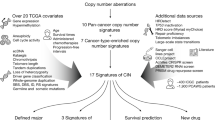
Abstract
Certain breast and ovarian cancers are characterised by high levels of chromosomal instability. We established a suite of eleven SNP array-based signatures of various forms of chromosomal instability. To understand what biological mechanisms might underpin these signatures, we generated and assembled genetic-feature data including allele-specific expression, fusion genes, gene expression, methylation, somatic coding mutations and protein expression. For each signature, we extracted a compendium of significantly associated genetic features using machine learning. We established an association between subchromosomal allelic imbalance-based measures and DNA repair genes. Numerical chromosomal instability and chromothripsis were found to have distinct genetic associations but shared a relationship to mitotic processes, while whole-genome doubling was characterised by TP53 mutation, and high AURKA and GINS1 expression. Furthermore, we identified two genetically distinct forms of tandem duplicator phenotypes. These findings identify potentially novel genomic targets for validation and drug development for specific subsets of breast and ovarian cancer.




Similar content being viewed by others
Code availability
R code (version 3.0.2) for the estimation of numerical CN and the generation of the SNCNA score is available upon request.
References
Wu S, Powers S, Zhu W, Hannun YA. Substantial contribution of extrinsic risk factors to cancer development. Nature. 2015;529:43–47.
Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78.
Coschi CH, Ishak CA, Gallo D, Marshall A, Talluri S, Wang J, et al. Haploinsufficiency of an RB-E2F1-condensin II complex leads to aberrant replication and aneuploidy. Cancer Discov. 2014;4:840–53.
Silk AD, Zasadil LM, Holland AJ, Vitre B, Cleveland DW, Weaver BA. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc Natl Acad Sci USA. 2013;110:E4134–41.
Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21.
Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller M-C, Shaikh N, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–6.
Watkins JA, Irshad S, Grigoriadis A, Tutt AN. Genomic scars as biomarkers of homologous recombination deficiency and drug response in breast and ovarian cancers. Breast Cancer Res. 2014;16:211.
Scheerens H, Malong A, Bassett K, Boyd Z, Gupta V, Harris J, et al. Current status of companion and complementary diagnostics: strategic considerations for development and launch. Clin Transl Sci. 2017;10:84–92.
Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40.
Cai H, Kumar N, Bagheri HC, von Mering C, Robinson MD, Baudis M. Chromothripsis-like patterns are recurring but heterogeneously distributed features in a survey of 22,347 cancer genome screens. BMC Genom. 2014;15:82.
Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40.
Schouten PC, Grigoriadis A, Kuilman T, Mirza H, Watkins JA, Cooke SA, et al. Robust BRCA1-like classification of copy number profiles of samples repeated across different datasets and platforms. Mol Oncol. 2015;9:1274–86.
Birkbak NJ, Wang ZC, Kim J-Y, Eklund AC, Li Q, Tian R, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2:366–75.
Watkins J, Weekes D, Shah V, Gazinska P, Joshi S, Sidhu B, et al. Genomic complexity profiling reveals that HORMAD1 overexpression contributes to homologous recombination deficiency in triple-negative breast cancers. Cancer Discov. 2015;5:488–505.
Popova T, Manié E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 Inactivation. Cancer Res. 2012;72:5454–62.
Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107:1776–82.
Menghi F, Inaki K, Woo X, Kumar PA, Grzeda KR, Malhotra A, et al. The tandem duplicator phenotype as a distinct genomic configuration in cancer. Proc Natl Acad Sci USA. 2016;113:E2373–E2382.
Watkins J, Tutt A, Grigoriadis A. Tandem duplications contribute to not one but two distinct phenotypes. Proc Natl Acad Sci USA. 2016;113:E5257–8.
Ng CKY, Cooke SL, Howe K, Newman S, Xian J, Temple J, et al. The role of tandem duplicator phenotype in tumour evolution in high-grade serous ovarian cancer. J Pathol. 2012;226:703–12.
Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54.
Morganella S, Alexandrov LB, Glodzik D, Zou X, Davies H, Staaf J, et al. The topography of mutational processes in breast cancer genomes. Nat Commun. 2016;7:11383.
Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16:110–20.
Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Wang W, Carvalho B, Miller ND, Pevsner J, Chakravarti A, Irizarry RA. Estimating genome-wide copy number using allele-specific mixture models. J Comput Biol. 2008;15:857–66.
