Abstract
The deleterious mutation model proposes that quantitative trait variation should be dominated by rare, partially recessive, deleterious mutations. Following artificial selection on a focal trait, the ratio of the difference in inbreeding effects between control and selected populations (ΔB), to the difference in trait means caused by directional selection (ΔM), can inform the extent to which deleterious mutations cause quantitative trait variation. Here, we apply the ΔB/ΔM ratio test to two quantitative traits (male mating success and body size) in Drosophila melanogaster. For both traits, ΔB/ΔM ratios suggested that intermediate-frequency alleles, rather than rare, partially recessive alleles (i.e. deleterious mutations), caused quantitative trait variation. We discuss these results in relation to viability data, exploring how differences between regimens in segregating (measured through inbreeding) and fixed (measured through population crosses) mutational load could affect the ratio test. Finally, we present simulations that test the statistical power of the ratio test, providing guidelines for future research.
Similar content being viewed by others
Introduction
Natural populations typically have high levels of genetic variation underlying most traits. Indeed, even life-history and sexually selected traits, which are subject to strong directional or stabilising selection and demonstrate low heritabilities, maintain ample additive genetic variation and have substantial evolutionary potential (Gustafsson 1986; Hansen et al. 2011; Houle 1992; Merilä and Sheldon 1999; Mousseau and Roff 1987; Pomiankowski and Møller 1995; Price and Schluter 1991). Therefore, understanding the mechanisms maintaining genetic variation in natural populations continues to be a focus of research in evolutionary biology (Barton and Keightley 2002; Josephs et al. 2017; Mitchell-Olds et al. 2007).
Mutation-selection balance is a key hypothesis for the maintenance of genetic variation (Haldane 1927). New alleles, that are, on average, partially recessive and mildly deleterious, are continuously introduced by mutation and transiently segregate at low frequency until selection acts to remove them (Charlesworth and Hughes 2000; Lynch and Walsh 1998). Since selection will be more efficient at removing dominant and additive mutations, standing quantitative genetic variation should be dominated by rare, partially and fully recessive alleles. For traits under directional selection, rare alleles should be consistent in the direction of their effects. For traits under stabilising selection, and assuming no mutation bias, approximately half of the rare alleles affecting the trait should decrease trait values, while the other half should increase trait values. Alternative explanations for the maintenance of genetic variation, such as negative frequency-dependent selection (e.g. Hughes et al. 2013) or overdominance (e.g. Johnston et al. 2013), rely on some form of balancing or fluctuating selection, where alleles responsible for quantitative trait variation (QTV) segregate at more intermediate-frequencies. Estimates from Drosophila melanogaster indicate that such alternative explanations are required to explain observed levels of standing QTV [i.e. mutation-selection balance is not sufficient to explain observed levels of QTV in most traits; (Charlesworth 2015; Charlesworth and Hughes 2000; Sharp and Agrawal 2018)]. Nevertheless, we require more studies that test the relative contributions of rare, partially recessive alleles and intermediate-frequency alleles to QTV (Josephs et al. 2017), since the nature of QTV will affect, among other things, how traits respond to selection and the effects of inbreeding.
Kelly (1999) introduced an approach to test their relative contribution to QTV through selection followed by inbreeding. Artificial selection changes allele frequencies at loci underlying the focal trait. Inbreeding then increases homozygosity (Charlesworth and Charlesworth 1999; Charlesworth and Willis 2009), such that recessive and partially recessive alleles have their full effect on the phenotype. Inbreeding effects can be calculated as the change in trait mean with inbreeding, the directional dominance (denoted B). Key to Kelly’s test is that partially recessive alleles contribute to both the response to selection and the effects of inbreeding. If selection increases the frequency of rare, partially recessive alleles, then the magnitude of B should also change with selection.
Quantitative traits, and particularly fitness-related traits, tend to show directional dominance (Table 10.2 in Lynch and Walsh 1998), and asymmetrical responses to selection are common (Frankham 1990). Consequently, if QTV is caused by rare, partially recessive alleles with biased effects, then bidirectional artificial selection should increase the frequency of these alleles in one direction of selection, thereby enhancing the magnitude of B, and decrease their frequency in the other direction of selection, reducing the magnitude of B.
To apply Kelly’s test, phenotypic data for the trait under artificial selection are required from four groups of individuals: outbred and inbred individuals from control and selected populations (MO, MI, MO* and MI*, respectively). The ratio of the difference in B (ΔB) to the difference in outbred means (ΔM) indicates the extent to which rare, partially recessive alleles cause QTV, and is calculated as (Kelly 1999; Kelly and Willis 2001):
where F is the inbreeding coefficient. Note that as F increases, homozygosity increases, and MI–MO should become more extreme. Dividing by F controls for more extreme B values expected with higher values of F; however, this assumes that B change linearly with F, which may not always hold. Evaluating ΔB/ΔM over a range of F coefficients may be favourable (Charlesworth et al. 2007); we have not adopted that approach here. If ΔB/ΔM ≥ 1, then QTV is caused by rare, partially recessive alleles (Fig. 1b). If ΔB/ΔM ~ 0 or negative, then QTV is caused by intermediate-frequency alleles (Fig. 1c). This approach has only been directly tested three times: for flower size in Mimulus guttatus (Kelly and Willis 2001), fecundity in Drosophila melanogaster (Charlesworth et al. 2007) and for combinations of cuticular hydrocarbons (CHCs) in D. serrata (Gosden et al. 2018). Intermediate-frequency alleles were found to cause QTV in flower size and fecundity (Charlesworth et al. 2007; Kelly and Willis 2001). However, some positive ratios were identified for CHC traits, where ratios also tended to be higher for males than females, indicating that rare, partially recessive alleles are more important to the genetic architecture of male than female traits (Gosden et al. 2018).
