal mesoderm. This would mean that i) the cranial/pharyngeal mesoderm has a lateral rather than paraxial origin, and partly shares a common developmental program with the amphioxus anterior somites and ventral part of posterior somites, and ii) the prechordal plate is in part homologous to the amphioxus anterior notochord.
Here we sought to explore the evolutionary origin of the vertebrate head mesoderm from a cell type perspective. In order to compare embryonic cell types between amphioxus and vertebrates, we conducted a scRNA-seq analysis of the Branchiostoma lanceolatum neurula (N3)24,25. The neurula stage shows the highest global transcriptional similarity with vertebrates26, corresponding to the chordate phylotypic stage27, and our cell atlas uncovers the gene expression signatures of most of the previously described embryonic territories at this stage. Concerning the mesodermal compartments, we found a cell population localized in the anterior part of the neurula and with a mixed profile between endoderm and notochord, supporting the existence of a transient prechordal plate-like structure in amphioxus22,28. We also show that cells of the first somite pair form a population with a transcriptomic profile different from the posterior somites, highlighting the peculiarity of this somitic pair. Moreover, these cells express orthologues of vertebrate genes expressed in both head and lateral plate mesoderm and their derivatives, bringing further support to our evolutionary scenario, and suggesting how, from pre-existing cell populations, new embryonic territories might have emerged in vertebrate anterior mesoderm. Finally, transgenic zebrafish experiments using regulatory regions of amphioxus Gata1/2/3, Tbx1/10 and Pitx also supports the lateral origin of the cranial/pharyngeal mesoderm and gives insights into how genes that were presumably not controlling muscle formation in the chordate ancestor were co-opted as master genes of the myogenesis program in the vertebrate head.
Results and discussion
A cell atlas of the amphioxus neurula stage embryo
To build a transcriptional cell atlas of the amphioxus neurula stage (N3), we applied MARS-seq29 to embryos at 21 hpf (hours post-fertilization, at 19 °C) (Fig. 1a). Briefly, cells were dissociated and alive single cells (calcein positive, propidium-iodide negative) were sorted into 384-well plates, followed by scRNA-seq library preparation. At this developmental stage, the embryo is made of around 3,000 cells and we sampled in total 14,586 single-cell transcriptomes, representing approximately a five-fold coverage. These cells were grouped into 176 transcriptionally coherent clusters (referred to as “metacells”30 (Fig. 1b, Supplementary Fig. 1a). Metacells were further assigned to a tissue/cell type by using transcriptional signatures of known marker genes: epidermis, endoderm, mesoderm, muscular somite and neural (Fig. 1c, Supplementary Fig. 2a). The proportion of cells assigned to each structure/germ layer was overall consistent with cell counting in 3D embryos reconstructed using confocal imaging of labeled nuclei followed by image segmentation, with more than half of the cells belonging to the epidermis (Fig. 1d).
a drawings of Mediterranean amphioxus developmental stages from the egg to the larva (with one open gill slit) stage. The developmental time (hours post fertilization, hpf) is given for embryos raised at 19 °C, and we highlight the neurula stage presented in this study (21 hpf). b Two-dimensional projection of cell clusters (metacells) using a force-directed layout based on the co-clustering graphs for individual cells (see Methods). Metacells are color-coded by cell type. c Normalized fold change expression of top variable genes (rows) per metacell (columns, grouped by cell type). For each metacell, we selected up to 30 markers with a minimum fold change ≥ 2. Selected gene names from known markers, used to annotate each cell type, are indicated to the right of the heatmap. Genes in bold case are shown in panel (d). d Pie charts depicting the fraction of cells mapped to each cell type among the cell transcriptomes and the cell counting experiment (top); and the 3D reconstruction with assignment of nuclei to each germ layer (bottom). A transverse section is shown on the left, and dorsal views with anterior to the top on the right (full, without epidermis nuclei, without epidermis and neural cells nuclei. e Expression profile of previously unknown marker genes for specific cell types (neural, endoderm, anterior epidermis, presumptive cerebral vesicle, notochord, and tailbud) analyzed by in situ hybridization (ISH, top, with anterior to the left and dorsal to the top in side views, n = 10 embryos) and corresponding two-dimensional expression maps (bottom, based on the same layout as panel b). Gene expression is shown as density maps representing UMI counts (per 10,000 UMIs) in each cell.
Gene expression signatures across epidermal metacells shows that this tissue is not homogenous. For example, we recognized anterior epidermal cells (i.e. expressing Arpd2, Fgfrl, Fzd5/8, Pax4/6)20,31,32,33, posterior cells (Cdx, Tbx6/16, Wnt3)34,35,36 and subpopulations of potential epidermal sensory cells (Delta, Elav, Tlx)37,38,39. Among the neural metacells, we identified several metacells corresponding to the presumptive cerebral vesicle (anterior central nervous system, Otx40). Concerning the mesodermal cell populations, we could assign several metacells to the notochord (Cola, Foxaa, Mnx, Netrin)41,42,43,44 and tailbud compartments (Nanos, Piwil1, Vasa, Wnt1)36,45,46. In the endoderm, one metacell could be assigned to the ventral endodermal region that later develops into the endostyle and the club-shaped gland (Foxe, Nkx2.5)12,47. We further validated our atlas by analyzing by in situ hybridization the expression of several genes with undescribed patterns at this stage, including genes enriched in neural plate (Tcf15-like), endoderm (PLAC8 motif-containing protein 1), anterior epidermal (Tmprss15), presumptive cerebral vesicle (Calcitonin Family Peptide 1 (Ctfp1)), notochord (Tenascin) or tailbud (Notum) populations (Fig. 1e and Supplementary Fig. 2a, b). We also verified the cell type identities of the metacells by identifying homologous cell populations in the neurula stage of B. floridae48, another amphioxus species, based on the pattern of shared transcription factors (TFs) (Supplementary Fig. 2c). Overall, our single-cell transcriptomic atlas uncovers the diversity of cell states associated to each major germ layer in the amphioxus neurula. This atlas can be browsed interactively in a dedicated web app: https://sebelab.crg.eu/amphioxus-neurula-21hpf/.
Cross-species comparison of neurula stage embryonic tissues
To gain insights into the evolutionary affinities of amphioxus neurula stage tissues, we compared aggregated expression profiles of the different structures and tissues with those of other chordates, using published developmental single-cell atlases for B. floridae48, Ciona intestinalis49, Xenopus tropicalis50, Danio rerio51 (Fig. 2a, b) and Mus musculus52 (Supplementary Fig. 3). We focused our comparative analysis on stages approximately corresponding to the amphioxus neurula stage26 and used single-cell expression profiles similarly grouped into embryonic tissues.
a Comparison between cell type transcriptomes of the amphioxus neurula stage (rows) and matched developmental time-points (rows) in the chordates Ciona intestinalis (initial to late tailbud stage), Danio rerio (14 hpf to 24 hpf), Xenopus tropicalis (S14 to S22 stages), and Branchiostoma floridae (including blastula [Bl], four gastrulation stages [G], three neurula [N] stages, and larva [Lv]). Cell type similarity was measured using SAMap scores based on all available pairwise markers (see Methods). Cell types are color-coded by cell type or developmental layer (endoderm, mesoderm/muscle, neuroectoderm, ectoderm, and other), and, in the case of the multi-stage chordate datasets, by developmental time-point (color intensity, legend to the side of each plot). b Most similar pairs of cell populations between the B. lanceolatum neurula (subpanels) and the B. floridae developmental stages (color-coded dots in each subpanel, from gastrula [G3] to larva [Lv]). Similarity is measured using Pearson’s correlation coefficient of cell type-level normalized fold change values, using genes with FC > 1.05 in at least one cell type. Only the top B. floridae cell type is displayed. Gray boxes highlight the neurula stages, where similarity typically peaks. c Graph representation of transcription factors (TFs, circular nodes) shared (i.e. connected by an edge) between amphioxus and homologous cell types in Ciona, Danio and Xenopus (square nodes). Specific TFs are considered to be shared between two cell types if they are significantly overexpressed in both. TFs in bold are shared between all species considered. For amphioxus, we required fold-change > 1.25, and BH-adjusted p-value < 0.05. For the matched cell types from other species, we required significant overexpression in at least one of the developmental time-points considered. A complete list of genes shared between all pairs of cell types is available in Supplementary Data 1. Di/mesenceph. prim= Di/mesencephalon primordium.
In general, we find strong similarities between the two amphioxus species (which diverged ca. 100 Mya53), with reciprocal matches for most cell types across species (Fig. 2a). As expected, the similarity between our B. lanceolatum neurula and the various cell types along the B. floridae time course is highest in the neurula stages of the latter (Fig. 2b). These results are in line with the validation of our cell type classification based on the identification of cell populations with matching TF usage across both species (Supplementary Fig. 2c).
In the pairwise comparisons with C. intestinalis, X. tropicalis and D. rerio, the notochord showed the strongest transcriptional similarity and shared expression of the TFs Brachyury2 (T), Foxaa and Foxab (Foxa1 and Foxa2) (Fig. 2c). Conversely, in spite of sharing the same core TFs, the mouse and the amphioxus notochords exhibited low global transcriptomic similarity (Supplementary Fig. 3). This lack of conservation compared to mouse probably reflects the different structural roles that the notochord can play during development in amniote and non-amniote vertebrates. Likewise, amphioxus differentiated muscular somites resemble muscle/skeletal muscle in tunicates and the three vertebrates (Fig. 2a), albeit with different sets of TFs between species (Fig. 2c). In contrast, the non-muscular part of amphioxus somites resembles vertebrate presomitic mesoderm and shares expression of the TFs Foxc (Foxc2), Snail (Snai2) and Hox3 (Hoxa3) (Fig. 2c).
Amphioxus neural cells also resemble vertebrate neural populations and co-express neural TFs like Soxb1c (Sox2), Soxc (Sox4) and Neurogenin (Neurog3). These same TFs are also shared by tunicate neural cells, but the overall transcriptome does not show similarity with amphioxus neurons. The opposite is true for the endodermal transcriptome: amphioxus endoderm transcriptome matches that of tunicates, but not vertebrate endoderm, although the TFs Foxaa and Foxab (Foxa1 and Foxa2) are expressed in all of them (Fig. 2c). Finally, the amphioxus anterior epidermis looks more distinct than the posterior one. Among vertebrate epidermal cells, its most similar pairs are secretory cells both in Danio and Xenopus (termed “Goblet cells” there). But it also hits different mesodermal tissues in Danio (e.g. the endothelium). On the other hand, the amphioxus posterior epidermis is broadly similar to many epidermal cell types of the two vertebrates, most notably the epidermal progenitors and ionocytes.