Knutsen T, Padilla-Nash HM, Wangsa D, Barenboim-Stapleton L, Camps J, McNeil N, et al. Definitive molecular cytogenetic characterization of 15 colorectal cancer cell lines. Genes Chromosomes Cancer. 2010;49:204–23.
SKY Karyotypes and Molecular Cytogenetics of Common Epithelial Cancers. 2012. http://www.path.cam.ac.uk/~pawefish. Accessed 21 November 2019.
Heinze G, Schemper M. A solution to the problem of separation in logistic regression. Stat Med. 2002;21:2409–19.
The Cancer Genome Atlas Research Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–54.
McBride DJ, Etemadmoghadam D, Cooke SL, Alsop K, George J, Butler A, et al. Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J Pathol. 2012;227:446–55.
Menghi F, Barthel FP, Yadav V, Tang M, Ji B, Tang Z, et al. The tandem duplicator phenotype is a prevalent genome-wide cancer configuration driven by distinct gene mutations. Cancer Cell. 2018;34:197–210.e5.
Manié E, Popova T, Battistella A, Tarabeux J, Caux-Moncoutier V, Golmard L, et al. Genomic hallmarks of homologous recombination deficiency in invasive breast carcinomas. Int J Cancer. 2015;138:891–900.
Schouten PC, van Dyk E, Braaf LM, Mulder L, Lips EH, de Ronde JJ, et al. Platform comparisons for identification of breast cancers with a BRCA-like copy number profile. Breast Cancer Res Treat. 2013;139:317–27.
Turner N, Lambros MB, Horlings HM, Pearson A, Sharpe R, Natrajan R, et al. Integrative molecular profiling of triple negative breast cancers identifies amplicon drivers and potential therapeutic targets. Oncogene. 2010;29:2013–23.
Telli ML, Timms KM, Reid JE, Hennessy B, Mills GB, Jensen KC, et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple negative breast cancer. Clin Cancer Res. 2016;22:3764–73.
Grigorova M, Staines JM, Ozdag H, Caldas C, Edwards PAW. Possible causes of chromosome instability: comparison of chromosomal abnormalities in cancer cell lines with mutations in BRCA1, BRCA2, CHK2 and BUB1. Cytogenet Genome Res. 2004;104:333–40.
Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36:1159–61.
Zhang C-Z, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–84.
Thompson L, Jeusset L, Lepage C, McManus K. Evolving therapeutic strategies to exploit chromosome instability in cancer. Cancers (Basel). 2017;9:151.
Willis NA, Frock RL, Menghi F, Duffey EE, Panday A, Camacho V, et al. Mechanism of tandem duplication formation in BRCA1-mutant cells. Nature. 2017;551:590–5.
Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91.
Neelsen KJ, Zanini IMY, Mijic S, Herrador R, Zellweger R, Ray Chaudhuri A, et al. Deregulated origin licensing leads to chromosomal breaks by rereplication of a gapped DNA template. Genes Dev. 2013;27:2537–42.
Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52.
Telli ML, Jensen KC, Vinayak S, Kurian AW, Lipson JA, Flaherty PJ, et al. Phase II study of gemcitabine, carboplatin, and iniparib as neoadjuvant therapy for triple-negative and BRCA1/2 mutation-associated breast cancer with assessment of a tumor-based measure of genomic instability: PrECOG 0105. J Clin Oncol. 2015;33:1895–901.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4.
Mayba O, Gilbert HN, Liu J, Haverty PM, Jhunjhunwala S, Jiang Z, et al. MBASED: allele-specific expression detection in cancer tissues and cell lines. Genome Biol. 2014;15:405.
Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph. 2014;20:1983–92.
Author information
Authors and Affiliations
Contributions
JW conceived and designed the study. JW collected and managed ‘omic and clinical data. JW performed copy-number analysis. JW generated ASE data. JW, MS and ER performed platinum treatment response prediction analysis. JW and MS performed elastic net regression. JW and MS wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Shafat, M.S., Rufaie, E.S. & Watkins, J. A survey of cancer genome signatures identifies genes connected to distinct chromosomal instability phenotypes. Pharmacogenomics J 21, 390–401 (2021). https://doi.org/10.1038/s41397-021-00217-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41397-021-00217-9
- Springer Nature Limited
This article is cited by
-
Recent advances in targeted drug delivery systems for resistant colorectal cancer
Cancer Cell International (2022)