Assessing the contribution of intermediate-frequency alleles and rare, partially recessive alleles to quantitative trait variation (QTV) through selection and inbreeding. a The ratio ΔB/ΔM, where ΔM is the change in trait mean with selection and ΔB is the difference in directional dominance (B) between the control (BC) and selected (BS) lines, is an indicator of the contribution of rare, partially recessive alleles to QTV (Kelly 1999). b When ΔB/ΔM ~ 1, QTV is caused by rare, partially recessive alleles. c When ΔB/ΔM ~ 0, QTV is caused by intermediate-frequency alleles. ΔB/ΔM was calculated assuming F = 0.5. Note that these equations are the same if the selected population was represented by the open circles, and the control population represented by the closed circles (i.e. selection for higher trait values)
Here, we apply the ratio test to two fitness-related traits, body size and male mating success, in D. melanogaster. Male mating success is under directional selection, has substantial inbreeding load (Ala‐Honkola et al. 2013; Hughes 1995b; Partridge et al. 1985; Pendlebury and Kidwell 1974; Sharp 1984; Valtonen et al. 2014), and some evidence suggests that mutation-selection balance may substantially contribute to QTV underlying mating success (Dugand et al. 2018; Hughes 1995a), although not all (Houle et al. 1994; Sharp and Agrawal 2018). In contrast, body size shows limited directional dominance (Table 10.2 in Lynch and Walsh 1998) and is likely to be under stabilising and/or balancing selection. For example, large flies have higher male mating success (e.g. Dugand et al. 2018; Partridge and Farquhar 1983; Partridge et al. 1987) and fecundity (e.g. Robertson 1957), while small flies have faster development times, particularly with larval crowding (Santos et al. 1994), and females re-mated to small males enjoy fertility benefits and avoid sexually conflictual fitness costs (Pitnick 1991; Pitnick and García–González 2002). Seasonal and clinal (altitudinal and latitudinal) variation is also likely to maintain balanced polymorphisms in natural populations through time and space (Gockel et al. 2001; Weeks et al. 2002), suggesting that alleles at more intermediate-frequencies will contribute to QTV.
In addition to applying Kelly’s ratio test to each trait, we employ other approaches designed to shed light on the same question. First, we test the symmetry of the response to selection, which should be asymmetrical if the distribution of effects of rare, partially recessive alleles is also asymmetrical (although there are alternative explanations; Falconer and Mackay 1996, pp 211–214), with a stronger response in the direction of lower fitness (Frankham 1990). Second, we quantify fixed mutational load in egg-to-adult viability. Fixed mutations cannot contribute to inbreeding depression, which may result in biased estimates of B, and consequently ΔB/ΔM, and may be particularly problematic if rates of fixation are regimen-specific. We also discuss the importance of inbreeding depression in egg-to-adult viability for the ratio test. Third, we use linear modelling approaches to test for differences in directional dominance between selection regimens (i.e. an interaction between the effects of selection and the effects of inbreeding). The intention is that the combination of these assays will provide a more complete picture of genetic architecture. Finally, we perform a series of simulations to test the statistical power of the ratio test (and linear models), and use these simulations to provide recommendations for future work.
Materials and methods
Kelly’s method
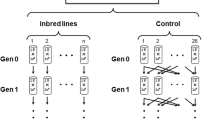
We applied the ratio test to data collected previously (see Dugand et al. 2018 for full details of the methods). Briefly, we conducted two separate artificial selection experiments; one where selection was applied to male mating success (success and failure selection), and the other where selection was applied to body size (large and small size selection). The selection lines were derived from the same stock population; a large, laboratory population derived from wild flies collected en masse from a single population in Innisfail, Queensland in 2012. The stock population had been in the laboratory for approximately 10 generations prior to artificial selection.
To apply artificial selection to male mating success, we exposed males to binomial mate choice trials where two males are presented to a single, non-virgin female. Males successful at mounting females in these choice trials were used to propagate four replicate success-selected lines (N = 25 successful males and 25 virgin females), while males that failed to mount females were used to propagate four failure-selected lines (N = 25 unsuccessful males and 25 virgin females). Four control lines were established with males not exposed to the mate choice protocol. Selection continued in this way for 14 rounds across 17 generations.
To select on body size, we applied 25% truncation selection in both directions. That is, we selected the 25 (out of 100) largest males and females for each of three large-selected lines, and the 25 smallest males and females for each of four small-selected lines. Selection was applied for 11 generations.
For all 19 selection lines, the grand-offspring of the flies from the final round of selection were reared at standard density (50 per vial) and collected upon emergence before reaching sexual maturity. Single-pair crosses (henceforth referred to as families) were then established by pairing males and females in vials for 24 h. Five families were established to assay body size [measured as wing size, which is tightly correlated with body size (Reeve and Robertson 1953)], while 10 families were used to assay mating success. Emerging adults from these crosses were collected as virgins and held in single-sex vials before crosses were established.