When examining the lists of shared markers between transcriptionally similar embryonic tissues/cell types (Supplementary Data 1), we observed a general overrepresentation of transcription factors (TFs) and chromatin factors compared with effector genes, as expected when comparing undifferentiated cell populations.
The accessible chromatin landscape of amphioxus neurula stage
To interrogate the regulatory logic underlying the observed cell-specific transcriptomes, we performed bulk ATAC-seq experiments in neurula-stage embryos. We defined a total of 51,028 ATAC-seq peaks and assigned them by proximity to 19,069 genes (median 2,05 peaks per expressed gene) (Supplementary Fig. 1b–h). We then grouped these peaks according to the expression pattern of the associated genes and conducted motif enrichment analysis on these regulatory element groups, using a combination of de novo inferred and known motifs (see Methods). This analysis revealed 317 distinct motifs with significant enrichments in specific cell populations (Fig. 3a).
a Heatmap representing the enrichment of specific TF binding motifs (columns) in the regulatory regions of genes in each cell type of the amphioxus neurula (rows). Names from selected motifs are indicated next to the heatmap (in bold those that also appear in panel b). The amphioxus motif library was obtained by merging experimentally determined vertebrate motifs from CIS-BP with de novo inferred motifs for each amphioxus cell type, and removing redundancy (see Methods). Therefore, motif names do not represent specific amphioxus TFs, but rather sequence similarity with motifs of vertebrate homologs. The regulatory regions associated with each gene were obtained from a bulk ATAC-seq experiment. b Examples of cell type-specific amphioxus TFs whose expression levels (vertical axis, as log2(FC)) match the enrichment of associated motifs (horizontal axis; shown below as information content logos). Circle size is proportional to the BH-adjusted −log10(p) binomial test of motif enrichment comparing motif occurrence near cell type-specific and unspecific genes (see Methods; exact p-values of these tests are shown for significant enrichment at p < 0.01, see Source Data for a complete list). Di/mesenceph. prim= Di/mesencephalon primordium.
The identified motifs are consistent with known TF regulators in amphioxus and other metazoans and, in addition, motif enrichments often parallel the expression of the associated TFs (Fig. 3b, Supplementary Fig. 4). For example, in peaks assigned to epidermal genes, we found enrichment for motifs like Dlx, Grhl, Klf1/2/4, Rfx1/2/3 or Tfap2, coincident with the expression of Dlx, Klf1/2/4 and Tfap2 in epidermal metacells (Fig. 3b). Interestingly, these TFs are part of the in silico reconstructed gene regulatory network controlling epidermis development described in amphioxus54 and are known epidermal fate determinants in vertebrates55,56,57,58 (Fig. 2c).
In endodermal cells, we found a slight but non-significant enrichment of a Fox motif in the regulatory regions of endodermal marker genes, possibly linked to the expression of Foxaa and Foxab in these tissues (Supplementary Fig. 4). Furthermore, in the endoderm and the endostyle, we also observed the coincident expression/motif enrichment of Gsc and Nkx2-5/6, respectively (Fig. 3b).
Neural cell types exhibited expression of various Sox and Pou family TFs and concomitant enrichment of their associated motifs, including Soxc (Sox4), Soxb2 (Sox14), Soxb1c (Sox2, although its motif enrichment is only significant in the anterior archencephalon at p < 0.01), and Pou3fl (Pou3f4, with significant motif enrichment in the Di-Mesencephalic primordium20 and neural tailbud cells; Fig. 3b). The neural specificities of these TFs appear to be conserved across vertebrates (Fig. 2c) and SoxB1 and Pou3f family factors have been proposed as potential major regulators of nervous system development in amphioxus54. The activity of Pou3fl (Pou3f4) in neural tailbud cells is also consistent with the function of TFs from these families in the maintenance of stemness in vertebrates59. This is also the case for the Myc/Max HLHs in the non-neural tailbud population, as observed in mouse60.
The peaks associated to genes overexpressed in non-muscular somite cell populations are enriched in T-box motifs, consistently with the expression in our dataset of various TFs of this family such as Eomes/Tbr1/Tbx21, Tbx15/18/22, and Brachyury2 (Fig. 3b) and with previously reported expression of these genes in forming somites61,62,63. The muscular somite population peaks are enriched in motifs shared with the non-muscular somite, but are also enriched in motifs for Myogenic Regulatory Factors (MRFs) such as Mrf4 (Myf6), which is also highly expressed in this cell type (Fig. 3b), in line with both the expression of the various amphioxus MRFs described by in situ hybridization64, and the role of their orthologues in vertebrate myogenesis65. The strongest TF-motif association concerns the previously reported notochordal marker Foxaa (Foxa2) (Fig. 3b)44, which is also shared with tunicates and vertebrates in our cross-species cell type comparisons (Fig. 2c). Overall, the accessible chromatin landscape of the neurula stage revealed the regulatory motif lexicons underlying amphioxus embryonic cell identities.
Characterization of neural, endodermal and somitic cell populations
We then focused on the detailed analysis of specific cell populations. To this end, we performed separate clustering of single cells classified as belonging to the endoderm and to the somites (muscular and non-muscular). We also performed such an analysis on single cells of the neural tissue (sensu stricto, derived from the neural plate) that is detailed in Supplementary Note and Supplementary Fig. 5.
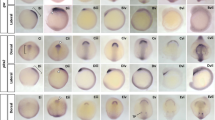
Concerning the endodermal compartment, we could recognize metacells corresponding to the main known territories (Fig. 4a, b and Supplementary Figs. 2 and 6). The expression of the ventral marker Nkx2.166, together with anteriorly expressed genes such as Dmbx, Fgfrl, Fzd5/8 and Sfrp1/2/520,31,33,67,68,69 (Supplementary Fig. 6) indicates that metacell 2 corresponds to the ventral anterior endoderm territory whereas metacells 3 and 7 show a combination of marker genes that are typical of the ventral endoderm that later develops into the club-shaped gland and the endostyle such as Foxe, Nkx2.5, Tbx1/10 and Pax1/912,47,70,71 (Fig. 4a, b and Supplementary Fig. 6). The expression of Pitx in metacell 3 suggests that metacells 3 and 7 correspond to the left and right part of this territory, respectively72.
a 2D projection of endodermal metacells on a side view scheme of an amphioxus neurula stage embryo with anterior to the left and dorsal to the top. b Gene expression as normalized fold changes of selected gene markers and corresponding ISH (n = 10 embryos). Gene expression is shown as density maps representing UMI counts (per 10,000 UMIs) in each cell, with cells positioned in the vicinity of their corresponding metacells. c Multiple in situ hybridization for Dmbx, Fgfrl and Brachyury2 (n = 5 embryos). Lateral views are shown on the left, with anterior to the left and dorsal to the top. Optical cross-sections at the level of the dotted line are also shown. The labeling intensity is stronger on one side due to the thickness of the embryo that was imaged on one side after mounting. The three genes are coexpressed in the anteriormost region of the dorsal mesendoderm (metacell 1). Normalized fold change expression of each marker is shown on the right. Scale bar: 25 µm.
Posterior to that, metacells 16 and 17 that are characterized by low or no expression of Soxf correspond to the first pharyngeal slit anlagen73 while metacell 14 expresses both Irxc and Foxaa, a combination specifically observed in a region that is just behind it44,74 (Fig. 4a, b and Supplementary Fig. 6). Metacells 5 and 11 express Pax1/9 but no ventral markers and could correspond to the dorsal mid endoderm region70 (Fig. 4a, b). Metacells 8 and 12 show a very similar profile with an enrichment in transcripts of mid/posterior endoderm markers such as Nkx2.2, Foxaa44,75 (Supplementary Fig. 6), and Fabp3/4/5/7/8/9/11/12, a marker described here for the first time (Fig. 4a, b). Metacell 12 additionally expresses Gata4/5/6, indicating that the corresponding cells are more ventral than those from metacell 814 (Supplementary Fig. 6). Metacell 9 has a transcriptional profile similar to that of metacells 8 and 12 combining expression of the mid/posterior marker Foxaa44 (Supplementary Fig. 6) and absence of Pax1/9 expression70 (Fig. 4a, b). The posterior marker Wnt876 is expressed in metacells 4 and 6 with metacell 4 also expressing the ventral marker Gata4/5/614, and, hence, representing the ventral posterior territory (Supplementary Fig. 6).
Finally, metacells 1, 10 and 13 are characterized by an enrichment in anterior markers Dmbx, Fgfrl, Fzd5/8 and Sfrp1/2/531,33,67,68,69 as well as Six3/6, Six4/5 and Zic77,78 (Fig. 4a, b and Supplementary Fig. 6). They show a transcriptional signature of the anterior dorsal mesendoderm, a region which is continuous with the notochord per se posteriorly, and which is continuous laterally with the endoderm per se. Metacell 1 is also expressing Thsd7 described here (Fig. 4a, b), together with Brachyury2, Pax3/7 and Zeb54,63,79 and lacks Nkx2.1 expression80 (Supplementary Fig. 6) suggesting it represents the axial part of this region, whereas metacells 10 and 13, expressing Nkx2.1, would correspond to the paraxial more ventral portion that latter form the left and right Hatschek’s diverticula80 (Supplementary Fig. 6). Therefore, metacell 1 represents a potential prechordal plate-like territory showing a transcriptomic profile characterized by anterior and axial markers together with endodermal markers.
We validated the existence of this cell population by undertaking multiple in situ hybridization for Dmbx, Fgfrl and Brachyury2, showing a clear overlap of expression in the anterior tip of the dorsal axial mesendoderm (Fig. 4c). Such territory was already proposed to exist in amphioxus based on both cell behavior and gene expression of several marker genes21,22,28 but our data highlight the strong difference in its transcriptomic profile compared to the other notochord cells, reinforcing the idea that ancestral chordates possessed a prechordal plate-like region that later evolved specific functions in vertebrates.
