Abstract
The dominance effect is considered to be a key factor affecting complex traits. However, previous studies have shown that the improvement of the model, including the dominance effect, is usually less than 1%. This study proposes a novel genomic prediction method called CADM, which combines additive and dominance genetic effects through locus-specific weights on heterozygous genotypes. To the best of our knowledge, this is the first study of weighting dominance effects for genomic prediction. This method was applied to the analysis of chicken (511 birds) and pig (3534 animals) datasets. A 5-fold cross-validation method was used to evaluate the genomic predictive ability. The CADM model was compared with typical models considering additive and dominance genetic effects (ADM) and the model considering only additive genetic effects (AM). Based on the chicken data, using the CADM model, the genomic predictive abilities were improved for all three traits (body weight at 12th week, eviscerating percentage, and breast muscle percentage), and the average improvement in prediction accuracy was 27.1% compared with the AM model, while the ADM model was not better than the AM model. Based on the pig data, the CADM model increased the genomic predictive ability for all the three pig traits (trait names are masked, here designated as T1, T2, and T3), with an average increase of 26.3%, and the ADM model did not improve, or even slightly decreased, compared with the AM model. The results indicate that dominant genetic variation is one of the important sources of phenotypic variation, and the novel prediction model significantly improves the accuracy of genomic prediction.
Similar content being viewed by others
Introduction
With the continuous growth of the global population, human demand for food and security continues to increase. Hybridization is an important tool for increasing meat and grain production. It has been successfully applied to the production of animals (such as chickens and pigs) and plants (such as corn and rice). The performance of hybrid individuals is affected by genetic factors such as additive, dominance, and epistatic genetic effects. Traditional genetic evaluation methods only consider additive genetic effects but not nonadditive genetic effects (dominance and epistatic effects). Many studies have shown that nonadditive genetic effects are an important component of phenotypic values (Costa-Neto et al. 2021; Mao et al. 2020; Su et al. 2012; Vitezica et al. 2017; Wang et al. 2020; Xu 2013). Li et al. (2017) reported that the dominant variance of broiler feed-related traits accounted for 29.5–58.4% of the genetic variance. There are three main reasons why traditional genetic evaluation methods only consider additive effects. (1) Additive genetic effects are the additive effects of a number of genes that can be accumulative and stably inherited, while nonadditive genetic effects are the effects of gene interactions that can change by allele recombination and thus cannot be stably inherited. (2) The traditional pedigree-based genetic evaluation method has difficulty accurately estimating nonadditive genetic effects since pedigree-based nonadditive genetic relationships between individuals are usually weak, and it is difficult to distinguish nonadditive genetic effects (such as dominance effects) and common environmental effects on siblings. (3) There is a high computational demand for computing the inverses of relationship matrices for non-additive genetic effects for large data.
Genomic prediction may overcome the limitation of conventional pedigree-based prediction in the estimation of nonadditive genetic effects. Because genomic prediction is based on genome information to predict the genetic value of individuals, it can provide a new approach to detect nonadditive genetic relationships between animals in the population and accurately distinguish the genotypic differences between sibling individuals (Meuwissen et al. 2001; Su et al. 2012). There is increasing interest in studies on genomic prediction considering nonadditive genetic effects, especially dominance effects (Su et al. 2012; Vitezica et al. 2017; Wang et al. 2020; Xiang et al. 2018). Genomic prediction models fitting dominance effects have been developed and constantly improved. Su et al. (2012) proposed a genomic model that fits the dominance effect at the unweighted scale of the dominance effect. Vitezica et al. (2013) presented a model fitting dominance effect on the scale of the gene substitution effect, which is consistent with the classical definition of quantitative genetics and breeding. Furthermore, Vitezica et al. (2017) proposed an expanded method from the natural and orthogonal interactions (NOIA) approach, which uses genotypic (not allelic) frequencies to construct incidence matrix. Xiang et al. (2018) proposed a model with a covariance matrix to account for the relationship between additive and dominance effects. However, using these models, many studies have reported that the improvement in accuracy of predicting genetic values is very small, mostly less than 1%, or even no improvement (Aliloo et al. 2016; Su et al. 2012; Vitezica et al. 2013; Xiang et al. 2018).