For mating success (10 families), females were paired with either a brother (inbred; F ~ 0.25; one per family) or an unrelated male from the same selection line (outbred; F ~ 0; one per family) in yeasted vials (one pair per vial) for 24 h. Ten males emerging from each vial (i.e. a total of 100 inbred males and 100 outbred males per selection line) were then individually competed against standard competitor males from the base population for access to standard females. These mating trials were conducted across 4 days, with one population from each regimen (success-selected, failure-selected and control) trialled on every day. The numbers of successful and unsuccessful experimental males per family were recorded. Families from which no males mated were excluded from analyses. The proportion of successful experimental males was then calculated for each family.
For body size (five families), pairs (brother-sister or unrelated; five of each cross per family) were held together for 48 h to mate and lay eggs before being transferred to fresh vials for 24 h. Five males and five females emerging from each of these vials had their wings measured. This involved removing right wings with forceps, dipping them in Histoclear (National Diagnostics, Inc) and mounting them on a glass slide with Aquamount (Thermo Fisher Scientific, Inc). Black and white TIFF images were then taken, and wing area was measured using landmark analysis (Gilchrist and Partridge 1999) with ObjectImage (Vischer et al. 1994). For comparison, we assayed the wing size of outbred and inbred stock flies from five replicate families in the same way. We used wing size as a proxy for body size because wing size can be easily and accurately measured on fresh or preserved flies.
Egg-to-adult viability
We have previously presented data on the egg-to-adult viability of outbred and inbred flies from all selection lines (Dugand et al. 2018), for which the comparison provides information about the segregating, recessive mutational load. The relevance of this is discussed in more detail below. For both selection experiments, viability was quantified for five brother-sister pairs and five unrelated pairs for each of five families per line (25 inbred and 25 outbred pairs per line).
Additionally, we quantified the egg-to-adult viability of between-line crosses to estimate fixed mutational load. If mutations have become fixed in some selection lines, then the viability of between-line crosses should be higher as deleterious, recessive mutations become masked. To estimate fixed load, we used the same five families as for quantifying segregating mutational load, additionally crossing five females from each family (sisters) to males from each of the other two (for large-selected lines) or three (for small-, success- and failure-selected lines) selection lines from within the same selection regimen. For example, females from L1F1 (large line one, family one) were paired with five random males from L2, five random males from L3, and five random males from L1 (excluding F1, which would represent an inbred cross). For each family, the number of eggs and adults were pooled for each cross type. Thus, we had three (for large-selected lines) or four (for success-, failure- and small-selected lines) viability scores for each family per line.
Statistical analyses
All statistical tests were performed using the R statistical platform (R Development Core Team 2010). Generalised/linear mixed models (G/LM/Ms) were performed using the lme4 package (Bates et al. 2014), and analysed with the car package (Fox and Weisberg 2011). Figures were created using ggplot2 (Wickham 2016).
ΔB/ΔM ratios
ΔB/ΔM ratios for male mating success were calculated by comparing each selection line to the control line that was assayed on the same day [since day affected mating success (Dugand et al. 2018)]. We used two approaches to test for significance. First, we used a resampling approach where, for 1000 iterations, we sampled (with replacement) n proportions for each cross/line (n is the number of families, typically 10). We then calculated MO, MI, MO* and MI* values for each cross/line as the mean of the n proportions, and calculated the ratio using Eq. 1 (F = 0.25). We used 95% confidence intervals to test whether ratios were significantly different to one or zero. Second, we tested the condition that 4MI* – 5MO* – 4MI + 5MO ≤ 0 when ΔB/ΔM ≥ 1 (Kelly and Willis 2001). This condition arises when ΔB/ΔM = 1 and F = 0.25 are substituted into Eq. 1. Note that the equation here differs slightly from Kelly and Willis (2001) because we measured the phenotypes of offspring from brother-sister pairings (F ~ 0.25), rather than from self-fertilisation (F ~ 0.5). We tested whether the value obtained was significantly less than zero using a one sample t-test, where the standard error was calculated as the square root of the sum of the variances of MO, MI, MO* and MI*, and the degrees of freedom was the total number of families minus four.
We repeated these analyses for male and female body size. Each selection line was compared to the stock population.
Finally, we calculated regimen-wide effects where, for each of the 1000 iterations, we effectively pooled all replicate lines within a regimen/cross (by taking the inbred and outbred means of the line means) and calculated 1000 ΔB/ΔM ratios for each regimen against the appropriate control (the base population for size-selected regimens, and the mean of the controls for the mating success-selected regimens).
G/LM/Ms for focal traits
If ΔB/ΔM ≥ 1, then B must be more negative in failure- and small-selected lines, or less negative in success- and large-selected lines, compared to control lines. Applying a linear modelling approach, differences in B between regimens should be manifest in a regimen-by-cross interaction. To test this for wing size, we performed linear models (LMs) for each sex/line (compared to the stock) on the five family means and analysed the effect of regimen, cross, and their interaction on male and female wing size. We recorded the P value of the interaction term for each line. We then performed the equivalent analyses for the mating success data using generalised linear models (GLMs), again, comparing each selection line to the control population assayed on the same day. For this analysis, we used the number of wins and losses for each family. Note that these G/LMs are not equivalent to the ratio test because any significance of an interaction effect is not directly affected by ΔM. In contrast, for a given value of ΔB, ΔB/ΔM decreases as ΔM increases. Nevertheless, a significant interaction effect would indicate that ΔB/ΔM > 0, provided that ΔB and ΔM were both positive (in success- and large-selected lines), or both negative (in failure- and small-selected lines).