Re-clustering of cells assigned to the somites resulted in 12 metacells (Fig. 5a, b and Supplementary Figs. 2 and 7). As expected, we found a population (metacell 8) with a profile typical of the muscular part of somites that starts to differentiate, characterized by the expression of Mef2, Lmo4, several MRFs, together with MLC-alk64,81,82 (Supplementary Fig. 7) and the Titin-like marker described here (Supplementary Figs. 2 and 7). Metacells 7 and 9 have similar profiles and also express Titin-like and several MRFs64 (Supplementary Figs. 2 and 7) together with Brachyury2, Delta39,63 and the newly described gene Twist-like (Supplementary Figs. 2 and 7). They hence correspond to the last somites that have just been formed, with metacell 7 more posterior as indicated by the expression of Wnt1 or Wnt476.
a 2D projection of somitic metacells on a side view scheme of an amphioxus neurula stage embryo, with anterior to the left and dorsal to the top. b Gene expression as normalized fold changes of selected gene markers and corresponding ISH (n = 10 embryos). c Multiple in situ hybridization for Ripply+Alx+Gata1/2/3 and Ripply+Tcf21/Msc+Alx (n = 5 embryos). Lateral views are shown on the left, with anterior to the left and dorsal to the top. Optical cross-sections at the level of the dotted line are also shown. The labeling intensity is stronger on one side due to the thickness of the embryo. Ripply, Alx and Gata1/2/3 are coexpressed in the non-muscular part of formed somites (metacells 2 and 11) whereas Ripply, Tcf21/Msc and Alx are coexpressed in the first somite pair (metacells 1 and 3). Scale bar: 25 µm.
More posteriorly, metacell 5 is characterized by the expression of newly described tailbud gene markers such as Bicc, and SF2 family helicase (Supplementary Figs. 2 and 7), together with Nanos, Otp, Vasa, and Wnt1, 4 and 620,46,76 but also expresses Brachyury2 and Mrf4, a combination corresponding to the tailbud somitic part63,64 (Supplementary Fig. 7). Metacells 4 and 6 also express tailbud markers but do not express MRF genes. Moreover, metacell 4 is characterized by an enrichment in transcripts of the ventral markers Gata1/2/3 and Vent1/Vent211,14,83 (Fig. 5a, b and Supplementary Fig. 7).
Concerning the four putative compartments of formed somites proposed by Young and colleagues17, we could only recognize, apart from the myotome region (clusters 7, 8 and 9), a non-myotome region (corresponding to cell clusters 2 and 11) enriched in markers expressed in territories described as lateral and/or ventral such as Alx, Gata1/2/3, Ripply and Vent1/Vent211,14,42,84 (Fig. 5a–c and Supplementary Fig. 7). We validated the existence of these cell populations using in situ hybridization for Alx, Gata1/2/3 and Ripply, which showed co-expression of the three genes in the non-myotome region of somites (Fig. 5c). To simplify, we will refer to the non-myotome portion of somites as the “ventral somite” throughout the remainder of the manuscript.
Finally, the most important novelty concerns the first somite pair, which clearly shows a transcriptomic profile divergent from the other pairs. Metacells 1 and 3 correspond to this first pair of somites, with metacell 1 representing the right somite, and metacell 3 the left one (Fig. 5a, b). Indeed, contrary to metacell 1, cells of the latter express the left side marker Pitx72 as well as Gremlin, which is expressed in the first left somite at this stage85 (Supplementary Fig. 7). Both metacells express the anterior marker Fgfrl31 (Supplementary Fig. 7), and three newly described markers: Erg/Fli1a, Tcf21/Msc and FReD containing protein (Fig. 5a, b and Supplementary Fig. 7). They also express the ventral somite marker genes Alx, Gata1/2/3, Ripply and Vent1/Vent211,14,42,83,84 (Fig. 5a, b and Supplementary Fig. 7). To note, no Wnt genes are expressed in these metacells, whereas the ventral markers are expressed together with Wnt1676 in metacells 2 and 11 that correspond to the ventral region of the formed somites posterior the the first pair (Supplementary Fig. 7). Interestingly, Erg/Fli1a is orthologous to Fli-1 which is implicated in vertebrate hemangioblast development together with Vegfr and Scl/Tal-186. It has been shown that the amphioxus orthologs of these genes are also expressed in the first pair of somites14, reinforcing the proposition of homology between this first pair and the embryonic hematopoietic/angiogenic field of vertebrates that derives from the lateral plate mesoderm. On the other hand, Tcf21/Msc is orthologous to Tcf21/Capsulin and Msc/MyoR are main regulators of head muscle myogenesis in vertebrates, upstream of MRFs87,88,89, suggesting that the first somite pair of amphioxus has a profile that resembles both vertebrate head and lateral plate mesoderm. We further validated the co-expression of Tcf21/Msc with Ripply and Alx in the first somite pair (Fig. 5c).
The evolution of the chordate anterior mesoderm
The most striking feature of the amphioxus neurula highlighted by our data is the presence of three cell populations with a peculiar transcriptional profile: cells of the first left and right somites (metacells 1 and 3, Fig. 5a, b), and cells that could correspond to a prechordal plate-like structure (metacell 1, Fig. 4a, b). The first somite pair in amphioxus has long been proposed as being distinct from the other pairs, and we previously showed that this somite pair is the only one whose formation is controlled by the FGF signaling pathway22,23,90. Our molecular atlas additionally shows that the cells of the first pair of somites transcriptionally resemble vertebrate head and lateral plate mesoderm (metacells 1 and 3, Fig. 5 and Supplementary Fig. 7), while the cells of the ventral part of amphioxus somites posterior to the first pair express orthologues of genes expressed in vertebrates lateral plate mesoderm or derivatives (metacells 2 and 11, Fig. 4 and Supplementary Fig. 6). These data support the homology we and others proposed between vertebrate lateral plate mesoderm and amphioxus ventral part of the somites, as well as the ventral origin of vertebrate cranial/pharyngeal mesoderm.
Such proposed homology based on transcriptomic profile should reflect a conserved regulatory logic. Considering homology at the level of gene expression regulation, we reasoned that if our scenario for vertebrate mesoderm evolution supported by our cell atlas is correct, regulatory regions of genes that are active in the ventral region of the somites at the neurula stage in amphioxus could drive expression of a reporter gene in the vertebrate lateral plate and head mesoderm, as a reminiscence of an ancestrally shared regulatory program. Among such genes, Gata1/2/3 is the transcription factor with the highest enrichment (fold change) in metacells 2 and 11 of the somite reclustering analysis (Fig. 5a, b), which we could assign to the ventral region of the somites. We first decided to test for the function of Gata1/2/3 in amphioxus embryogenesis to validate its role in the development of the ventral region of the amphioxus somites.
We showed that overexpression of a constitutive repressor form (Gata1/2/3-engrailed) induced somite defects clearly visible at the N4 stage (Fig. 6). Indeed, although the dorsal MLC-alk expressing region, which later form the muscles, is little affected, the ventral Alx-positive part, which normally elongates at this stage, has formed anarchically organized epithelial spheres, surrounded by a basal lamina, which have colonized the ventral region in embryos with the strongest phenotype (Fig. 6). These data demonstrate that Gata1/2/3 plays a major role in the development of this somite compartment and that repression of expression of its target genes induces a loss of coordinated cell-shape changes and migration.
Control embryos (n = 5), injected embryos with a mild phenotype (n = 5/10) and injected embryos with a strong phenotype (n = 5/10) from two independent injection experiments were processed for double in situ hybridization for MLC-alk and Alx followed by immunostaining against Laminin. Side views of 3D reconstructions of the nuclei and Laminin labeling are shown on the top, with anterior to the left and dorsal to the top. Sections in side views showing all the chanels are shown below, as well as transverse optic sections at the level indicated by the white lines. MLC-alk is expressed in the myotome region of the somites that is little affected. Alx is expressed in the ventral region of the somites that is completely disorganized in injected embryos, as revealed by anti-Laminin labeling. Scale bar: 25 µm.
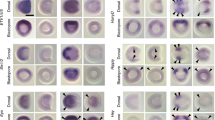
We hence decided to test whether the regulatory elements controlling the expression of amphioxus Gata1/2/3 at the neurula stage are recognized by any tissue/cell type specific regulatory state in zebrafish, which would point at evolutionary conservation (at least partially) of Gata1/2/3 regulation. We show that one of the putative regulatory regions selected using ATAC-seq data (Supplementary Fig. 8a) was able to drive the expression of the eGFP reporter gene in zebrafish embryonic regions that partly overlap with the expression of Prrx1a in the head mesoderm and finbuds (Supplementary Fig. 8b and Supplementary Fig. 9a). This suggests that Gata1/2/3 in the chordate ancestor probably already had the potentiality to be recruited during vertebrate evolution for expression in the pharyngeal and lateral plate mesoderm. However, these data still need to be supported by a more precise analysis of reporter expression domains at different developmental stages.
In vertebrates, both the anterior axial (prechordal plate) and pharyngeal/cranial mesoderm structures develop into different muscle populations: the extraocular muscles, and several facial/branchial muscles, respectively10. Interestingly, myogenesis in these cells, although it is mediated by the activity of members of the MRF family, is controlled by the upstream factors Pitx2 (extraocular muscles) and Tbx1 (pharyngeal muscles) and not by Pax3/7 and Six1/2 factors as it is the case for muscles deriving from the somites10,89,91. In amphioxus, we previously showed that all the somites form under the control of Pax3/7, Six1/2 and/or Zic23. Moreover, Pitx, the ohnologue of vertebrate Pitx1, Pitx2 and Pitx3, has been shown by in situ hybridization to be expressed on the left side of the embryo and in few neurons and is controlling left/right asymmetry36,72,92, while we observed in our data its expression only in two metacells (3 and 11) in the somite subclustering atlas (Supplementary Fig. 7). On the other hand, Tbx1/10 has been shown to be expressed long after MRFs in the amphioxus somites23,71 and we showed in our data a reduced expression in metacell 8 in the somite subclustering atlas (Supplementary Fig. 7), metacell we assigned to the muscular part of the trunk somites, while its expression was not detected in the other metacells expressing MRFs. Furthermore, functional analysis by TALEN mutagenesis or morpholino injection for Pitx93 and Tbx1/1094 respectively, showed that knockout or knockdown of these genes did not alter somite development.
If our scenario of head mesoderm evolution is correct, it implies that Pitx2 and Tbx1 were co-opted for the control of myogenesis in the vertebrate head. In order to test this co-option, we investigated the activity of putative regulatory regions of amphioxus Pitx and Tbx1/10 in zebrafish reporter assays. In the case of Tbx1/10, one genomic region tested was able to drive the expression of the reporter gene in head and finbud regions that partly overlap with Prrx1a expression territories (Supplementary Fig. 8c, d, and Supplementary Fig. 9b). This suggests that Tbx1/10 in the chordate ancestor probably contained regulatory information that allowed its later recruitment in the vertebrate head mesoderm for a new function as a myogenesis controlling factor. In the case of Pitx, one region drove a restricted reporter expression in the zebrafish hatching gland (Supplementary Fig. 8e, f). Interestingly, this structure derives from the anterior prechordal plate and expresses Pitx295,96,97,98, suggesting that the Pitx gene in the chordate ancestor already had the potentiality to be recruited in this mesoderm region during vertebrate evolution. However again, these data still need to be supported by a more precise analysis of reporter expression domains at different developmental stages.