Although many studies have shown that models considering dominance effects do not lead to a clear improvement in predicting breeding values and total genetic values, many studies have shown that dominance variances are considerable in complex traits of animals and plants (Su et al. 2012; Vitezica et al. 2017; Xu et al. 2014). The universal phenomenon of heterosis in animals and plants also indicates that dominance effects are substantial. Therefore, there is a need for alternative statistical methods and models that can efficiently estimate genetic values, including dominance effects. This study proposed a novel method to account for genomic dominance effects, which integrated additive and dominance effects through locus-specific weights on heterozygous genotypes to construct a genomic relationship matrix. The model was tested by validation of prediction accuracy based on chicken and pig data.
Materials and methods
The novel prediction model
The model developed in this study is based on the assumption that the degree of dominance is different at different loci, and thus, the model accounts for the difference in degrees of dominance by locus-specific weights on heterozygous genotypes. The approach includes two main steps: (1) estimating the dominant degree based on the deviation between the heterozygous and homozygous marker genotypes; and (2) using the estimated degree of dominance as weights on heterozygous genotypes to construct a genomic relationship matrix. The details of the approach are as follows.
Let the A1 homozygous (A1A1), heterozygous (A1A2), and A2 homozygous (A2A2) genotypes be coded as 0, 1, and 2, respectively. The prior degree of dominance at a locus is defined as
where \(\overline x _{A_1A_1}\), \(\overline x _{A_1A_2}\) and \(\overline x _{A_2A_2}\) are the means of corrected phenotypes of the three genotypes A1A1, A1A2, and A2A2, respectively.
The weighted heterozygous genotype (Cd) is defined as:
At the same time, if \(\overline x _{A_1A_1}\, > \,\overline x _{A_2A_2}\), A1A1 and A2A2 are recoded as 2 and 0, respectively.
The boundaries of 0 and 2 for Cd are to restrict the degree of dominance in the range from zero to complete dominance. Relaxation of the boundaries will allow overdominance.
The above modified genotype coefficients are then applied to construct a genomic relationship matrix in the best linear unbiased predictor (GBLUP) model for estimating the genetic values, including additive and dominance effects. The genomic relationship matrix can be built using the same method as those for the additive genomic relationship matrix (e.g., VanRaden 2008), applying a genotype matrix built with Cd values. A simple simulation in R language was shown to illustrate the calculation of genotype matrix (Supplementary File 1).
Data
The novel model was tested using two published datasets from chickens (Liu et al. 2014) and pigs (Cleveland et al. 2012). The chicken dataset contained 511 F2 birds from two outbred lines. All birds had phenotypic records. The phenotypes were body weight at the 12th week (W12), eviscerating percentage (EP), which is percentage of eviscerated weight to body weight before slaughter, and breast muscle percentage (BMP), which is percentage of breast muscle weight to eviscerated weight. The phenotypes in the chicken data (from one herd) were corrected for fixed effects (sex and batch), and the fixed effects were estimated using linear least squares approach by linear fixed effects model. The birds were genotyped using the Illumina Chicken 60K SNP BeadChip. SNP markers with a minor allele frequency (MAF) lower than 0.01 were deleted. After editing, 46,672 SNPs were used to build the relationship matrix.
The pig dataset consisted of 3534 animals from a single PIC nucleus pig line. The data concealed the actual trait names but used the symbols T1, T2, T3, T4, and T5 instead. Three traits (T1, T2, and T3) were analyzed in this study, and the traits had 2804, 2715, and 3141 phenotypic records, respectively. The three traits were either corrected for environmental factors (e.g., year of birth or farm) and rescaled by correcting for the overall mean (T3) or was a rescaled, weighted mean of corrected progeny phenotypes (T1 and T2), for which many pigs have no individual phenotype (Cleveland et al. 2012). All 3534 animals were genotyped with the Illumina PorcineSNP60 BeadChip. SNP markers with a MAF lower than 0.01 were also deleted. After editing, 52,842 SNPs were used for calculating the relationship matrix.
Statistical analysis
The data were analyzed using three statistical models, i.e., the novel model proposed in this study, a typical GBLUP model considering additive genetic effects only (VanRaden 2008), and a typical additive and dominance GBLUP model (Su et al. 2012).