Similar to the regimen-wide tests presented above, we then performed generalised/linear mixed effects models (G/LMMs) for each regimen against the appropriate control. That is, for wing size, we performed one LMM for each regimen/sex, analysing the fixed effects of regimen, cross and their interaction on the mean wing size of the five families for each line. Line (nested within regimen) was included as a random effect. For male mating success, we performed GLMMs for each regimen against the control, testing the fixed effects of regimen, cross, and their interaction, and the random effects of day and line (nested within regimen), on male mating success. We recorded the P value of the interaction term from each analysis.
GLMMs for egg-to-adult viability
Analyses for segregating mutational load are described in (Dugand et al. 2018). To assess fixed mutational load, all regimens were analysed independently. We analysed the effects of female ID (i.e. the line the female was from), male ID and their interaction (fixed effects) on the number of live flies and undeveloped eggs. Family was included as a random effect and, for each model, we fit an observation-level random effect to account for overdispersion (Harrison 2014). These analyses test if crossing between lines influences viability. A significant interaction might suggest that within-line crosses have reduced viability compared to between-line crosses, which would be indicative of fixed mutational load.
Power simulations
Above, we described two statistical approaches (G/LMs and resampling) for testing the significance of ΔB/ΔM > 0. In this section, we performed simulations to evaluate the power of these approaches under different sets of parameters. Specifically, we tested how the response to selection, changes in inbreeding effects, and changes in sample size influenced the power of the two statistical approaches and the robustness of the ratio test.
For each set of parameters, we generated four normal distributions (1000 values) with means equal to MO, MI, MO* and MI* (i.e. one control and one selected population), and standard deviations (SDs) equal to the observed (from female wing size data) SDs of outbred and inbred stock and small-selected fly wing sizes. MO and MI were the observed values for the stock population; these values do not change for any of the simulations (nor do SDs). MO* was calculated as one, two, three or four SDs smaller than outbred control mean. MI* was calculated by setting ΔB/ΔM to be one or three and solving for MI* using Eq. 1. After systematically changing ΔB/ΔM (one and three) and ΔM (one, two, three and four SDs), we had a total of eight data sets. We then performed the following on all eight data sets.
We randomly sampled n samples from each of the four distributions. This random sampling reflects sampling effort; for example, the number of families assayed from each cross/line. While at a population level selection reduced sized and increased inbreeding effects, n random samples from the four distributions could produce values that do not reflect this, particularly when n is small. Therefore, we recorded the proportion of times that the n samples did not conform to the conditions: ΔB < 0 and ΔM < 0 (i.e. the proportion of times that sampled values for the response to selection and inbreeding effects were not in the simulated direction). We then analysed the data sets that did conform to these conditions using the methods described above. That is, we (i) resampled (with replacement) from the n samples from each distribution 1000 times, generating 1000 ratios and recorded the 0.025 confidence interval, and (ii) performed a LM on the sampled values and recorded the P value of the interaction term. We repeated this 1000 times for each value of n (three to 100) and recorded the number of times that the ratio was significantly >0, the number of times that the P value of the interaction was <0.05, and the number of times that ΔB ≥ 0 or ΔM ≥ 0.
Results
ΔB/ΔM ratios and G/L(M)Ms for focal traits
Male mating success experiment
Selection and inbreeding responses for male mating success are presented in Table 1. All eight selection lines responded in the direction of selection and most lines showed some directional dominance (Table 1). On average, the response to selection was symmetrical; however, responses were more consistent among success-selected lines. We found a range of positive and negative ratios. Moreover, confidence intervals were very large and mostly included zero and one, making it difficult to draw conclusions from line effects. This was not surprising given that the SDs for each line were very large, and responses to selection were mostly <1 SD (Table 1). The generalised linear models identified one significant interaction, but this was in the wrong direction (BS > BC for a failure-selected line). Regimen-wide ratios were negative for both regimens, but were not significantly different to one, and regimen-by-cross interactions were both non-significant. These results align with our previous GLMM analysis where we showed a significant response to selection, but no significant regimen-by-cross interaction (Dugand et al. 2018).
Wing size experiment
All lines responded to selection on body size and most showed some directional dominance (Table 2). The response was, on average, symmetrical relative to the stock population, but responses varied substantially across lines (Table 2). We found a range of positive and negative ratios, although confidence intervals were large and always included zero for males and females. We found no significant interaction terms for any LM. Regimen-wide effects were non-significant as identified by both resampling and LMMs.
GLMMs for egg-to-adult viability
Male mating success experiment
Fixed load was not evident in either success- or failure-selected lines since there was no significant female ID-by-male ID interaction that might have suggested that within-line crosses have lower viability than between-line crosses (Table 3). There was a significant effect of female ID on egg-to-adult viability in the success-selected lines, which was apparently driven by the offspring of females from line three having low viability (Supplementary Fig. 1). There were no significant female ID, male ID or interaction effects on egg-to-adult viability in the failure-selected lines (Table 3).