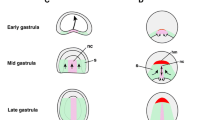
To conclude, our results support a scenario for the emergence of the vertebrate lateral plate mesoderm and cranial/pharyngeal mesoderm through the segregation of pre-existing cell populations (homologous to amphioxus ventral part of the somites, first pair and posterior, respectively), which, by becoming partly independent from the somites, could evolve new structures in the trunk and in the head (Fig. 7). We also provide new evidence of the existence of a prechordal plate-like territory in amphioxus and give insights into how the appearance of vertebrate head muscles developing from the prechordal plate and cranial/pharyngeal mesoderm might have been achieved through the co-option of Pitx2 and Tbx1 for the control of myogenesis.
Schemes of putative embryos in dorsal views with anterior to the top are shown, with transverse sections at the level of the anterior and trunk regions on the left. Diagrams on the right represent mesoderm cell populations that were inferred at each step. We propose that the chordate ancestor possessed a mesoderm organized in an axial domain with two cell populations: a prechordal-plate like region in the anterior part (dark purple), and a notochord (light purple) more posteriorly; and a paraxial domain completely segmented into somites (green), containing in the ventral part a cell population homologous to the ventral part of amphioxus somites (orange), and showing heterogeneity between the anterior (dark orange/green) and trunk (light orange/green) regions. During the first step of evolution, we propose that the ventral somite cell populations became independent from the paraxial mesoderm to give rise to the unsegmented lateral plate mesoderm (lateral plate mes., orange). In a second step, the anterior paraxial mesoderm would have been lost (dark green), and we previously proposed that this could be due to a change in the function of the FGF signaling pathway22,23. This loss would have led to a relaxation of the developmental constraints imposed by the segmented paraxial mesoderm in the anterior region, enabling remodeling of the tissues of the anterior axial mesoderm (dark purple) and anterior lateral mesoderm (dark orange), which could have evolved into the prechordal plate (pink) and pharyngeal/cranial mesoderm (blue/green). The ability of these new embryonic structures, derived from non-myogenic cell populations, to form muscles, would have been associated with the co-option of Pitx2 and Tbx1/10 as master genes of the myogenesis program.
Methods
Ethics approval
All the experiments were performed following the Directive 2010/63/EU of the European parliament and of the council of 22 September 2010 on the protection of animals used for scientific purposes. Ripe adults from the Mediterranean invertebrate amphioxus species (B. lanceolatum) were collected at the Racou beach near Argelès-sur-Mer, France, (latitude 42° 32′ 53′ ′ N and longitude 3° 3′ 27′ ′ E) with specific permission from the Prefect of Region Provence Alpes Côte d’Azur. Zebrafish embryos were obtained from AB and Tübingen strains, and manipulated following protocols approved by the Ethics Committee of the Andalusia Government and the national and European regulation established.
Cell suspension preparation
Adult amphioxus (Branchiostoma lanceolatum) were collected at the Racou beach near Argelès-sur-Mer, France. Gametes were obtained by heat stimulation as previously described in ref. 99. Embryos ( ~ 100) at 21 hours post-fertilization (hpf, at 19 °C) were washed 2 times in Ca2 + /Mg2 + -free and EDTA-free artificial seawater (CMFSW: 9 mM KCl, 449 mM NaCl, 33 mM Na2SO4, 2,15 mM NaHCO3, 10 mM Tris-HCl). CMFSW was replaced by CMFSW with Liberase TM at 250 µg/mL (Roche, 05401119001). Cells were then dissociated by a series of pipetting and vortexing during 25 minutes at room temperature. The reaction was stopped by the addition of 1/10th volume of 500 mM EDTA. The cell suspension was centrifuged at max speed for 1 min. The pellet was resuspended in CMFSW containing Calcein violet (InvitrogenTM, 65085439) and Propidium iodide (PI, 1 µg/mL, InvitrogenTM, P3566).
MARS-seq
Live single cells were selected using a FACSAria II cell sorter. To this end, we sorted only Calcein positive/PI negative cells, and doublet/multiplet exclusion was performed using FSC-W versus FSC-H. Cells were distributed into 384-wells capture plates containing 2 µl of lysis solution: 0.2% Triton and RNase inhibitors plus barcoded poly(T) reverse-transcription (RT) primers for single-cell RNA-seq. Single-cell libraries were prepared using MARS-seq29. First, using a Bravo automated liquid handling platform (Agilent), mRNA was converted into cDNA with an oligo containing both the unique molecule identifiers (UMIs) and cell barcodes. 0.15% PEG8000 was added to the RT reaction to increase efficiency of cDNA capture. Unused oligonucleotides were removed by Exonuclease I treatment. cDNAs were pooled (each pool representing the original 384-wells of a MARS-seq plate) and linearly amplified using T7 in vitro transcription (IVT) and the resulting RNA was fragmented and ligated to an oligo containing the pool barcode and Illumina sequences, using T4 ssDNA:RNA ligase. Finally, RNA was reverse transcribed into DNA and PCR amplified. The size distribution and concentration of the resulting libraries were calculated using a Tapestation (Agilent) and Qubit (Invitrogen). scRNA-seq libraries were pooled at equimolar concentration and sequenced to saturation (median 6 reads/UMI) on an Illumina NextSeq 500 sequencer and using high-output 75 cycles v2.5 kits (Illumina), obtaining 483 M reads in total.
To quantify single-cell gene expression, MARS-seq reads were first mapped onto Branchiostoma lanceolatum genome (GCA_927797965.1, annotation version 3) using STAR v2.7.3100 (with parameters: –outFilterMultimapNmax 20 –outFilterMismatchNmax 8) and associated with exonic intervals. Mapped reads were further processed and filtered as previously described29. Briefly, UMI filtering includes two components, one eliminating spurious UMIs resulting from synthesis and sequencing errors, and the other eliminating artefacts involving unlikely IVT product distributions that are likely a consequence of second strand synthesis or IVT errors. The minimum FDR q-value required for filtering in this study was 0.02.
Single cell transcriptome clustering
We used Metacell 0.3730 to select gene features and construct high-granularity cell clusters (metacells), which were further annotated into cell types (see below). First, we selected informative genes using the mcell_gset_filter_multi function in the metacell R library, including genes fulfilling these criteria: a total gene UMI count > 30 and >2 UMI in at least three cells, a size correlation threshold of -0.1, and a normalized niche score threshold of 0.01. This resulted in the selection of 844 genes to be used for downstream clustering. Second, we used these genes to build a K-nearest neighbors cell graph with K = 100 (mcell_add_cgraph_from_mat_bknn function), which was the basis to define metacells with an additional K-nearest neighbor procedure (mcell_coclust_from_graph_resamp and mcell_mc_from_coclust_balanced functions) using K = 30, minimum metacell size of 15 cells, and 1000 iterations of bootstrap resampling (at 75% of the cells); and a threshold α = 2 to remove edges with low co-clustering weights. Third, we removed one metacell which exhibited low transcriptomic information ( > 50 cells with a median UMI/cell <500). This resulted in 176 metacell clusters, which were annotated to known cell types (Supplementary Data 4) based on the expression level of known markers (Supplementary Fig. 1). This same metacell clustering procedure has been applied to each of the neurula stages of B. floridae48.
We recorded gene expression in cell clusters (metacells or cell types) by computing a regularized geometric mean within each cluster and dividing this value by the median across clusters. This normalized gene expression can be interpreted as an expression fold change (FC) for a given metacell or cell type.
Two-dimensional projection of the metacells were created using a force-directed layout based on the metacell co-clustering graph (mcell_mc2d_force_knn function).
Gene expression profiles across cell clusters were visualized with heatmaps, using the ComplexHeatmap 2.10.0 R library101. Cell cluster ordering was fixed according to annotated cell types; and gene order was determined using the highest FC value per cluster. Genes were selected based on minimum differential expression per metacell/cell type, with a maximum number of markers per clusters selected in each case (the actual thresholds used in each heatmap are specified in the corresponding figure legends).
Finally, we selected cells belonging to the endoderm, neural and somitic metacells (Supplementary Data 4), and reclustered them using the same metacell-based approach as described for the whole dataset (except that in this case we allowed for smaller metacells, with 10 cells; (Supplementary Data 4). The two-dimensional arrangement of the resulting metacells was curated based on the expression of cell type-specific known markers of various cell subtypes (Supplementary Figs. 5–7).
ATAC-seq library preparation
For ATAC-seq library construction, 25 embryos at the 21 hpf (19 °C) were transferred in a 1.5 ml tube, in four replicates. We then followed the method described in ref. 102. After a one minute centrifugation at 13 000 rpm, seawater was carefully removed. 50 μl of cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% Igepal) were added and cells were lysed by gentle pipetting. While 25 μl of the lysate was centrifuged at 500 g for 10 minutes at 4 °C, the other 25 μl were used to count nuclei after DNA labeling with DAPI and around 50 000 nuclei were used per transposition reaction. The supernatant was removed, the nuclei resuspended in the reaction mix (25 μl 2x TD buffer (Illumina), 2,5 μL Tn5 transposase (Illumina), 22,5 μL nuclease free H2O) and incubated at 37°C for 30 minutes. Following transposition, 3 μl of 3 M AcoNa (pH5.3) were added to the reaction to adjust the pH, and the DNA was purified using the MinElute PCR purification Kit (Qiagen), following the manufacturer’s instructions. The transposed DNA was eluted in 10 μL elution buffer preheated at 37 °C. To amplify the library, the following components were combined: 10 μL of transposed DNA, 10 μL of nuclease free H2O, 2,5 μL Nextera PCR primer 1 (25 μM), 2.5 μL Nextera PCR primer 2 (25 μM) and 25 μL NEBNext® high-fidelity 2x PCR master mix (NEB). We used the following conditions for PCR amplification: 72 °C for 5 minutes, 98 °C for 30 seconds, followed by 13 cycles at 98 °C for 10 seconds, 63 °C for 30 seconds and 72 °C for 1 minute. Following PCR amplification, 3 μl of 3 M AcoNa (pH5.3) were added to the reaction to adjust the pH, and the library was purified using the MinElute PCR purification Kit (Qiagen), following the manufacturer’s instructions using 20 μL of elution buffer preheated at 37 °C.
Analysis of neurula regulatory regions
We used the ATAC-seq data from the 21 hpf embryo to build a catalog of neurula regulatory regions. For comparison, we also used previously published26 ATAC-seq libraries of 15 hpf and 36 hpf embryos (the closest developmental timepoints available in that study; NCBI SRA accession numbers SRR6245277 to SRR6245279), as well as H3K4me3 ChIP-seq libraries from these same timepoints (SRA accession numbers SRR6245317 to SRR6245320).