Genomic BLUP model combining additive and dominance genetic effects
The novel model proposed in this study (CADM) integrates additive and dominance genetic effects and can be written as:
where y is the vector of phenotypic trait records, μ is the overall mean, 1 is a vector of 1s, Zg is incidence matrix of g, g is the vector of the genotype value combining additive and dominance effects, and e is the vector of random residuals. It is assumed that g ~ N(0, \({{{\mathbf{G}}}}_{{{{\boldsymbol{ad}}}}}\sigma _g^2\)) and e ~ N(0, \({{{\mathbf{I}}}}\sigma _e^2\)), where Gad is the additive-dominance genomic relationship matrix, I is an identity matrix, \(\sigma _g^2\) is the additive-dominance genetic variance, and \(\sigma _e^2\) is the residual variance.
The combined additive and dominance genomic relationship matrix Gad was constructed using the modified marker genotype coefficients (Cd), where locus-specific weights were exerted on heterozygous genotypes, and a method similar to the additive genomic relationship matrix was applied (VanRaden 2008). Briefly, \({{{\mathbf{G}}}}_{{{{\boldsymbol{ad}}}}} = \frac{{{{{\mathbf{M}}}}_{{{{\boldsymbol{ad}}}}}{{{\mathbf{M}}}}_{{{{\boldsymbol{ad}}}}}^\prime }}{{{\sum} {2p_i\left( {1 - p_i} \right)} }}\), where Mad is a (n × m) matrix (n = number of individuals, m = number of marker loci) that specifies SNP genotype coefficients at each locus. The coefficients of the ith column in matrix Mad are (0–2pi) for A1A1, (Cd–2pi) for A1A2 where Cd was as defined earlier, and (2–2pi) for A2A2, where pi is the frequency of allele 1 (A1) at locus i.
CADM can be considered as a (co)variance matrix, but the definition is not exactly the same as the one in conventional breeding model. In CADM, we considered dominance effect as 0 for homozygous and non-zero for heterozygous, i.e., a genomic model (Su et al. 2012; Vitezica et al. 2013).
Genomic BLUP model for additive genetic effects
The linear mixed model with additive genetic effects (AM) is:
where the definitions of y and e are the same as those in the CADM model, where a is the vector of additive genetic effects, Za is the corresponding incidence matrix. It was assumed that a ~ N (0, \({{{\mathbf{G}}}}\sigma _a^2\)), where G is the additive relationship matrix and \(\sigma _a^2\) is the additive genetic variance.
The additive genomic relationship matrix G was constructed using the original SNP marker information (VanRaden 2008), in which elements of M are coded as (0–2pi) for A1A1, (1–2pi) for A1A2, and (2–2pi) for A2A2 at locus i.
Genomic BLUP model including additive and dominance genetic effects as two components
The linear mixed model including additive and dominance genetic effects (ADM) is:
where d is the vector of dominance effects, Zd is the corresponding incidence matrix. It is assumed that d ~ N (0, \({{{\mathbf{D}}}}\sigma _d^2\)), where D is the dominance relationship matrix, and \(\sigma _d^2\) is the dominance genetic variance.
The method for constructing the additive genomic relationship matrix G is the same as in the AM model. The dominance genomic relationship matrix D is constructed by \({{{\mathbf{D}}}} = \frac{{{{{\mathbf{HH}}}}^\prime }}{{{\sum} {\left( {2p_iq_i} \right)^2} }}\) (Vitezica et al. 2013) where H is a (n × m) matrix of heterozygosity coefficients. The coefficients of the ith column in matrix H are (−2pi2) for A1A1, (2piqi) for A1A2, and (−2qi2) for A2A2, where qi and pi are the frequencies of allele 1 (A1) and allele 2 (A2) at locus i, respectively.
The variance-covariance components were estimated using the average information restricted maximum likelihood (AIREML) (Gilmour et al. 1995). The estimation of variance components and the prediction of genetic effects were carried out using the DMU package (Madsen et al. 2010).
Cross-validation
The accuracy of genomic prediction was verified by five-fold cross-validation. The entire dataset was randomly split into five subsets of similar size. In each fold of cross-validation, one of the five subsets was used as the validation data in which the phenotypic values were masked during genomic prediction; the remaining four subsets were used as the training data. For each validation, the data used to obtain weights on heterozygous genotypes in the CADM method were in line with the training population (i.e., the records in the validation population were excluded from the data to calculate the degree of dominance). The cross-validation was repeated 10 times.
Prediction accuracy was defined as the correlation between the predictions and corrected phenotypic values. Differences between the correlations from different models were tested by bootstrapping (Efron 1979) and Bonferroni’s method (Bland and Altman 1995). Bootstrap and Bonferroni’s methods were performed with the sample() function and “agricolae” package in R (R Core Team 2020).