Wing size experiment
We identified significant main effects of male ID and female ID on the egg-to-adult viability of large-selected flies, but no significant interaction (Table 3). There were no significant effects on the viability of small-selected flies (Table 3).
Power simulations
The results from the simulations are presented in Fig. 2 and are discussed in detail below.
Power of resampling approach (solid black) and linear models (solid grey) for identifying significant Kelly ratios or interaction terms. The figures show the relationship between the number of samples (e.g. the number of plants for the Kelly and Willis (2001) study, or the number of families in this study) and the power to detect significant results when the response to selection (1–4 SDs from the control mean) and the ΔB/ΔM ratio (one or three) are varied. The dashed line represents the proportion of times that ΔB ≥ 0 or ΔM ≥ 0 (color figure online)
Discussion
Artificial selection followed by inbreeding can reveal the relative contributions of rare, partially recessive alleles and intermediate-frequency alleles to QTV. If QTV is dominated by rare, partially recessive alleles, then selection should change the frequency of these rare alleles, which should directly change the inbreeding load in selected populations such that ΔB/ΔM > 1. In contrast, if QTV is caused by intermediate-frequency alleles, then selection should have little or no effect on inbreeding load, and ΔB/ΔM ~ 0 (Kelly 1999).
Male mating success
ΔB/ΔM ratios for mating success were highly variable, making it difficult to draw firm conclusions about the relative contribution of rare, partially recessive alleles and intermediate-frequency alleles to QTV. Regimen-wide ratios were negative and suggested a major contribution by intermediate-frequency alleles, in line with previous research (Sharp and Agrawal 2018). Furthermore, the response to selection was relatively symmetrical, which is again suggestive of intermediate-frequency alleles. However, we recently sequenced the genomes of these populations and found that, at loci associated with male mating success, the frequency of the minor (rarer) allele tended to be rarer than average in the base population [i.e. compared to the frequency of the minor allele across the rest of the genome (Dugand et al. 2019)]. That is, rare alleles were largely responsible for QTV underlying male mating success. Therefore, the negative experiment-wide ratios that we identified are either inaccurate (see points below), or show that the rare alleles under selection had largely additive effects on mating success (Kelly 1999). If the latter, then a different set of (recessive) alleles must have been responsible for the inbreeding depression in male mating success, but this set of alleles did not contribute to the selection response. We highlight four points that suggest that our ratios may be inaccurate.
First, we previously demonstrated that inbreeding depression in egg-to-adult viability was evident in failure-selected, but not success-selected lines (Dugand et al. 2018). Our viability data are not only useful for understanding the pleiotropic effects of alleles under selection (i.e. that alleles that increase mating success have recessive, pleiotropic effects on viability), but also outline an important consideration for the ratio test. Males excluded by developmental selection (Polak and Tomkins 2013) were counted in the viability assay, but not exposed to the mating success assay. Under the reasonable assumption that inviable males would have had poor mating success, then not being able to assay these males would cause us to underestimate B in failure-selected lines, thereby underestimating ΔB/ΔM.
Second, some models of sexual selection predict increased inbreeding effects with increasing trait value as condition-dependent traits recruit new loci (Rowe and Houle 1996; Wilkinson and Taper 1999), which could explain the continued inbreeding depression in success-selected lines.
Third, SDs for line means were very large and ΔM was consistently small (mostly <0.5 SDs) and varied greatly across lines, making the ratio tests weak. Most notably, two failure-selected lines (three and four) had very small ΔM. Consequently, ΔB/ΔM ratios were unreliable, which is depicted by the vastly different ratios generated by the two different methods for calculating ΔB/ΔM. Given the small ΔM, resampling would cause ΔM to be both positive and negative across the 1000 iterations. Hence, while B is more negative in these two failure-selected lines compared to control lines, generating positive values for ΔB/ΔM 1, overlap between MO* and MO substantially affects the resampling approach and generated a vastly different picture for ΔB/ΔM 2.
Finally, applying the ratio test to a binomial trait may be statistically problematic. For example, consider a sigmoidal relationship between male attractiveness (continuous) and male mating success (binomial). For a given change in attractiveness, the extent of the change in mating success depends on the overall level of attractiveness. Hence, B and ΔM would be directly affected by the position on the curve, distorting the ratio test.
Wing size
We found evidence for both positive and negative ΔB/ΔM ratios for male and female size. As with mating success, ΔM varied across lines and confidence intervals for ΔB/ΔM were large and often included zero and one, making it difficult to draw any firm conclusions.
For females, when ΔM > 4 SDs, then ΔB/ΔM 2 < 0; however, when ΔM < 3 SDs, then ΔB/ΔM 2 > 0. A similar pattern is evident for males. These results indicate that we may have ‘overshot’ the ideal range in which to apply the ratio test in some selection lines; Kelly (1999) recommended a 1–3 SD ΔM. Hence, our data suggest that rare, partially recessive alleles may have been an important component of genetic architecture in the base population.