The ATAC-seq libraries corresponding to the 15, 21 and 36 hpf embryos were mapped separately to the B. lanceolatum genome using bwa 0.7.17 (mem algorithm103). The resulting BAM files were (i) filtered using alignmentSieve (from the deeptools 3.5.1 package104) to exclude weak alignments MAPQ > 30), (ii) corrected to shift the left and right ends of reads, to account for ATAC mapping biases ( + 4/ − 5 bp in the positive and negative strands, using the --ATACshift flag in alignmentSieve), and (iii) filtered to only include nucleosome-free alignments (--maxFragmentLength 120 with alignmentSieve). Duplicated reads were marked with biobambam2 2.0.87105, coordinate-sorted, and removed to produce filtered BAM files. Then, we concatenated the BAM files stage-wise. Normalized coverage for each stage was reported as bins per million mapped reads (BPM), calculated using the bamCoverage tool in deeptools. The ChIP-seq libraries for 15 and 36 hpf were processed in the same way (except for the ATAC mapping bias correction step and the filtering of nucleosome-free alignments).
For the 21 hpf ATAC-seq experiment, we used MACS2 2.2.7.1106 to identify regulatory elements with the callpeak utility, starting from the nucleosome-free filtered BAM file, with the following options: (i) an effective genome size equal to the ungapped amphioxus genome length, (ii) keeping duplicates from different libraries (--keep-dup all flag), (iii) retaining peaks with a q-value < 0.01, (iv) enabling multiple summit detection (--call-summits flag), and (v) disabling the modeling of peak extension for ChIP-seq libraries (--nomodel flag).
We then assigned the MACS2-predicted regulatory elements to their proximal genes, based on their distance to each gene’s transcription start site (TSS). Specifically, we selected well-supported MACS2 regulatory elements (q-value < 1 × 10−6), standardized their lengths to 250 bp (125 bp to each side of the predicted peak summit), and assigned each peak to nearby genes based on distance to their TSS (excluding genes further away than 20 kbp, and genes located beyond a more proximal gene). Peaks overlapping the promoter region of a particular gene (defined based on TSS coordinates +/– 50/200 bp or coincidence with H3K4me3 ChIP-seq peaks for the 15 and 36 hpf datasets) were not assigned to any other gene. The peak sets were reduced to non-overlapping sets to avoid redundant regions. These genome coordinate operations were done using the GenomicRanges 1.46 and IRanges 2.28 packages in R107. We used these gene-regulatory element assignments to define lists of cell type-specific regulatory elements, based on the expression specificity of each gene (expression fold change ≥ 1.5 in a given cell type). In parallel, we also defined a set of background regulatory regions for each cell type (consistent of regulatory regions linked to non-overexpressed genes, at fold change ≤ 1). In total, we assigned 51,028 regulatory regions (ATAC peaks) to 19,069 genes (out of 27,102), with a median of 2 peaks per gene.
We used the cell type-specific sets of active regulatory elements (and their corresponding background sets) to identify motifs de novo using the findMotifsGenome.pl utility in homer 4.11108 Specifically, we set a constant peak size of 250 bp and attempted to identify motifs for each cell type, using k-mers of length 8, 10, 12, and 14; and tolerating up to four mismatches in the global optimization step.
In order to build a final motif collection for amphioxus, we concatenated the cell type-specific de novo motifs with known TF binding motifs from the CIS-BP database (as available the 3rd of March, 2023)109. Specifically, we used 3547 experimentally determined motifs (with SELEX or PBMs), corresponding to vertebrate or tunicate species (Homo sapiens, Mus musculus, Xenopus tropicalis, Xenopus laevis, Danio rerio, Tetraodon nigroviridis, Meleagris gallopavo, Gallus gallus, Anolis carolinensis, Takifugu rubripes, Ciona intestinalis, and Oikopleura dioica). We reduced the redundancy of this extensive de novo + known motif collection based on motif-motif sequence similarity, as follows: (i) we removed motifs with homer enrichment p-values < 1 × 10−9; (ii) we retained with high contiguous information content (IC), defined as having IC ≥ 0.5 for at least four consecutive bases or IC ≥ 0.5 for two or more blocks of at least three bases; (iv) for each of the remaining motifs, we measured their pairwise sequence similarity by calculating the weighted Pearson correlation coefficient of the position probability matrices of each motif, using the merge_similar function in the universalmotif 1.12.4110 R library with a similarity threshold = 0.95 for hierarchical clustering and a minimum overlap of 6 bp between two motifs in the motif alignment step. Finally, we selected the best motif per cluster based on its IC (highest). This resulted in a final, non-redundant collection of 1,595 motifs.
Then, we calculated the enrichment of each motif among the sets of regulatory regions specific to each cell type. To that end, we used the calcBinnedMotifEnrR function in the monalisa 1.0 R library111 to count motif occurrences in three sets of regulatory regions (bins) defined based on the expression levels of their associated genes: highly cell type-specific genes (FC ≥ 1.5), mildly cell type-specific genes (FC ≥ 1.1 and <1.5), and non-cell type-specific genes (FC < 1). Motif occurrences were defined as motif alignments with scores above 80% of that motif’s maximum alignment score (defined from the corresponding position weight matrices). Motif enrichment in each bin was then calculated using the fold change of occurrence relative to randomly sampled genomic regions (matched by GC content and length, using twice as many regions for background as for the foreground), and its significance assessed using a binomial test followed by Benjamini-Hochberg p-value adjustment. We retained the fold change and p-values for th set of highly cell type-specific regulatory regions (i.e. from genes with FC ≥ 1.5) for further analysis (Fig. 3 and Supplementary Data 1).
Finally, we scanned the B. lanceolatum genome to identify discrete occurrences of each of the 1595 motifs across the 51,028 MACS2-defined regulatory regions. We used the findMotifHits function in monalisa. In order to define bona fide motif alignments, we calculated an empirical p-value for each motif alignment (only best alignment per regulatory region) based on the rank of its alignment score when compared to a background distribution of randomly sampled genomic regions of similar sequence composition (only best alignment score per random background bin). Specifically, we divided the foreground regions into 10 equal-size sets based on their GC content, and matched each set with random genomic background sequences (not in the foreground) of similar GC content (same category) and equal length (set to 250 bp). These motif aligments were used to identify enhancer-specific motifs in Fig. 5 (complete list in Supplementary Data 2).
Cross-species cell type comparison
We used SAMap 1.0.2112 to evaluate the similarity between B. lanceolatum cell types and the previously published developmental single-cell transcriptomes of Danio rerio51 (reference gene set in original study: GRCz10 v1), Xenopus tropicalis50 (reference gene set in original study: Xenbase version 9.0), Ciona intestinalis49 (reference gene set in original study: KH2012 from the Ghost Database (http://ghost.zool.kyoto-u.ac.jp/download_kh.html), and Mus musculus52.
For each query species, we used the UMI tables corresponding to the timepoints closest to the B. lanceolatum 21hpf developmental stage (12 in total): 14 hpf, 18 hpf and 24 hpf for D. rerio (GEO accession: GSE112294); S14, S16, S18, S20 and S22 for X. tropicalis (GSE113074); the initial, early, middle and late tailbud stages for C. intestinalis (GSE131155) and the stages E7.00, E7.25, E7.50, E7.75, E8.00, E8.25 and E8.50 for M. musculus. For C. intestinalis, we used the cell type annotations used in the original paper. For D. rerio and X. tropicalis, we used the consensus cell annotations employed by Tarashansky et al.112.
To run SAMap, we first created a database of pairwise alignments with blastp 2.5.0 (comparing B. lanceolatum peptides to each query species separately; in the case of Danio rerio we used blastx/tblastn instead of blastp as the original gene set51 was only available as un-translated transcripts). Second, we used the cell-level UMI counts of each gene to calculate the SAMap mapping scores for each pair of cell types (between B. lanceolatum and each of the 12 query developmental datasets in other species), using all cells within each cluster for score calculation. SAMap identifies the best cross-species markers from the BLAST-based homology graph using an iterative procedure based on gene-gene expression correlation and cross-species embedding of the transcriptomic manifolds112.
Finally, we identified shared marker genes between cell types of B. lanceolatum and the query chordate species by identifying sets of cell type-overexpressed genes with the scanpy 1.9.3113 rank_genes_groups function to calculate cell type-level fold change values and overexpression significance (Wilcoxon rank-sum tests followed by BH p-value adjustment). For each species, cell type-specific genes were then determined based on fold change and overexpression significance (at adjusted p < 0.05 and FC ≥ 1).
For cross-species comparisons, genes were linked based on shared orthology group membership. Orthology groups between genes of the the four species were determined using Broccoli 1.1114 (using predicted peptides as input; disabling the k-mer clustering step; using up to 10 hits per species for maximum-likelihood phylogenetic tree calculations. If available, we gave preference to phylogeny-derived annotation of orthology groups (see below) over the Broccoli orthology assignments. The orthology group assignments of all genes are available in Supplementary Data 3.
We also performed a more detailed analysis of shared TFs between amphioxus and the other three chordates, selecting cell type-specific amphioxus TFs (p < 0.05 and FC ≥ 1.25; see below details on TF annotation) and evaluating whether their orthologs in chordates were also over-expressed in cell types homologous to the amphioxus endoderm (in this case, it was compared to endodermal tissues in the other chordates), endostyle (to other endodermal tissues), muscular somites (to vertebrate skeletal muscle and tunicate muscle/heart), somites (to vertebrate presomitic mesoderm or tunicate muscle/heart), notochord (to other notochordal tissues) hypothalamus and neurons (each of which was compared to vertebrate neurons, hindbrain, forebrain/midbrain, notoplate and neuroendocrine cells; and to the tunicate nervous system), and the anterior and posterior epidermis (each compared to epidermal progenitors, ionocytes, small secretory epidermal cells, goblet cells, and hatching gland).