Results
Genetic variances
Table 1 shows the variances in proportion to the phenotypic variance for the three quantitative traits (BMP, EP, and W12) from the chicken dataset. For W12, the composite genetic variance (composites of additive and dominance genetic variances) estimated from the CADM model was lowest, and the total genetic variance estimated from the ADM model was highest among the three models. The additive genetic variance estimated from the ADM model was lower than that from the AM model. For EP, the composite genetic variance estimated from the CADM model was similar to the total genetic variance estimated from the ADM model, and the additive genetic variance estimated from the ADM model was lower than that from the AM model. For BMP, the composite genetic variance estimated from the CADM model was highest, the ADM model did not capture dominance genetic variance, and the additive genetic variance was consistent with that from the AM model.
Similar to the situations with chicken traits, the variance component values estimated from the three genomic models were different in pig traits (Table 2). For T1 and T3, the composite genetic variances estimated from the CADM model were highest among the three models. For T1, the ADM model did not capture the dominance genetic variance, and the variances estimated from the ADM and AM models were consistent. For T3, the ADM model captured the dominance genetic variance, but the cumulative genetic variance of additive and dominance effects was lower than the composite genetic variance from the CADM model. For T2, the composite genetic variance estimated from the CADM model was similar to the total genetic variance estimated by the ADM model, the ADM model captured the dominance genetic variance, and the additive variance estimated from the ADM model was similar to that from the AM model.
Genomic predictive ability
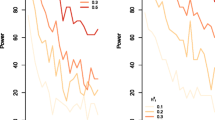
As shown in Table 3, in chickens, fivefold cross-validation revealed that the CADM model had the highest genomic predictive ability among the three models (more results of each of the replicates are shown in Supplementary File 2, Tables S1 and S2). Compared with the AM model, the genomic prediction accuracy was substantially improved for all three traits, with an average increase of 27.1%, and BMP (with the lowest heritability) had the largest increase, 46.1%. Compared with the AM model that considered additive effects only, the ADM model did not increase the genomic predictive ability. As shown in Table 4, the fivefold cross-validation for the pig dataset revealed that among the three genomic models, the CADM model had the highest genomic predictive ability for all three traits, similar to the results for the chicken data. Compared with the AM model, the accuracy of genomic prediction for the three traits increased by 57.9%, 5.4%, and 15.6%, respectively. T1 with the lowest heritability has the highest increase. The predictive ability of the ADM model did not improve or even slightly decreased compared with that of the AM model. In chicken dataset, the regression coefficients for the CADM and AM models were similar, ADM model had the largest bias for all the three traits (Table S3 in Supplementary File 2). In pig dataset, the regression coefficients for the CADM and AM models were similar for T2 and T3, CADM model had the less bias for T1 (Table S4 in Supplementary File 2).
Discussion
The novel CADM model
The phenotypic performance of a trait is affected not only by additive genetic effects but also by dominance and epistatic effects (Radoev et al. 2008). Dominance effects include incomplete dominance, complete dominance and overdominance, and these effects vary for different traits. Typical genomic prediction models with dominance genetic effects assume that dominance genetic effects of all loci have the same variance and directly use genomic markers to construct a dominance genetic relationship matrix between individuals using the same matrix for all traits (Alves et al. 2020; Liu and Chen 2018; Su et al. 2012). In the present study, the SNP loci are weighted according to the degree of deviation between heterozygous and homozygous loci, which differentiates the degrees of dominance effects on the trait between loci. The assumption of the CADM model is closer to the true distribution of dominance effects where dominance effects could be large for some QTLs, and small or no effect for the others. This could be one of the reasons that the CADM improves the accuracy of predictions. To the best of our knowledge, this is the first study of weighting dominance genetic effects for genomic prediction. In addition, the method included the dominance effects by integrating dominance and additive genetic effects into one component without increasing the number of unknowns to be estimated. Therefore, it is hypothesized that the model can improve the accuracy of predictions. As expected, based on the chicken and pig data examined here, our method greatly improved the genomic predictive ability.
In this study, overdominance was not considered in the CADM model. This may not completely match the actual situation. In fact, the CADM model is available for overdominance by relaxing the limitation (0 ≤ Cd ≤ 2) of heterozygous genotype coefficients. However, according to our experience, relaxation sometimes leads to problems with convergence or abnormal solutions caused by the uncertainty of the estimated degree of dominance. Lower MAF together with linkage disequilibrium will strongly affect the estimates. In this study, we used MAF ≥ 0.01 because it is popularly used threshold for editing marker genotype data in genomic prediction. For calculating difference between three genotypes of a locus, how to handle the low MAF loci is a challenge.