Small-selected flies were less viable than large-selected flies (Dugand et al. 2018). One explanation for this could be that selection caused an accumulation of unconditionally deleterious mutations with pleiotropic effects on viability in small-selected lines. However, viability-affecting mutations should be at least partially recessive to remain in populations. We found that both large- and small-selected lines suffered similar levels of inbreeding depression for viability (i.e. no regimen-by-cross interaction; Dugand et al. 2018), indicating that any load was not primarily caused by recessive mutations. Therefore, we suggest that a more likely explanation for differences in viability is that females from small-selected lines simply produced smaller eggs that were less viable (Azevedo et al. 1997). It is unclear whether (or how) the lower viability of small-selected flies would directly influence the ratio test, perhaps in underestimating ΔM and, therefore, overestimating ΔB/ΔM.
Our analyses found no evidence to support fixed load in any regimen. However, one large-selected line and one small-selected line had lower viability for within-line crosses compared to between-line crosses, the pattern expected if deleterious mutations have become fixed in these lines. Genetic drift or selection could have caused deleterious alleles to become fixed; if the latter, this has implications for genetic architecture and for applying the ratio test. For example, fixed load in the large-selected line suggests that large-selected individuals carry alleles that increase size and have deleterious, pleiotropic effects on viability. Given that the same is true for a small-selected line, this result suggests that size would be under stabilising selection, where more extreme phenotypes carry a larger burden of mutations that reduce fitness. Hence, quantifying fixed load in this way could prove useful for understanding genetic architecture. Fixed load could also directly affect the ratio test since deleterious recessive alleles no longer contribute to inbreeding depression. Interestingly, the small-selected line with (apparent) fixed load showed no directional dominance for wing size, in other small-selected lines, inbreeding decreased wing size. Taken together, these results suggest that recessive mutations that reduce size and have pleiotropic effects on viability have become fixed in this line, thereby causing us to underestimate B and, consequently, ΔB/ΔM. This result highlights the potential problem of selecting for too long or too strongly, and the benefit of quantifying fixed load.
Power simulations
Using simulations, we sought to evaluate the power of the ratio test and the effects of sampling. Sample size and the magnitude of the response to selection can, to some extent, be under the control of the experimenter. In this section, we discuss how these simulations could be informative for future studies applying the ratio test.
At a population level, ΔM was simulated to be 1–4 SD and ΔB/ΔM was one or three. The dashed line in Fig. 2 indicates the proportion of times that a random sample of n values from each of the four distributions generated values where ΔB ≥ 0 and/or ΔM > 0 (hereafter, we refer to this as the proportion that failed). Hence, the inverse of this line indicates the proportion of times that ΔB/ΔM > 0, thereby correctly identifying an important contribution of rare, partially recessive alleles to QTV. From the iterations that did not fail, we tested for significance using a resampling approach and LMs.
When ΔM = 1 SD, ΔB/ΔM = 1, and n was small, nearly 70% of the 1000 samples failed. This highlights a key point; if ΔM is not sufficiently large, then resampling is prone to generating positive and negative values for ΔM across iterations. This is exemplified by the conservative nature of the ratio test in identifying significant results under these parameters. The proportion of significant results identified by LMs increases somewhat linearly as n increases. In contrast, there was a substantial lag in the proportion of significant results identified by resampling because, again, resampling 1000 times from the n values would generate both positive and negative ΔM values. Hence, resampling to fit 95% confidence intervals suffers from the same problem as sampling from a population, making the ratio test conservative. This problem was overcome when ΔM = 2 SD. Kelly (1999) recommend a 1–3 SD ΔM to accurately test genetic architecture in the base population. We suggest that a 2–3 SD ΔM may be a good compromise between power and accuracy.
Experimenters have control of n more than ΔM. Kelly and Willis (2001) demonstrated that large values of n (>100) were achievable using M. guttatus. However, experiments conducted on dioecious species may be relatively limited because families need to be established in order to inbreed. Our simulations indicated that, regardless of ΔM, increasing n to be approximately 25 could substantially reduce the proportion of failures, to a negligibly small proportion when ΔB/ΔM > 1. This is encouraging because 25 families could be a realistic goal under many circumstances; for example, n was mostly in the range of 20–40 for measures of fecundity (Charlesworth et al. 2007). While the proportion of significant results remains low at n = 25, failures are, at least, largely removed and ΔB/ΔM ratios should be considered to be relatively accurate.
Conclusions
Our ΔB/ΔM ratios largely indicated a contribution of intermediate-frequency alleles to QTV underlying both male mating success and wing size. However, we have outlined a number of caveats to these conclusions. Most notably, confidence intervals were large and often included both zero and one, a feature of all studies that have applied the ratio test using species of Drosophila (Charlesworth et al. 2007; Gosden et al. 2018). We found assays of viability to be useful adjuncts to the ratio test, both for achieving a deeper understanding of genetic architecture (pleiotropic effects), and for testing whether the ratio test might be affected by developmental selection or fixed mutational load.