Gene family annotation
We ran gene phylogenies to refine the orthology assignments of TF gene families. We used translated peptide sequences from 32 metazoan (longest isoforms per gene, Supplementary Data 3, which were scanned using hmmsearch (HMMER 3.3.2115) to identify hits of TF-specific HMM profiles (from Pfam 33.0116) representing their corresponding DNA-binding regions. For each gene family, the collection of homologous proteins was aligned to itself using diamond blastp v0.9.36117 and clustered into low-granularity homology groups using the Markov Cluster Algorithm MCL v14.137118 (using alignment bit-scores as weights, and a gene family-specific inflation parameter; Supplementary Data 3). Then, each homology group was aligned using mafft 7.475119 (E-INS-i mode, up to 10,000 refinement iterations). The alignments were trimmed with clipkit 1.1.3120 (kpic-gappy mode and a gap threshold = 0.7) and used to build phylogenetic trees with IQ-TREE v2.1121 (running each tree for up to 10,000 iterations until convergence threshold of 0.999 is met for 200 generations; the best-fitting evolutionray model was selected with ModelFinder122; statistical supports were obtained using the UFBoot procedure with 1,000 iterations123). Outlier genes were removed from each tree using treeshrink v1.3.363 (gene-wise mode using the centroid rooting algorithm; scaling factors set to a = 10 and b = 1); and the trees were recalculated if necessary if any outgroup needed to be removed. Finally, we used Possvm 1.1124 to identify orthology groups from each gene tree (with up to 10 steps of iterative gene tree rooting), and annotated the orthogroups and the B. lanceolatum TFs with reference human gene names.
For genes used to assign metacells to known amphioxus embryonic territories and named in the manuscript, we either used the previously published amphioxus gene names when they exist, or a name based on fine orthology analysis. Amino acid sequences from B. lanceolatum were used to search Genbank for putative homologs by blastp. Sequences were aligned using ClustalX125. Alignments were manually corrected in SeaView126. Maximum Likelihood phylogenetic trees were reconstructed using IQ-TREE v2.1121 with default parameters (fast bootstraping and automatic best model search). Genes with no clear orthology signal were named based on the presence of known protein domains.
In situ hybridization and immunostaining
DIG labeled probes were synthesized from fragments cloned into pBKS, or from PCR amplified DNA fragments purchased at Integrated DNA Technologies, Inc (IDT), using the appropriate RNA polymerase (T7, T3 or SP6; Roche, RPOLT7-RO, RPOLT3-RO, RPOLSP6-RO) and the DIG-labeling Mix (Roche, 11277065910). Amphioxus embryos at 21 hpf (19 °C) were fixed in paraformaldehyde (PFA) 4% in MOPS buffer, dehydrated in 70% ethanol and kept at -20 °C. Colorimetric In situ hybridization was undertaken as previously described in36. HCR in situ hybridization was performed as described in127. Ten embryos were used for each experiment. The probes were designed using the following generator (https://github.com/rwnull/insitu_probe_generator) as published in128 and obtained from IDT, and HCR amplifiers (B1-Alexa647 for Brachyury2, Gata1/2/3, and Tcf21/Msc; B2-Alexa488 for Dmbx, MLC-alk and Ripply; B3-Alexa594 for Alx and Fgfrl) were obtained from Molecular Instruments, Inc. Immunostaining after HCR was undertaken as described in14. We used anti-laminin antibody produced in rabbit at 1:50 dilution (Sigma, L9393), and the Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor” 680 at 1:500 dilution (InvitrogenTM, A-21076). For imaging, embryos were embedded into ProLong™ Diamond Antifade Mountant with DAPI (InvitrogenTM, P36961) and confocal stacks were acquired on whole embryos on a Leica SP8 Confocal microscope using a 40X oil-immersion objective. Images were processed using ImageJ (Fiji) and IMARIS v9.7 (Bitplane). Zebrafish embryos were raised until the desired stage, fixed in 4% PFA and dehydrated in methanol. HCR technique was performed as previously described in129 and Molecular Instrument’s web page (https://www.molecularinstruments.com/hcr-rnafish-protocols). 20 embryos were used for each HCR experiment. Oligo pools used as probes were designed using Easy_HCR130 and obtained from IDT. HCR amplifiers (B5-Alexa Fluor-647 for Prrx1a and B1-Alexa Fluor-546 for GFP) were obtained from Molecular Instruments, Inc. The accession numbers/sequences used for probe synthesis or as HCR probes are given in Supplementary Data 5.
Zebrafish transgenesis
The putative regulatory regions were cloned after PCR amplification on genomic DNA in the PCR8/GW/TOPO vector (Life Technologies). Using Gateway technology (Life Technologies), the inserts were then shuttled into an enhancer detection vector composed of a gata2 minimal promoter, an enhanced GFP reporter gene, and a strong midbrain enhancer (z48) that works as an internal control for transgenesis in zebrafish131. Transgenic embryos were generated using the Tol2 transposase system132. Briefly, 1-cell stage embryos were injected with 2 nl of a mix containing 25 ng/µL of Tol2 transposase mRNA, 20 ng/µL of purified vector, and 0.05% of phenol red. Zebrafish embryos were obtained from AB and Tübingen strains at the fish facility of Centro Andaluz de Biología del Desarrollo (Seville, Spain). Each crossing implied 10 females and 10 males aged 4 months. The sex of the embryos was not determined. 300 embryos were injected at one-cell stage for the generation of each transgenic line. We then raised the embryos that exhibited a strong GFP expression in the midbrain (approximately a 15-20% of them), driven by the z48 enhancer. When these animals (F0) reached adulthood (4 months), they were outcrossed with wild type animals in order to identify founders and generate stable transgenic lines (F1). Around 30% of the selected animals resulted to be founders, generating a stable transgenic offspring. An element was considered negative when at least three founders transmitted GFP expression only in the midbrain. On the contrary, when other domains besides the midbrain were observed in the offspring of at least three independent founders, and presented no (or very subtle) variation between them, this element was considered positive. From all the constructs injected and screened for F1, 3 showed 3 founders with similar expression (see Supplementary Data 6). The other animals screened showed the expression of the internal control in the midbrain, but not a consistent and specific expression in other domains in 3 independent founders. Embryos were raised until the desired stage, visualized under an Olympus SZX16 fluorescence stereoscope and photographed with an Olympus DP71 camera or fixed in PFA for in situ hybridization. The transgenic lines are no longer available.
Constitutive repressor mRNA injection
Constitutive repressor form of Gata1/2/3 was created by fusing the coding sequence of the repressor domain of the engrailed protein133 to the N-terminal side of the full-length sequence of Gata1/2/3. The sequence (see Supplementary Data 5) was inserted into pCS2 + . The vector was linearized and in vitro transcription was performed using the mMESSAGE mMACHINE® SP6 Transcription Kit (InvitrogenTM, AM1340). Microinjections were carried out as described in134 with some minor changes. Briefly, eggs were fertilized and dechorionated by pipetting. They were placed in an Petri dish coated with agarose and microinjected with a mix containing 1.5 µg of Gata1/2/3-Engrailed mRNA, 0,5 µg of mCherry mRNA, 18% of glycerol and 18% of Fast Green. 10 hours after fertilization, the embryos showing red fluorescence were placed in a new Petri dish without coating. The embryos were then fixed when desired for subsequent in situ hybridization experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The B. lanceolatum sequencing data generated in this study have been deposited in the GEO database under accession code GSE255742, and the corresponding processed gene expression data are available in this same database and the Source Data file. The gene expression data from other chordates, generated in previous studies, are available in the following databases: for Ciona intestinalis GEO database GSE131155; M. musculus, D. rerio and X. tropicalis, TOME database (http://tome.gs.washington.edu/); B. floridae, the publication-specific database (https://lifeomics.shinyapps.io/shinyappmulti/). Accession numbers of sequences used for in situ hybridization probe synthesis are given in Supplementary Data 5. Source data are provided with this paper.
References
Annona, G., Holland, N. D. & D’Aniello, S. Evolution of the notochord. Evodevo 6, 30 (2015).
Delsuc, F., Brinkmann, H., Chourrout, D. & Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968 (2006).
Lemaire, P. Evolutionary crossroads in developmental biology: the tunicates. Development 138, 2143–2152 (2011).
Holland, L. Z. Genomics, evolution and development of amphioxus and tunicates: The Goldilocks principle. J. Exp. Zool. Part B: Mol. Developmental Evolution 324, 342–352 (2015).
Bertrand, S. & Escriva, H. Evolutionary crossroads in developmental biology: amphioxus. Development 138, 4819–4830 (2011).
Escriva, H. My Favorite Animal, Amphioxus: Unparalleled for Studying Early Vertebrate Evolution. BioEssays 40, 1800130 (2018).
Holland, L. Z. & Onai, T. Early development of cephalochordates (amphioxus). WIREs Developmental Biol. 1, 167–183 (2012).
Prummel, K. D., Nieuwenhuize, S. & Mosimann, C. The lateral plate mesoderm. Development 147, dev175059 (2020).
Diogo, R. et al. A new heart for a new head in vertebrate cardiopharyngeal evolution. Nature 520, 466–473 (2015).
Sambasivan, R., Kuratani, S. & Tajbakhsh, S. An eye on the head: the development and evolution of craniofacial muscles. Development 138, 2401–2415 (2011).
Kozmik, Z. et al. Characterization of Amphioxus AmphiVent, an evolutionarily conserved marker for chordate ventral mesoderm. Genesis 29, 172–179 (2001).
Holland, N. D., Venkatesh, T. V., Holland, L. Z., Jacobs, D. K. & Bodmer, R. AmphiNk2-tin, an amphioxus homeobox gene expressed in myocardial progenitors: insights into evolution of the vertebrate heart. Dev. Biol. 255, 128–137 (2003).
Onimaru, K., Shoguchi, E., Kuratani, S. & Tanaka, M. Development and evolution of the lateral plate mesoderm: comparative analysis of amphioxus and lamprey with implications for the acquisition of paired fins. Dev. Biol. 359, 124–136 (2011).
Pascual-Anaya, J. et al. The evolutionary origins of chordate hematopoiesis and vertebrate endothelia. Dev. Biol. 375, 182–192 (2013).
Prummel, K. D. et al. A conserved regulatory program initiates lateral plate mesoderm emergence across chordates. Nat. Commun. 10, 1–15 (2019).
Mansfield, J. H., Haller, E., Holland, N. D. & Brent, A. E. Development of somites and their derivatives in amphioxus, and implications for the evolution of vertebrate somites. EvoDevo 6, 1–30 (2015).
Yong, L. W. et al. Somite compartments in amphioxus and its implications on the evolution of the vertebrate skeletal tissues. Front. Cell Develop. Biol. 9, 607057 (2021).
Holland, N. D. Formation of the initial kidney and mouth opening in larval amphioxus studied with serial blockface scanning electron microscopy (SBSEM). Evodevo 9, 16 (2018).
Langeland, J. A., Holland, L. Z., Chastain, R. A. & Holland, N. D. An amphioxus LIM-homeobox gene, AmphiLim1/5, expressed early in the invaginating organizer region and later in differentiating cells of the kidney and central nervous system. Int. J. Biol. Sci. 2, 110 (2006).
Albuixech-Crespo, B. et al. Molecular regionalization of the developing amphioxus neural tube challenges major partitions of the vertebrate brain. PLoS Biol. 15, e2001573 (2017).