Genomic predictive ability
Since the dominance effect is an important factor influencing phenotype, previous studies (Aliloo et al. 2016; Garcia-Baccino et al. 2019; Heidaritabar et al. 2016; Su et al. 2012; Vitezica et al. 2018) have used models containing the dominance effect to improve genomic predictive ability, but the improvements were very small. Su et al. (2012) studied the dominance effect of pig average daily gain and found that the genomic predictive ability increased from 0.319 to 0.330 after the dominance effect was included in the model. Garcia-Baccino et al. (2019) analyzed three growth traits in American Angus males and found that the model contained a dominance effect that did not affect genomic predictive ability. Aliloo et al. (2016) studied genomic predictions using models with dominance effects for four traits in Holstein and Jersey cattle and found that only fat yield in Holstein showed a slight increase in genomic predictive ability (from 0.270 to 0.273). Heidaritabar et al. (2016) studied eight traits in a population of laying hens and found that genomic predictive ability did not improve after including dominance effects in the model. In contrast to these studies, the present study proposed a novel approach that employs a modification of heterozygous genotype coefficients to integrate additive and dominance effects. The method was validated using chicken and pig datasets, and the average genomic predictive ability was improved by more than 20%.
For different traits, the dominance effects on phenotypic variation are different, so the combined additive and dominance relationship matrix (Gad). Some traits may be mainly affected by additive genetic effects, and some traits may be mainly affected by nonadditive genetic effects (Xu 2013). The CADM model could benefit for the traits in which dominance effects are largely different between different QTLs. Currently, breeding programs usually consider only additive genetic effects, i.e., breeding values. However, for many economically important traits, especially those related to fitness, which usually have low heritability, dominance effects may play a substantial role. For these traits, an improvement in the total genetic values of a population is important. For this purpose, the model proposed in this study would be a good alternative for genomic prediction of total genetic values. The genetic effect estimated from the CADM may be not suitable for breeding based on additive genetic effect, but can be used for optimizing the mating scheme. In commercial populations, such as chicken, pig, and cattle, it is important to make use of both additive genetic effect and heterosis. A matting design based on results from the CADM would be a good alternative to improve performance of a commercial population. Every method has its limitations, the CADM model has to calculate genotype matrix for each trait. In contrast, the AM and ADM models only need to calculate the genotype matrix once and can be used for all traits. The comparison with corrected phenotype is unfavorable for AM model for which the predictions do not include dominance effect. We include AM model in this study, because AM is a classic genomic prediction model.
Conclusion
With our CADM model that combines additive and dominance genetic effects through defining heterozygous genotype coefficients, the abilities of genomic prediction are greatly improved. The results from this study indicate that dominant genetic variation is one of the important sources causing phenotypic variation, and the novel prediction model integrating dominance effects can significantly improve the accuracy of genomic prediction.
References
Aliloo H, Pryce JE, Gonzalez-Recio O, Cocks BG, Hayes BJ (2016) Accounting for dominance to improve genomic evaluations of dairy cows for fertility and milk production traits. Genet Sel Evol 48:8
Alves K, Brito LF, Baes CF, Sargolzaei M, Robinson JAB, Schenkel FS (2020) Estimation of additive and non-additive genetic effects for fertility and reproduction traits in North American Holstein cattle using genomic information. J Anim Breed Genet 137(3):316–330
Bland JM, Altman DG (1995) Multiple significance tests: the Bonferroni method. BMJ (Clin Res ed) 310(6973):170
Cleveland MA, Hickey JM, Forni S (2012) A common dataset for genomic analysis of livestock populations. G3: Genes|Genomes|Genet 2(4):429–435
Costa-Neto G, Fritsche-Neto R, Crossa J (2021) Nonlinear kernels, dominance, and envirotyping data increase the accuracy of genome-based prediction in multi-environment trials. Heredity 126(1):92–106
Efron B (1979) Bootstrap methods: another look at the jackknife. Ann Stat 7(1):1–26
Garcia-Baccino CA, Lourenco DAL, Miller S, Cantet RJC, Vitezica ZG (2019) Estimating dominance genetic variances for growth traits in American Angus males using genomic models. J Anim Sci 98:1
Gilmour AR, Thompson R, Cullis BR (1995) Average information REML: an efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 51(4):1440–1450
Heidaritabar M, Wolc A, Arango J, Zeng J, Settar P, Fulton JE et al. (2016) Impact of fitting dominance and additive effects on accuracy of genomic prediction of breeding values in layers. J Anim Breed Genet 133(5):334–346
Li Y, Hawken R, Sapp R, George A, Lehnert SA, Henshall JM et al. (2017) Evaluation of non-additive genetic variation in feed-related traits of broiler chickens. Poult Sci 96(3):754–763
Liu H, Chen G-B (2018) A new genomic prediction method with additive-dominance effects in the least-squares framework. Heredity 121(2):196–204
Liu T, Qu H, Luo C, Shu D, Wang J, Lund M et al. (2014) Accuracy of genomic prediction for growth and carcass traits in Chinese triple-yellow chickens. BMC Genet 15(1):110
Madsen P, Su G, Labouriau R, Christensen OF (2010) 9th World Congress on Genetics Applied to Livestock Production. Leipzig, Germany, p 732.