In conclusion, we remain encouraged by the potential of Kelly’s ratio test. Despite the difficulties faced so far, male mating success (this study), fecundity (Charlesworth et al. 2007) and multivariate attractiveness (Gosden et al. 2018) may be among the more ambitious (complex and noisy) traits to test for genetic architecture. It remains to be seen whether the ratio test will be more robust when applied to more straightforward traits (such as flower size) with larger sample sizes. Artificial selection experiments are common, and it would be beneficial if Kelly’s ratio test was applied more frequently and in conjunction with molecular genetic data (Dugand et al. 2019) or other biometric approaches (Curtsinger and Ming 1997; Kelly 2008) that, together, provide a robust test of genetic architecture (e.g. in distinguishing between QTV caused by rare additive alleles versus intermediate-frequency alleles). Very few empirical tests exist for quantifying the relative contribution of rare, partially recessive alleles and intermediate-frequency alleles to QTV (Kelly 1999; Sharp and Agrawal 2018), yet this has important implications to a swathe of fields, including evolutionary genetics, sexual selection, and conservation. Finally, to evaluate how genetic architecture might change over space and time, these biometric tests should be applied under a range of conditions, for example, when populations are near to (e.g. long-standing laboratory populations), or far from (e.g. new laboratory populations), their adaptive peak.
Data archiving
The data that support the findings of this study are openly available in Dryad (datadryad.org) at https://doi.org/10.5061/dryad.42p6759.
References
Ala‐Honkola O, Hosken DJ, Manier MK, Lüpold S, Droge‐Young EM, Berben KS et al. (2013) Inbreeding reveals mode of past selection on male reproductive characters in Drosophila melanogaster. Ecol Evol 3(7):2089–2102
Azevedo RBR, Partridge L, French V (1997) Life‐history consequences of egg size in Drosophila melanogaster. Am Natur 150(2):250–282
Barton NH, Keightley PD (2002) Understanding quantitative genetic variation. Nat Rev Genet 3(1):11–21
Bates D, Mächler M, Bolker B, Walker S (2014) Fitting linear mixed-effects models using lme4. J Stat. Softw 67(1):1–48
Charlesworth B (2015) Causes of natural variation in fitness: evidence from studies of Drosophila populations. Proc Natl Acad Sci USA 112(6): 1662-1669.
Charlesworth B, Charlesworth D (1999) The genetic basis of inbreeding depression. Genet Res 74(3):329–340
Charlesworth B, Hughes KA (2000) The maintenance of genetic variation in life-history traits. In: Singh RS, Krimbas CB (eds) Evolutionary Genetics: From Molecules to Morphology. Cambridge University Press, Cambridge, p 369–392
Charlesworth B, Miyo T, Borthwick H (2007) Selection responses of means and inbreeding depression for female fecundity in Drosophila melanogaster suggest contributions from intermediate-frequency alleles to quantitative trait variation. Genet Res 89(02):85–91
Charlesworth D, Willis JH (2009) The genetics of inbreeding depression. Nat Rev Genet 10(11):783–796
Curtsinger JW, Ming R (1997) Non-linear selection response in Drosophila: a strategy for testing the rare-alleles model of quantitative genetic variability. Genetica 99(1):59–66
Dugand RJ, Kennington WJ, Tomkins JL (2018) Evolutionary divergence in competitive mating success through female mating bias for good genes. Sci Adv 4(5):eaaq0369
Dugand RJ, Tomkins JL, Kennington WJ (2019) Molecular evidence supports a genic capture resolution of the lek paradox. Nat Commun 10(1):1359
Falconer D, Mackay T (1996) Introduction to quantitative genetics. Longman Scientific & Technical, Essex
Fox J, Weisberg S (2011) An R Companion to Applied Regression, 2nd edn. Sage, Thousand Oaks
Frankham R (1990) Are responses to artificial selection for reproductive fitness characters consistently asymmetrical? Genet Res 56(01):35–42
Gilchrist AS, Partridge L (1999) A comparison of the genetic basis of wing size divergence in three parallel body size clines of Drosophila melanogaster. Genetics 153(4):1775–1787
Gockel J, Kennington WJ, Hoffmann A, Goldstein DB, Partridge L (2001) Nonclinality of molecular variation implicates selection in maintaining a morphological cline of Drosophila melanogaster. Genetics 158(1):319–323
Gosden TP, Reddiex AJ, Chenoweth SF (2018) Artificial selection reveals sex differences in the genetic basis of sexual attractiveness. Proc Natl Acad Sci 115(21):5498–5503
Gustafsson L (1986) Lifetime reproductive success and heritability: empirical support for Fisher’s fundamental theorem. Am Natur 128(5):761–764
Haldane JBS (1927) A mathematical theory of natural and artificial selection, part V: selection and mutation. Math Proc Camb Philos Soc 23(7):838–844
Hansen TF, Pélabon C, Houle D (2011) Heritability is not evolvability. Evol Biol 38(3):258–277
Harrison XA (2014) Using observation-level random effects to model overdispersion in count data in ecology and evolution. PeerJ 2:e616
Houle D (1992) Comparing evolvability and variability of quantitative traits. Genetics 130(1):195–204
Houle D, Hughes KA, Hoffmaster DK, Ihara J, Assimacopoulos S, Charlesworth B (1994) The effects of spontaneous mutation on quantitative traits. I. Variances and covariances of life history traits. Genetics 138(3):773–785
Hughes KA (1995a) The evolutionary genetics of male life-history characters in Drosophila melanogaster. Evolution 49(3):521–537
Hughes KA (1995b) The inbreeding decline and average dominance of genes affecting male life-history characters in Drosophila melanogaster. Genet Res 65:41–52