Andrews, T. G. R., Pönisch, W., Paluch, E. K., Steventon, B. J. & Benito-Gutierrez, E. Single-cell morphometrics reveals ancestral principles of notochord development. Development 148, dev199430 (2021).
Meister, L., Escriva, H. & Bertrand, S. Functions of the FGF signalling pathway in cephalochordates provide insight into the evolution of the prechordal plate. Development 149, dev200252 (2022).
Aldea, D. et al. Genetic regulation of amphioxus somitogenesis informs the evolution of the vertebrate head mesoderm. Nat. Ecol. Evol. 3, 1233–1240 (2019).
Bertrand, S. et al. The ontology of the amphioxus anatomy and life cycle (AMPHX). Front Cell Dev. Biol. 9, 668025 (2021).
Carvalho, J. E. et al. An updated staging system for cephalochordate development: one table suits them all. Front Cell Dev. Biol. 9, 668006 (2021).
Marletaz, F. et al. Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 564, 64–70 (2018).
Duboule, D. Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development 1994, 135–142 (1994).
Ferran, J. L., Irimia, M. & Puelles, L. Is there a prechordal region and an acroterminal domain in amphioxus? Brain, Behav. Evol. 96, 334–352 (2022).
Keren-Shaul, H. et al. MARS-seq2.0: an experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing. Nat. Protoc. 14, 1841–1862 (2019).
Baran, Y. et al. MetaCell: analysis of single-cell RNA-seq data using K-nn graph partitions. Genome Biol. 20, 1–19 (2019).
Bertrand, S., Somorjai, I., Garcia-Fernandez, J., Lamonerie, T. & Escriva, H. FGFRL1 is a neglected putative actor of the FGF signalling pathway present in all major metazoan phyla. BMC Evolut. Biol. 9, 226 (2009).
Glardon, S., Holland, L. Z., Gehring, W. J. & Holland, N. D. Isolation and developmental expression of the amphioxus Pax-6 gene (AmphiPax-6): insights into eye and photoreceptor evolution. Development 125, 2701–2710 (1998).
Qian, G., Li, G., Chen, X. & Wang, Y. Characterization and embryonic expression of four amphioxus Frizzled genes with important functions during early embryogenesis. Gene Expr. Patterns 13, 445–453 (2013).
Belgacem, M. R., Escande, M.-l, Escriva, H. & Bertrand, S. Amphioxus Tbx6/16 and Tbx20 embryonic expression patterns reveal ancestral functions in chordates. Gene Expr. Patterns 11, 239–243 (2011).
Brooke, N. M., Garcia-Fernàndez, J. & Holland, P. W. H. The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 392, 920–922 (1998).
Somorjai, I., Bertrand, S., Camasses, A., Haguenauer, A. & Escriva, H. Evidence for stasis and not genetic piracy in developmental expression patterns of Branchiostoma lanceolatum and Branchiostoma floridae, two amphioxus species that have evolved independently over the course of 200 Myr. Dev. Genes Evol. 218, 703–713 (2008).
Benito-Gutierrez, E., Illas, M., Comella, J. X. & Garcia-Fernandez, J. Outlining the nascent nervous system of Branchiostoma floridae (amphioxus) by the pan-neural marker AmphiElav. Brain Res Bull. 66, 518–521 (2005).
Kaltenbach, S. L., Yu, J. K. & Holland, N. D. The origin and migration of the earliest‐developing sensory neurons in the peripheral nervous system of amphioxus. Evolution Dev. 11, 142–151 (2009).
Rasmussen, S. L., Holland, L. Z., Schubert, M., Beaster‐Jones, L. & Holland, N. D. Amphioxus AmphiDelta: evolution of Delta protein structure, segmentation, and neurogenesis. Genesis 45, 113–122 (2007).
Williams, N. A. & Holland, P. W. Old head on young shoulders. Nature 383, 490–490 (1996).
Ferrier, D. E., Brooke, N. M., Panopoulou, G. & Holland, P. W. The Mnx homeobox gene class defined by HB9, MNR2 and amphioxus AmphiMnx. Dev. Genes Evol. 211, 103–107 (2001).
Meulemans, D. & Bronner-Fraser, M. Insights from amphioxus into the evolution of vertebrate cartilage. PLoS One 2, e787 (2007).
Shimeld, S. An amphioxus netrin gene is expressed in midline structures during embryonic and larval development. Dev. Genes Evol. 210, 337–344 (2000).
Shimeld, S. M. Characterisation of amphioxus HNF-3 genes: conserved expression in the notochord and floor plate. Dev. Biol. 183, 74–85 (1997).
Wu, H. R. et al. Asymmetric localization of germline markers Vasa and Nanos during early development in the amphioxus Branchiostoma floridae. Dev. Biol. 353, 147–159 (2011).
Zhang, Q. J., Luo, Y. J., Wu, H. R., Chen, Y. T. & Yu, J. K. Expression of germline markers in three species of amphioxus supports a preformation mechanism of germ cell development in cephalochordates. Evodevo 4, 17 (2013).
Mazet, F. The Fox and the thyroid: the amphioxus perspective. Bioessays 24, 696–699 (2002).
Ma, P. et al. Joint profiling of gene expression and chromatin accessibility during amphioxus development at single-cell resolution. Cell Rep. 39, 110979 (2022).
Cao, C. et al. Comprehensive single-cell transcriptome lineages of a proto-vertebrate. Nature 571, 349–354 (2019).
Briggs, J. A. et al. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780 (2018).
Wagner, D. E. et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019).
Huang, Z. et al. Three amphioxus reference genomes reveal gene and chromosome evolution of chordates. Proc. Natl Acad. Sci. USA 120, e2201504120 (2023).
Leon, A. et al. Gene Regulatory Networks of Epidermal and Neural Fate Choice in a Chordate. Mol. Biol. Evol. 39, msac055 (2022).
Li, L. et al. TFAP2C- and p63-dependent networks sequentially rearrange chromatin landscapes to drive human epidermal lineage commitment. Cell Stem Cell 24, 271–284 e278 (2019).
Miles, L. B. et al. Mis-expression of grainyhead-like transcription factors in zebrafish leads to defects in enveloping layer (EVL) integrity, cellular morphogenesis and axial extension. Sci. Rep. 7, 17607 (2017).
Pera, E., Stein, S. & Kessel, M. Ectodermal patterning in the avian embryo: epidermis versus neural plate. Development 126, 63–73 (1999).
Segre, J. A., Bauer, C. & Fuchs, E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 22, 356–360 (1999).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Mastromina, I., Verrier, L., Silva, J. C., Storey, K. G. & Dale, J. K. Myc activity is required for maintenance of the neuromesodermal progenitor signalling network and for segmentation clock gene oscillations in mouse. Development 145, dev161091 (2018).
Beaster‐Jones, L., Horton, A. C., Gibson‐Brown, J. J., Holland, N. D. & Holland, L. Z. The amphioxus T‐box gene, AmphiTbx15/18/22, illuminates the origins of chordate segmentation. Evolution Dev. 8, 119–129 (2006).
Horton, A. C. & Gibson‐Brown, J. J. Evolution of developmental functions by the Eomesodermin, T‐brain‐1, Tbx21 subfamily of T‐box genes: insights from amphioxus. J. Exp. Zool. 294, 112–121 (2002).
Holland, P. W., Koschorz, B., Holland, L. Z. & Herrmann, B. G. Conservation of Brachyury (T) genes in amphioxus and vertebrates: developmental and evolutionary implications. Development 121, 4283–4291 (1995).
Aase-Remedios, M. E., Coll-Lladó, C. & Ferrier, D. E. More than one-to-four via 2R: evidence of an independent amphioxus expansion and two-gene ancestral vertebrate state for MyoD-related myogenic regulatory factors (MRFs). Mol. Biol. Evol. 37, 2966–2982 (2020).
Hernández-Hernández, J. M., García-González, E. G., Brun, C. E. & Rudnicki, M. A. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Dev. Biol. 72, 10–18 (2017).
Venkatesh, T. V., Holland, N. D., Holland, L. Z., Su, M.-T. & Bodmer, R. Sequence and developmental expression of amphioxus AmphiNk2–1: insights into the evolutionary origin of the vertebrate thyroid gland and forebrain. Dev. genes Evol. 209, 254–259 (1999).
Kong, W., Yang, Y., Zhang, T., Shi, D. L. & Zhang, Y. Characterization of s FRP 2‐like in amphioxus: insights into the evolutionary conservation of W nt antagonizing function. Evol. Dev. 14, 168–177 (2012).
Takahashi, T. & Holland, P. W. Amphioxus and ascidian Dmbx homeobox genes give clues to the vertebrate origins of midbrain development. Development 131, 3285–3294 (2004).
Yu, J. K. et al. Axial patterning in cephalochordates and the evolution of the organizer. Nature 445, 613–617 (2007).
Holland, N. D., Holland, L. Z. & Kozmik, Z. An amphioxus Pax gene, AmphiPax-1, expressed in embryonic endoderm, but not in mesoderm: implications for the evolution of class I paired box genes. Mol. Mar. Biol. Biotechnol. 4, 206–214 (1995).
Mahadevan, N. R., Horton, A. C. & Gibson-Brown, J. J. Developmental expression of the amphioxus Tbx1/10 gene illuminates the evolution of vertebrate branchial arches and sclerotome. Dev. genes Evol. 214, 559–566 (2004).
Boorman, C. J. & Shimeld, S. M. Pitx homeobox genes in Ciona and amphioxus show left–right asymmetry is a conserved chordate character and define the ascidian adenohypophysis. Evol. Dev. 4, 354–365 (2002).
Cattell, M. V., Garnett, A. T., Klymkowsky, M. W. & Medeiros, D. M. A maternally established SoxB1/SoxF axis is a conserved feature of chordate germ layer patterning. Evol. Dev. 14, 104–115 (2012).
Kaltenbach, S. L., Holland, L. Z., Holland, N. D. & Koop, D. Developmental expression of the three iroquois genes of amphioxus (BfIrxA, BfIrxB, and BfIrxC) with special attention to the gastrula organizer and anteroposterior boundaries in the central nervous system. Gene Expr. Patterns 9, 329–334 (2009).
Holland, L. Z., Venkatesh, T. V., Gorlin, A., Bodmer, R. & Holland, N. Characterization and developmental expression of AmphiNk2-2, an NK2 class homeobox gene from amphioxus (Phylum Chordata; Subphylum Cephalochordata). Dev. Genes Evol. 208, 100 (1998).