Mao X, Sahana G, Johansson AM, Liu A, Ismael A, Løvendahl P et al. (2020) Genome-wide association mapping for dominance effects in female fertility using real and simulated data from Danish Holstein cattle. Sci Rep. 10(1):2953
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157(4):1819–1829
R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Radoev M, Becker HC, Ecke W (2008) Genetic analysis of heterosis for yield and yield components in Rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179(3):1547
Su G, Christensen OF, Ostersen T, Henryon M, Lund MS (2012) Estimating additive and non-additive genetic variances and predicting genetic merits using genome-wide dense single nucleotide polymorphism markers. PLoS ONE 7(9):e45293
VanRaden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91(11):4414–4423
Vitezica ZG, Legarra A, Toro MA, Varona L (2017) Orthogonal estimates of variances for additive, dominance, and epistatic effects in populations. Genetics 206(3):1297
Vitezica ZG, Reverter A, Herring W, Legarra A (2018) Dominance and epistatic genetic variances for litter size in pigs using genomic models. Genet Sel Evol 50(1):71
Vitezica ZG, Varona L, Legarra A (2013) On the additive and dominant variance and covariance of individuals within the genomic selection scope. Genetics 195(4):1223–1230
Wang D, Tang H, Liu JF, Xu S, Zhang Q, Ning C (2020) Rapid epistatic mixed-model association studies by controlling multiple polygenic effects. Bioinformatics 36(19):4833–4837
Xiang T, Christensen OF, Vitezica ZG, Legarra A (2018) Genomic model with correlation between additive and dominance effects. Genetics 209(3):711
Xu S (2013) Mapping quantitative trait loci by controlling polygenic background effects. Genetics 195(4):1209
Xu S, Zhu D, Zhang Q (2014) Predicting hybrid performance in rice using genomic best linear unbiased prediction. Proc Natl Acad Sci 111(34):12456
Acknowledgements
This work was supported by the Key-Area Research and Development Program of Guangdong Province (2018B020203001 and 2020B020222002), Natural Science Foundation of Guangdong Province (2018B030311044), Guangdong Basic and Applied Basic Research Foundation (2020A1515011277), Technical System of Poultry Industry of Guangdong Province (2021KJ128), and China Agriculture Research System of MOF and MARA (CARS-41). The first author acknowledges financial support provided by the China Scholarship Council (CSC).
Author information
Authors and Affiliations
Contributions
Conception and design of the study: TL, CL, HQ, and GS. Analyzing data: TL and GS. Interpreting the results: TL, CL, JM, YW, DS, HQ, and GS. Writing of the manuscript: TL, CL, JM, YW, DS, HQ, and GS. All authors gave necessary suggestions, revised and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, T., Luo, C., Ma, J. et al. Including dominance effects in the prediction model through locus-specific weights on heterozygous genotypes can greatly improve genomic predictive abilities. Heredity 128, 154–158 (2022). https://doi.org/10.1038/s41437-022-00504-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-022-00504-6
- Springer Nature Switzerland AG
This article is cited by
-
Investigating the impact of non-additive genetic effects in the estimation of variance components and genomic predictions for heat tolerance and performance traits in crossbred and purebred pig populations
BMC Genomic Data (2023)
-
Dominance is common in mammals and is associated with trans-acting gene expression and alternative splicing
Genome Biology (2023)