Hughes KA, Houde AE, Price AC, Rodd FH (2013) Rare male mating advantage in wild guppy populations. Nature 503:108–110
Johnston SE, Gratten J, Berenos C, Pilkington JG, Clutton-Brock TH, Pemberton JM et al. (2013) Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature 502:93–5
Josephs EB, Stinchcombe JR, Wright SI (2017) What can genome‐wide association studies tell us about the evolutionary forces maintaining genetic variation for quantitative traits? N Phytol 214:21–33
Kelly JK (1999) An experimental method for evaluating the contribution of deleterious mutations to quantitative trait variation. Genet Res 73(3):263–273
Kelly JK (2008) Testing the rare-alleles model of quantitative variation by artificial selection. Genetica 132(2):187–198
Kelly JK, Willis JH (2001) Deleterious mutations and genetic variation for flower size in Mimulus guttatus. Evolution 55(5):937–942
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer Associates, Sunderland
Merilä J, Sheldon BC (1999) Genetic architecture of fitness and nonfitness traits: empirical patterns and development of ideas. Heredity 83(2):103–109
Mitchell-Olds T, Willis JH, Goldstein DB (2007) Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet 8(11):845–856
Mousseau TA, Roff DA (1987) Natural selection and the heritability of fitness components. Heredity 59(Pt 2):181–197
Partridge L, Farquhar M (1983) Lifetime mating success of male fruitflies (Drosophila melanogaster) is related to their size. Anim Behav 31(3):871–877
Partridge L, Hoffmann A, Jones JS (1987) Male size and mating success in Drosophila melanogaster and D. pseudoobscura under field conditions. Anim Behav 35(2):468–476
Partridge L, Mackay TFC, Aitken S (1985) Male mating success and fertility in Drosophila melanogaster. Genet Res 46(3):279–285
Pendlebury W, Kidwell J (1974) The effect of inbreeding on male mating ability in Drosophila melanogaster. Theor Appl Genet 44(3):128–132
Pitnick S (1991) Male size influences mate fecundity and remating interval in Drosophila melanogaster. Anim Behav 41(5):735–745
Pitnick S, García–González F (2002) Harm to females increases with male body size in Drosophila melanogaster. Proc R Soc Lond B Biol Sci 269(1502):1821–1828
Polak M, Tomkins JL (2013) Developmental selection against developmental instability: a direct demonstration. Biol Lett 9(2):20121081
Pomiankowski A, Møller A (1995) A resolution of the lek paradox. Proc R Soc Lond B Biol Sci 260(1357):21–29
Price T, Schluter D (1991) On the low heritability of life-history traits. Evolution 45(4):853–861
R Development Core Team (2010) R Foundation for Statistical Computing. R Development Core Team, Vienna
Reeve ECR, Robertson FW (1953) Studies in quantitative inheritance. 2. Analysis of a strain of Drosophila melanogaster selected for long wings. J Genet 51:276–316
Robertson FW (1957) Studies in quantitative inheritance XI. Genetic and environmental correlation between body size and egg production in Drosophila Melanogaster. J Genet 55(3):428–443
Rowe L, Houle D (1996) The lek paradox and the capture of genetic variance by condition dependent traits. Proc R Soc B Biol Sci 263(1375):1415–1421
Santos M, Fowler K, Partridge L (1994) Gene-environment interaction for body size and larval density in Drosophila melanogaster: an investigation of effects on development time, thorax length and adult sex ratio. Heredity 72(5):515–521
Sharp NP, Agrawal AF (2018) An experimental test of the mutation-selection balance model for the maintenance of genetic variance in fitness components. Proc R Soc B 285(1890):20181864
Sharp PM (1984) The effect of inbreeding on competitive male-mating ability in Drosophila melanogaster. Genetics 106(4):601–612
Valtonen TM, Roff DA, Rantala MJ (2014) The deleterious effects of high inbreeding on male Drosophila melanogaster attractiveness are observed under competitive but not under non-competitive conditions. Behav Genet 44(2):144–154
Vischer NOE, Huls PG, Woldringh CL (1994) Object-Image: an interactive image analysis program using structured point collection. Binary 6:160–166
Weeks AR, McKechnie SW, Hoffmann AA (2002) Dissecting adaptive clinal variation: markers, inversions and size/stress associations in Drosophila melanogaster from a central field population. Ecol Lett 5(6):756–763
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer-Verlag, New York
Wilkinson GS, Taper M (1999) Evolution of genetic variation for condition-dependent traits in stalk-eyed flies. Proc R Soc Lond B Biol Sci 266(1429):1685–1690
Acknowledgements
We thank Michael Jennions, Tracey Chapman, David Hosken, John Kelly, Louise Johnson, Genevieve Hayes, and three anonymous reviewers for providing valuable feedback on previous versions of this manuscript, and the Centre for Evolutionary Biology at The University of Western Australia for support. This work was funded by the School of Biological Sciences (University of Western Australia) and an Australian Research Council Future Fellowship to J.L.T. (FT110100500). R.J.D. was supported by an Australian Postgraduate Award Scholarship through The University of Western Australia.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Dugand, R.J., Kennington, W.J. & Tomkins, J.L. Evaluating the genetic architecture of quantitative traits via selection followed by inbreeding. Heredity 123, 407–418 (2019). https://doi.org/10.1038/s41437-019-0219-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41437-019-0219-x
- Springer Nature Switzerland AG