Somorjai, I. M. L. et al. Wnt evolution and function shuffling in liberal and conservative chordate genomes. Genome Biol. 19, 98 (2018).
Gostling, N. J. & Shimeld, S. M. Protochordate Zic genes define primitive somite compartments and highlight molecular changes underlying neural crest evolution. Evol. Dev. 5, 136–144 (2003).
Kozmik, Z. et al. Pax-Six-Eya-Dach network during amphioxus development: conservation in vitro but context specificity in vivo. Dev. Biol. 306, 143–159 (2007).
Holland, L. Z., Schubert, M., Kozmik, Z. & Holland, N. D. AmphiPax3/7, an amphioxus paired box gene: insights into chordate myogenesis, neurogenesis, and the possible evolutionary precursor of definitive vertebrate neural crest. Evolution Dev. 1, 153–165 (1999).
Venkatesh, T. V., Holland, N. D., Holland, L. Z., Su, M. T. & Bodmer, R. Sequence and developmental expression of amphioxus AmphiNk2-1: insights into the evolutionary origin of the vertebrate thyroid gland and forebrain. Dev. Genes Evol. 209, 254–259 (1999).
Holland, L. Z., Pace, D. A., Blink, M. L., Kene, M. & Holland, N. D. Sequence and expression of amphioxus alkali myosin light chain (amphimlc-alk) throughout development: implications for vertebrate myogenesis. Developmental Biol. 171, 665–676 (1995).
Zhang, Y., Wang, L., Shao, M. & Zhang, H. Characterization and developmental expression of AmphiMef2 gene in amphioxus. Sci. China Ser. C: Life Sci. 50, 637–641 (2007).
Kozmikova, I., Candiani, S., Fabian, P., Gurska, D. & Kozmik, Z. Essential role of Bmp signaling and its positive feedback loop in the early cell fate evolution of chordates. Dev. Biol. 382, 538–554 (2013).
Li, X. et al. Expression of a novel somite-formation-related gene, AmphiSom, during amphioxus development. Dev. genes evolution 216, 52–55 (2006).
Le Petillon, Y., Oulion, S., Escande, M.-L., Escriva, H. & Bertrand, S. Identification and expression analysis of BMP signaling inhibitors genes of the DAN family in amphioxus. Gene Expr. Patterns 13, 377–383 (2013).
Xiong, J.-W. Molecular and developmental biology of the hemangioblast. Developmental Dyn. 237, 1218–1231 (2008).
Moncaut, N. et al. Musculin and TCF21 coordinate the maintenance of myogenic regulatory factor expression levels during mouse craniofacial development. Development 139, 958–967 (2012).
Mundhada, A., Kulkarni, U., Swami, V., Deshmukh, S. & Patil, A. Craniofacial Muscles-differentiation and Morphogenesis. Annu. Res. Revi. Bio. 9, 1–9 (2016).
Schubert, F. R., Singh, A. J., Afoyalan, O., Kioussi, C. & Dietrich, S. To roll the eyes and snap a bite – function, development and evolution of craniofacial muscles. Semin. Cell Developmental Biol. 91, 31–44 (2019).
Bertrand, S. et al. Amphioxus FGF signaling predicts the acquisition of vertebrate morphological traits. Proc. Natl Acad. Sci. USA 108, 9160–9165 (2011).
Tzahor, E. Heart and craniofacial muscle development: A new developmental theme of distinct myogenic fields. Developmental Biol. 327, 273–279 (2009).
Li, G. et al. Cerberus-Nodal-Lefty-Pitx signaling cascade controls left-right asymmetry in amphioxus. Proc. Natl Acad. Sci. USA 114, 3684–3689 (2017).
Xing, C. et al. Pitx controls amphioxus asymmetric morphogenesis by promoting left-side development and repressing right-side formation. BMC Biol. 19, 166 (2021).
Koop, D. et al. Roles of retinoic acid and Tbx1/10 in pharyngeal segmentation: amphioxus and the ancestral chordate condition. EvoDevo 5, 36 (2014).
Essner, J. J., Branford, W. W., Zhang, J. & Yost, H. J. Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development 127, 1081–1093 (2000).
Faucourt, M., Houliston, E., Besnardeau, L., Kimelman, D. & Lepage, T. The Pitx2 homeobox protein is required early for endoderm formation and nodal signaling. Developmental Biol. 229, 287–306 (2001).
John, L. B., Trengove, M. C., Fraser, F. W., Yoong, S. H. & Ward, A. C. Pegasus, the ‘atypical’ Ikaros family member, influences left–right asymmetry and regulates pitx2 expression. Developmental Biol. 377, 46–54 (2013).
Wang, H., Holland, P. W. H. & Takahashi, T. Gene profiling of head mesoderm in early zebrafish development: insights into the evolution of cranial mesoderm. EvoDevo 10, 14 (2019).
Fuentes, M. et al. Insights into spawning behavior and development of the European amphioxus (Branchiostoma lanceolatum). J. Exp. Zool. B Mol. Dev. Evol. 308, 484–493 (2007).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2012).
Gu, Z., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Magri, M. S. et al. Assaying chromatin accessibility using ATAC-seq in invertebrate chordate embryos. Front Cell Dev. Biol. 7, 372 (2019).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Tischler, G. & Leonard, S. biobambam: tools for read pair collation based algorithms on BAM files. Source Code Biol. Med. 9, 13 (2014).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS computational Biol. 9, e1003118 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Weirauch, M. T. et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443 (2014).
WEBSITE: Tremblay BJ. universalmotif: Import, Modify, and Export Motifs with R. R package version 1.22.0, https://bioconductor.org/packages/universalmotif/ (2024).
Machlab, D. et al. monaLisa: an R/Bioconductor package for identifying regulatory motifs. Bioinformatics 38, 2624–2625 (2022).
Tarashansky, A. J. et al. Mapping single-cell atlases throughout Metazoa unravels cell type evolution. eLife 10, e66747 (2021).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Derelle, R., Philippe, H. & Colbourne, J. K. Broccoli: Combining Phylogenetic and Network Analyses for Orthology Assignment. Mol. Biol. Evolution 37, 3389–3396 (2020).
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A. & Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41, e121 (2013).
Punta, M. et al. The Pfam protein families database. Nucleic acids Res. 40, D290–D301 (2012).
Buchfink, B., Reuter, K. & Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 18, 366–368 (2021).
Enright, A. J., Van Dongen, S. & Ouzounis, C. A. An efficient algorithm for large-scale detection of protein families. Nucleic acids Res. 30, 1575–1584 (2002).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evolution 30, 772–780 (2013).
Steenwyk, J. L., Buida III, T. J., Li, Y., Shen, X.-X. & Rokas, A. ClipKIT: a multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biol. 18, e3001007 (2020).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Grau-Bové, X. & Sebé-Pedrós, A. Orthology clusters from gene trees with possvm. Mol. Biol. Evol. 38, 5204–5208 (2021).
Larkin, M. A. et al. Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Gouy, M., Guindon, S. & Gascuel, O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010).
Andrews, T. G. R., Gattoni, G., Busby, L., Schwimmer, M. A. & Benito-Gutiérrez, È. Hybridization Chain Reaction for Quantitative and Multiplex Imaging of Gene Expression in Amphioxus Embryos and Adult Tissues. Methods Mol. Biol. 2148, 179–194 (2020).
Kuehn, E. et al. Segment number threshold determines juvenile onset of germline cluster expansion in Platynereis dumerilii. J. Exp. Zool. Part B: Mol. Dev. Evol. 338, 225–240 (2022).
Schwarzkopf, M. et al. Hybridization chain reaction enables a unified approach to multiplexed, quantitative, high-resolution immunohistochemistry and in situ hybridization. Development 148, dev199847 (2021).
Elagoz, A. M. et al. Optimization of whole mount RNA multiplexed in situ hybridization chain reaction with immunohistochemistry, clearing and imaging to visualize octopus embryonic neurogenesis. Front Physiol. 13, 882413 (2022).
Gehrke, A. R. et al. Deep conservation of wrist and digit enhancers in fish. Proc. Natl Acad. Sci. USA 112, 803–808 (2015).
Kawakami, K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 8, S7 (2007).
Jaynes, J. B. & O’Farrell, P. H. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 10, 1427–1433 (1991).
Hirsinger, E. et al. Expression of fluorescent proteins in Branchiostoma lanceolatum by mRNA injection into unfertilized oocytes. J. Vis. Exp. 52042, https://doi.org/10.3791/52042 (2015).
Acknowledgements
We would like to thank Jose Luis Ferran for advices on the revision. This work benefited from access to the Observatoire Océanologique de Banyuls-sur-Mer, an EMBRC-France and EMBRC-ERIC site. Embryo imaging experiments were undertaken using the material of the BIOPIC platform. The laboratory of H.E. and S.B. was supported by the CNRS, and by the “Agence Nationale de la Recheche” under the grants ANR-19-CE13-0011 to H.E. and ANR-21-CE13-0034 to S.B. Research in A.S-P. group was supported by the European Research Council (ERC-StG 851647 to A.S-P.) and the Spanish Ministry of Science and Innovation (PID2021-124757NB-I00 to A.S-P.). X.G-B. is supported by the European Union’s H2020 research and innovation program under Marie Sklodowska-Curie grant agreement 101031767 to X.G-B. A.E. was supported by FPI PhD fellowships from the Spanish Ministry of Science and Innovation. J.J.T. was supported by the Spanish Ministerio de Economía y Competitividad (grant PID2019-103921GB-I00 and PID2022-141288NB-I00 to J.T.). M.I. laboratory research has been funded by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (ERCCoG-LS2-101002275 to M.I.), by the Spanish Ministry of Economy and Competitiveness (PID2020-115040GB-I00 to M.I.) and by the ‘Centro de Excelencia Severo Ochoa 2013-2017’(SEV-2012-0208 to M. I.).
Author information
Authors and Affiliations
Contributions
Conceptualization of this study was done by J.L. G-Z., M.I., S.B., A.S.P.and H.E.; the study was carried out by X.G.B., L.S., L.M., A.S, A.N., A.E., S.N., O.F., M.I., S.B., A.S.P. and H.E.; writing of the original draft was done by X.G.B., S.B., A.S.P. and H.E.; funding was acquired by A.S.B., J.T., M.I., S.B., A.S.P. and H.E.; this study was supervised by J.L. G-Z, J.T., M.I., S.B., A.S.P. and H.E.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Grau-Bové, X., Subirana, L., Meister, L. et al. An amphioxus neurula stage cell atlas supports a complex scenario for the emergence of vertebrate head mesoderm. Nat Commun 15, 4550 (2024). https://doi.org/10.1038/s41467-024-48774-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-48774-4
- Springer Nature Limited